In the last few years, new drugs able to inhibit the B-cell receptor (BCR) pathway have been approved by the US and European drug control agencies for the treatment of chronic lymphocytic leukemia (CLL) and some indolent B lymphomas. Among them, ibrutinib and idelalisib, respectively, target the Bruton tyrosine kinase (BTK) and the phosphatidylinositol 3-kinase δ (PI3Kδ), essential molecules for transmitting the activation signals from BCR that sustain malignant B cells.1 These inhibitors were found to be particularly effective in CLL cases with relapsed/refractory disease or presenting poor prognostic factors and/or unfit for the best existing immunotherapies.2 After the initial excitement, the progressive occurrence of unwanted toxicities within the trials using ibrutinib or idelalisib has led to a more cautious use of these drugs. The case of idelalisib is the most striking. In fact, a high rate of serious adverse events, including deaths related to infections including cytomegalovirus (CMV) and pneumocystis jirovecii pneumonias (PJP), has caused some clinical trials to be stopped.3 Now, after a careful review by the drug agencies, a good risk-benefit balance has been confirmed for this inhibitor, and strong recommendations have been introduced to monitor treated patients for the appearance of infection symptoms, and to manage administration of PJP prophylaxis and regular CMV monitoring.4
Why idelalisib gives rise to these infections is still unknown. We therefore decided to study how the drug acts on the non-malignant counterpart of the immune system in CLL patients, focusing on the T compartment. PI3Kδ is known to be an intracellular transducer of T-cell receptor (TCR) signaling and there is much evidence to suggest it has an important role in the correct functioning of T cells.5 Indeed, research studies conducted on mice with p110δ deficiency found an impairment of T-cell proliferation, differentiation, and cytokine production.6,7 In accordance with this, experiments blocking PI3Kδ in healthy human T cells showed an impaired production of T-helper and inflammatory cytokines even if no changes in survival were seen.7,8
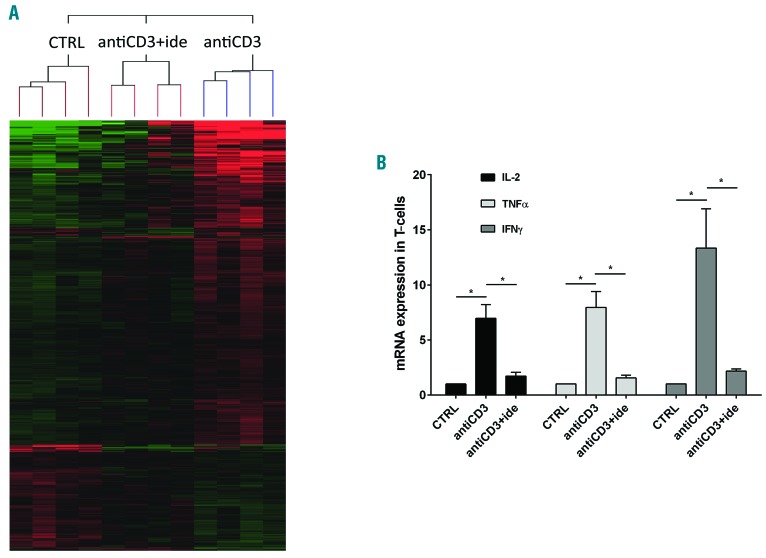
By hybridization on 4X44K whole human genome microarray, we performed a gene expression profile (GEP) analysis on total RNA of T cells purified from peripheral blood mononuclear cells (PBMCs) of untreated CLL patients. T cells were incubated in the presence or absence of 5 mM idelalisib for 1 hour (h) and stimulated with antiCD3 antibody for 4 h. Experiments are described in the Online Supplementary Methods and in Online Supplementary Figure S1. Comparing the GEP between control (no treatment/no stimulation), antiCD3 (antiCD3 stimulation alone) and antiCD3+ide (antiCD3 stimulation with drug pre-treatment), we first identified the differentially expressed genes with Anova (P≤0.01 was considered significant). Among these, we focused on the 65 genes with fold-change [FC]≥2 between the “antiCD3” and “antiCD3+ide” conditions (Figure 1A). Of the 61 genes down-regulated by idelalisib, the most represented categories were chemokines involved in T-cell migration (such as CXCL9-10-11), inflammatory and T-helper cytokines (such as IL-2, TNFα, IFNγ, IL-13, IL-21), mitogenic signal transducers (such as IRF4, STAT1, EGR1-2-3-4) and receptors (such as CD69, CD25) commonly induced during activation of T cells (Online Supplementary Table S1). These data indicate that idelalisib reduces the expression of important genes for T-cell-mediated immunity in CLL, so we searched for possible drug interferences with cytokine production, migration and proliferation of T cells.
Figure 1.
Idelalisib reduces the cytokine expression in activated T cells of chronic lymphocytic leukemia (CLL) patients. (A) Supervised hierarchical clustering of gene expression profiles in RNA samples of T cells purified from 4 CLL patients, each cultured with the three following conditions: i) control (CTRL, no treatment/no stimulation); ii) antiCD3 (stimulation with antiCD3 antibody); iii) antiCD3+ide (5 mM idelalisib pre-treatment followed by antiCD3 stimulation). Shown are genes differentially expressed between the described conditions, compared by Anova test; P≤0.01. Green squares: low levels of transcript; red squares: high levels of transcript; black squares: equal level of transcript. Complete microarray data are deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO accession number GSE108224. (B) Expression levels of IL-2, TNFα and IFNγ were measured by real-time PCR in RNA samples of T cells purified from 5 CLL patients and cultured with the same three conditions described in (A). Values are represented by the histograms as mean±Standard Error and normalized to CTRL values, defined as 1. IL-2, TNFα and IFNγ are significantly up-regulated by antiCD3 stimulation but idelalisib strongly abrogates these inductions (*P<0.05, Student t-test).
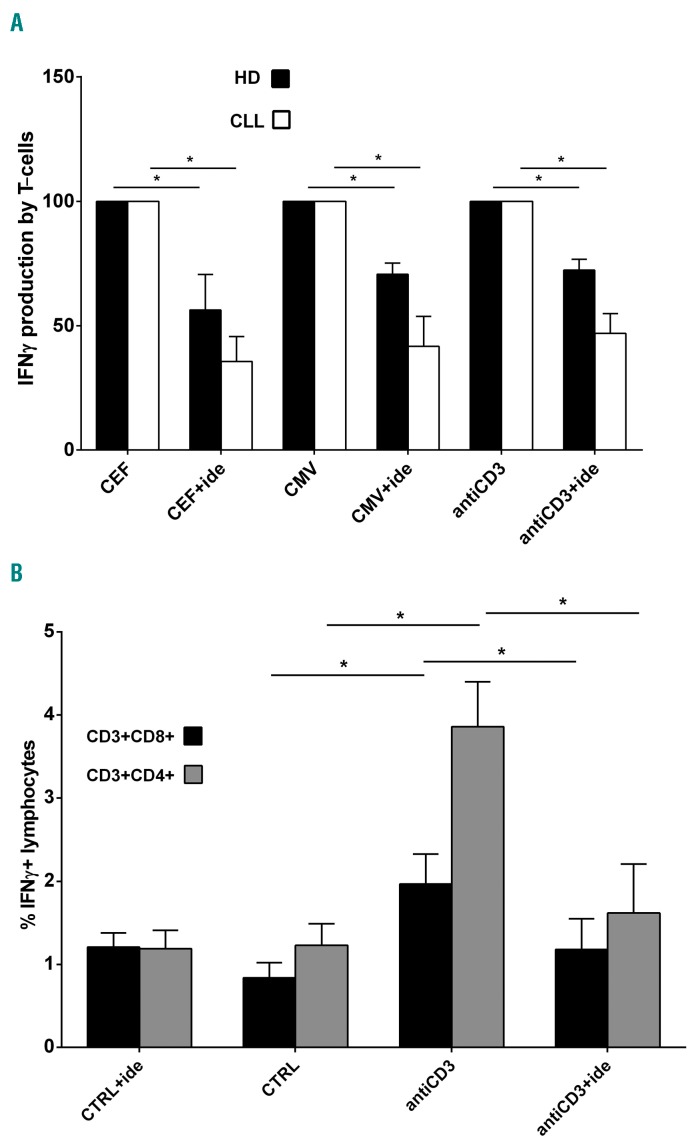
We used real-time PCR to evaluate the mRNA expression of some inflammatory cytokines in T cells purified from CLL patients, pre-blocked or not with idelalisib, and stimulated with antiCD3 antibody, as previously described for GEP experiments. We observed that amounts of IL-2, TNFα and IFNγ increased 6.9, 7.9 and 13.3-fold in T cells stimulated by antiCD3 (P=0.009, P=0.008 and P=0.026, respectively) in comparison to non-stimulated controls (Figure 1B). In the presence of idelalisib, we found that IL-2, TNFα and IFNγ expressions decreased 4.0, 5.2 and 6.3 times if compared to antiCD3 stimulation alone (P=0.008, P=0.007 and P=0.033, respectively) (Figure 1B). Moreover, we performed ELISpot analyses using Human IFN-γ ELISpotPRO (ALP) to verify the effect of idelalisib on IFNγ protein release by T cells of CLL patients, stimulated or not with antiCD3 antibody for 18 h. In parallel, we cultured T samples with 100 ng/mL PepMIX HCMVA (pp65) (peptide mix of CMV phosphoprotein 65) or 100 ng/mL CEF [peptide mix of CMV, Epstein Barr virus (EBV) and influenza virus] for 18 h, in the presence or absence of 5 mM idelalisib, looking for any modulation in T-cell responses to these viruses. We confirmed that the IFNγ release by T samples promoted by antiCD3 stimulation is reduced by 53.2% in the presence of idelalisib (P<0.001) (Figure 2A). Interestingly, the drug decreased cytokine production also in T cells stimulated by CMV (reduced by 58.4%, P=0.009) or CEF peptides (reduced by 64.4%, P=0.003) (Figure 2A). Performing similar ELISpot tests on T samples collected from healthy donors (HD), we found that idelalisib diminishes the IFNγ release induced by each stimulation (reduced by 27.7% for antiCD3 antibody, by 29.4% for CMV peptides, and by 43.7% for CEF peptides, respectively; all P<0.05) (Figure 2A). In the absence of stimulation, the drug did not modify the number of IFNγ spots for either CLL or HD T-cell samples. Together these show that idelalisib impairs IFNγ production by activated T cells, supporting the crucial role of PI3Kδ in this process, both in CLL and in healthy donors. Since the drug inhibits T cells not only in the case of generic TCR stimulation, but also in response to virus-specific stimulation, it could favor a higher rate of CMV reactivations in those CLL patients who are treated with it.
Figure 2.
IFNγ release from stimulated T cells is impaired by idelalisib in chronic lymphocytic leukemia (CLL). (A) ELISpots for IFNγ release were performed on T samples obtained from 5 CLL patients and 4 healthy donors (HD), stimulated or not with antiCD3 antibody, cytomegalovirus (CMV) or CEF peptides, in the presence or absence of 5 μM idelalisib. In the graph, the spot number obtained with the drug is indicated as mean percentage±Standard Error (SE) of the spot number observed with the correspondent stimulation, defined as 100%. Black squares: HD; white squares: CLL. Since control (CTRL, no treatment/no stimulation) and CTRL+ide (idelalisib treatment/no stimulation) conditions showed the same IFNγ spot number (usually zero) for all CLL and HD T samples, they have not been included in the graph. The increased production of IFNγ induced in T cells by each stimulation is significantly decreased by idelalisib, both for CLL and HD (*P<0.05, Student t-test). (B) IFNγ cytokine secretion assays (CSA) were performed on T samples collected from 5 CLL patients, pre-treated or not with 5 μM idelalisib and stimulated or not with antiCD3 antibody. We acquired approximately 100,000 events inside the “lymphocytes” gate and the analysis identified the IFNγ+ lymphocytes inside the CD3+CD8+ or CD3+CD4+ gates. In the graph, the mean percentages±SE of CD3+CD8+IFNγ+ and CD3+CD4+IFNγ+ lymphocytes are represented with black squares and with gray squares, respectively. Control (CTRL, no treatment/no stimulation) and CTRL+ide (idelalisib treatment/no stimulation) show substantially the same percentages of CD3+CD8+IFNγ+ and CD3+CD4+IFNγ+ lymphocytes (P=n.s., Student t-test). With antiCD3 stimulation, these percentages become respectively 2.3 and 3.1-fold higher in comparison to CTRL (*P<0.05, Student t-test). The presence of idelalisib under stimulation reduces the percentages of CD3+CD8+IFNγ+ and CD3+CD4+IFNγ+ lymphocytes by 1.7 and 2.4 times, respectively, if compared to antiCD3 stimulation alone (*P<0.05, Student t-test). n.s.: not significant.
We also studied the possible different action of idelalisib towards the CD4 or the CD8 T-cell subset, by performing IFNγ cytokine secretion assays (CSA) on T samples collected from CLL patients, pre-treated or not with 5 μM idelalisib and stimulated or not with antiCD3 anti body for 1 h. CD3+CD8+IFNγ+ and CD3+CD4+IFNγ+ populations were significantly increased by antiCD3 stimulation but were significantly reduced by idelalisib (both P<0.05) (Figure 2B). Therefore, the drug seems to affect IFNγ production in both CD4 and CD8 T-cell subsets.
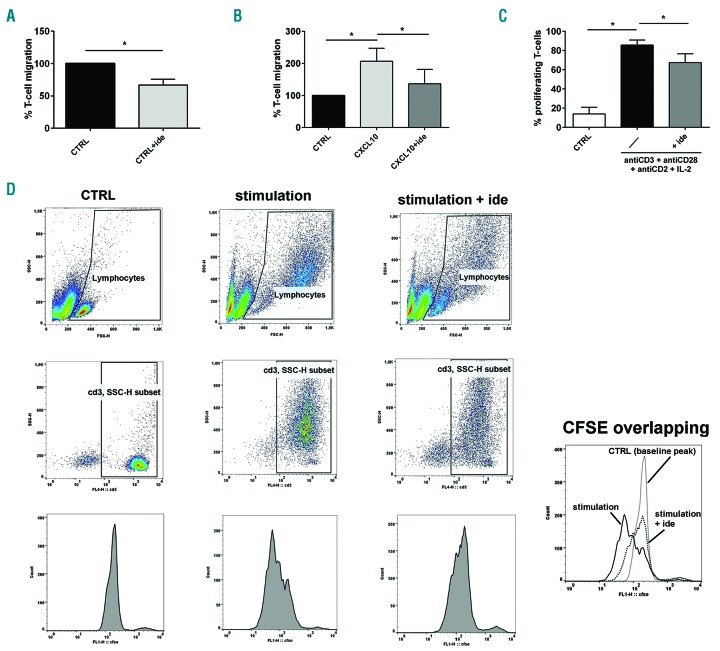
We then investigated the effect of idelalisib on the migration ability of T cells of CLL patients in vitro. T cells purified from CLL cases were pre-treated or not with 5 μM idelalisib and were left to migrate for 2 h in the presence or absence of 100 ng/mL CXCL10. We found that the PI3Kδ inhibitor reduced the percentage of T cells that spontaneously migrated from 100% to 66.7±9.0% (P=0.022) (Figure 3A). When we added CXCL10 as chemoattractant, T-cell migration increased from 100% to 206.9%±40.5% but it was reduced to 136.7%±44.8% in the presence of the drug (both P<0.05) (Figure 3B). These data suggest that idelalisib is able to hamper the migration of T cells of CLL patients, even when they are attracted by proper chemokines.
Figure 3.
Idelalisib inhibits T-cell migration and proliferation in chronic lymphocytic leukemia (CLL). (A) Migration experiments were performed on T cells purified from 5 CLL patients, in presence or in absence of 5 μM idelalisib [Control (CTRL)+ide and CTRL, respectively]. The histograms represent the mean percentages±Standard Error (SE) of migrated cells normalized to CTRL, defined as 100%. The drug strongly reduced the number of T cells that spontaneously migrated (*P<0.05, Student t-test). (B) T cells purified from 6 CLL patients were stimulated with CXCL10 in the presence or absence of 5 μM idelalisib and tested for their migration. The mean percentage±SE of migrated T cells was normalized to CTRL (no treatment/no stimulation), defined as 100%. The number of migrated cells is increased by CXCL10 stimulation but is significantly decreased by idelalisib (*P<0.05, Student t-test). (C) Proliferation ability was evaluated by flow cytometry by analyzing the CFSE dilution peaks in T cells purified from 3 CLL patients, pre-treated or not with 5 μM idelalisib and stimulated with a mix of antiCD3+antiCD28+antiCD2 antibodies and IL-2. Not treated/non-stimulated cells were used as controls (CTRL). The mean percentages±SE of proliferating T cells are represented by the histograms. The increased T-cell proliferation obtained with stimulation is significantly reduced by the drug (*P<0.05, Student t-test). (D) Strategy example for the flow-cytometry analysis of T-cell proliferation in the three tested conditions: i) CTRL (not treated/non-stimulated cells), ii) stimulation (stimulation with a mix of antiCD3+antiCD28+antiCD2 and IL-2), iii) stimulation+ide (pre-treatment with 5 μM idelalisib followed by stimulation with a mix of antiCD3+antiCD28+antiCD2 and IL-2). We acquired approximately 10,000 events inside the “lymphocytes” gate based on FSC-H and SSC-H features. For the analysis, inside this first gate we drew a “CD3, SSC-H subset” gate, including only CD3+ cells. Inside this last gate, we evaluated the CFSE dilution peaks of proliferating cells, located to the left of the baseline CFSE peak identified based on the CTRL. For each studied case, we drew the same gates for all three conditions. In the single box on the bottom right, the CSFE dilution peaks of T cells are overlapped and compared between the described conditions.
Finally, we evaluated the action of idelalisib on T-cell proliferation in CLL. T cells purified from CLL cases were stained with CFSE, pre-treated or not with 5 μM idelalisib and then stimulated with a mix of antiCD3+antiCD28+antiCD2 antibodies and 300 U/mL IL-2 for 72 h. The percentage of proliferating T cells increased 6.15-fold after stimulation (P<0.001) but decreased 1.27 times in the presence of the drug (P<0.05) (Figure 3C and D). We also measured T-cell vitality in PBMCs of CLL patients treated or not with 1 μM or 5 μM idelalisib in vitro for 24 and 48 h and found no significant changes when compared to controls (data not shown), in accordance with existing literature.6 The PI3Kδ inhibitor can, therefore, reduce the proliferative capacity of T cells of CLL patients in response to strong stimulations, without affecting their survival.
In conclusion, our study has demonstrated the inhibitory effect of idelalisib towards several important functions of T cells of CLL patients, in particular cytokine production, migration and proliferation, in response to stimulation. Interestingly, the drug-blocking action was observed also in T cells stimulated with CMV, EBV or influenza antigens. CMV-specific immunity has previously been demonstrated to play a crucial role in controlling infection by this virus.9 In immunocompromised hosts, CMV infections are often found to accompany PJP, possibly sharing some immunogenic mechanism, although the precise biological reasons are unknown.10 Our in vitro data suggest that idelalisib can impair T-cell-mediated responses to CMV, giving one possible explanation for the high frequency of CMV-reactivations and PJP observed in CLL cases treated with this drug. A very recent paper has observed an increased percentage of Tregs by immunohistochemistry in biopsies of patients who received idelalisib and experienced gastrointestinal symptoms, sometimes in association with PCR positivity for infectious pathogens.11 These findings, together with our results, support the hypothesis that a T-cell impairment induced by idelalisib, able to favor infections and/or viral reactivations, can be responsible for some important side-effects of the drug. Certainly, the role of Tregs in this context remains to be elucidated. Future functional experiments conducted on T samples collected from CLL patients in the course of treatment with idelalisib will be helpful to better clarify the drug effect on the different T-cell subsets in vivo. Similarly, the impact of ibrutinib on either innate or adaptive immunity warrants further investigation, given the recent reports of invasive fungal infections in treated patients.12,13 Increasing the knowledge of biological effects of such TK inhibitors on non-malignant cells may promote their proper and appropriate use, through the early recognition, monitoring and prevention of those infectious complications that could possibly be drug-related.
Supplementary Material
Footnotes
Funding: this work was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC IG14376), (TRIDEO 16923), (AIRC/FIRC #16430] and Fondazione Umberto Veronesi and Ricerca Finalizzata Giovani Ricercatori 2011-2012, Ministero della Salute (GR-2011-02349282) and PRIN 2015, Ministero dell’Istruzione, dell’Università e della Ricerca (PRIN 2015ZMRFEA_002).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Burger JA, Chiorazzi N. B cell receptor signaling in chronic lymphocytic leukemia. Trends Immunol. 2013;34(12):592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanford DS, Wierda WG, Burger JA, Keating MJ, O’Brien SM. Three newly approved drugs for chronic lymphocytic leukemia: incorporating ibrutinib, idelalisib, and obinutuzumab into clinical practice. Clin Lymph Myel Leuk. 2015;15(7):385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Weerdt I, Koopmans SM, Kater AP, van Gelder M. Incidence and management of toxicity associated with ibrutinib and idelalisib: a practical approach. Haematologica. 2017;102(10):1629–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.http://www.ema.europa.eu/docs/en_GB/document_library/EPAR__Product_Information/human/003843/WC500175377.pdf
- 5.Okkenhaug K, Ali K, Vanhaesebroeck B. Antigen receptor signalling: a distinctive role for the p110delta isoform of PI3K. Trends Immunol. 2007;28(2):80–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okkenhaug K, Patton DT, Bilancio A, Garçon F, Rowan WC, Vanhaesebroeck B. The p110delta isoform of phosphoinositide 3-kinase controls clonal expansion and differentiation of Th cells. J Immunol. 2006;177(8):5122–5128. [DOI] [PubMed] [Google Scholar]
- 7.Soond DR, Bjørgo E, Moltu K, et al. PI3K p110delta regulates T-cell cytokine production during primary and secondary immune responses in mice and humans. Blood. 2010;115(11):2203–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herman SE, Gordon AL, Wagner AJ, et al. Phosphatidylinositol 3-kinase-δ inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals. Blood. 2010;116(12):2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barron MA, Gao D, Springer KL, et al. Relationship of reconstituted adaptive and innate cytomegalovirus (CMV)-specific immune responses with CMV viremia in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2009;49(12):1777–1783. [DOI] [PubMed] [Google Scholar]
- 10.Kim T, Park SY, Lee HJ, et al. Assessment of cytomegalovirus and cell-mediated immunity for predicting outcomes in non-HIV-infected patients with Pneumocystis jirovecii pneumonia. Medicine (Baltimore). 2017;96(30):e7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeung CC, Hockenbery DM, Westerhoff M, et al. Pathological assessment of gastrointestinal biopsies from patients with idelalisib-associated diarrhea and colitis. Future Oncol. 2018. March 23 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 12.Chamilos G, Lionakis MS, Kontoyiannis DP. Call for Action: Invasive Fungal Infections Associated With Ibrutinib and Other Small Molecule Kinase Inhibitors Targeting Immune Signaling Pathways. Clin Infect Dis. 2017. September 27 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grommes C, Younes A. Ibrutinib in PCNSL: The Curious Cases of Clinical Responses and Aspergillosis. Cancer Cell. 2017;31(6):731–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.