Abstract
New achievements in the field of cancer treatment are results of recent advances in molecular medicine and gene therapy. Usage of microRNAs (miRNAs) which are small noncoding RNAs is one of the molecular research lines for the diagnosis and treatment of cancers. miRNAs have an important role in post-transcriptional regulation of the gene expression and are involved in cellular activities such as growth, differentiation, cell death and cancer development. One of the miRNAs that showed downregulation in human acute erythroleukemia is hsa-let-7c-5p. Down-regulation of hsa-let-7c-5p has been reported in in vitro studies of different cancers. In the present study, upregulation of hsa-let-7c-5p is performed in human acute erythroleukemia cell line (KG-1) using miRNA mimic. qRT-PCR, MTT assay, Annexin-V, and propidium iodide staining at different time points after miRNA mimic transfection were accomplished to assess the expression level of hsa-let-7c-5p, cell viability, apoptosis and late apoptosis. In addition, the expression level of PBX2 oncogene, a validated target gene of hsa-let-7c-5p, is evaluated by RT-qPCR to show the effectivity of this approach on erythroleukemia cancer cells. Our results can be used in translational medicine for future investigation in acute erythroleukemia and to approach treatment based on miRNA mimic therapy.
Keywords: Erythroleukemia, hsa-let-7c-5p, miRNA mimic, Proliferation, Apoptosis
Introduction
MicroRNAs (miRNAs) have been recognized as highly conserved post-transcriptional regulators of gene expression and they are involved in many biological, cellular and pathological processes. Initially discovered in 1993 in Caenorhabditis elegans, miRNAs were later shown to have roles in gene expression post-transcriptionally. miRNA genes are transcribed by RNA polymerase ΙI that yields to primary miRNAs (Pri-miRNA), then Drosha (an RNAase Ш enzyme) produces pre-miRNA. Later, pre-miRNA is transported into the cytoplasm by exportin 5 wherein, it is cut by Dicer to generate miRNA/miRNA duplex. Strands of the duplex are transmitted into RNA-induced silencing complex (RISC). miRNA binds to complementary target sites of 3´ UTR (untranslated region) of mRNAs and prevents the production of proteins (Dong et al. 2009; Drakaki and Iliopoulos 2009; Reddy et al. 2010). miRNAs are involved in biological processes, including apoptosis, cell cycle, proliferation, differentiation, aging, metabolism, development, and growth (Gammell 2007). The human genome has about ~ 2500 mature miRNAs [miRBase, Kozomara and Griffiths-Jones, 2014; URL: http://www.mirbase.org] that regulate the expression of more than 60% of all protein-coding genes in humans (Bartel 2004; Stoicea et al. 2016).
According to previous studies, miRNAs are involved in a wide range of diseases including, infections, metabolism, inflammatory disorders, heart diseases, and cancers (Bartel 2004; Kloosterman and Plasterk 2006). Dysregulation of miRNAs including up and down-regulation of their biogenesis, mimics the activities related to oncogenes and tumor suppressor genes, depending on the target mRNAs regulated by particular miRNAs (Ruan et al. 2009). A number of miRNAs have increasing or decreasing expression in cancer cells which can change important regulators of cellular processes such as PTEN, MYC, Ras, TNK4 etc. (Amini-Farsani et al. 2018; Ruan et al. 2009). The roles of miRNAs in some cancers are obvious. For example, in breast cancer the expression level of miR-21 is increased (Gong et al. 2011) while let-7 family has a decreased expression in lung cancer. Moreover, miR-221 and miR-140 have an increased and a decreased expression in the brain tumors, respectively (George and Mittal 2010). According to the evidences, regulation of miRNAs expression can be an option for treatment of certain cancers. Thus, miRNAs can be used in treatment of malignancies, that may lead to major changes in cancer treatment. Hsa-let-7c-5p miRNA, a member of the highly conserved let-7 family, is an intronic miRNA located on human chromosome 21q21.1 within the LINC00478 gene (http://www.mirbase.org). This miRNA has reduced expression and acts as a tumor suppressor gene in several cancers including prostate, colorectal adenocarcinoma, and acute myeloid leukemia (Hertel et al. 2012; Nadiminty et al. 2012).
Let-7c acts as a tumor suppressor by regulation of cancer-related genes such as N-RAS, C-MYC, MMP1, PBX2, PBX3, etc. (Johnson et al. 2005; Nadiminty et al. 2012). The PBX2 gene is located on the 6p21.32 chromosome, within the major histocompatibility complex (MHC). This gene encodes a member of the TALE/PBX homeobox family which is a transcriptional activator, whose aberrant expression enhances HOXA9-dependant leukemogenesis and consequently it is involved in the AML phenotype. PBX2 is an oncogene target of hsa-let-7c in acute erythroleukemia (Monica et al. 1991; Moskow et al. 1995; Pelosi et al. 2013).
AML is one of the most common and fatal types of hematopoietic malignancies with high heterogeneity in the affected population in terms of morphology, immunophenotype, cytogenetics and molecular abnormalities (Döhner et al. 2017, 2015; Kavianpour et al. 2016; Marcucci et al. 2005; Mazzella et al. 2000; Rowley 2008). AML is the most common acute leukemia affecting adults whose incidence increases with age. Despite different treatment strategies, the majority of patients will die and only 35–40% of the younger patients (aged < 60) and 5–15% of older ones (aged > 60) with AML can survive over 5 years (Döhner et al. 2010, 2015). Current treatment of AML involves chemotherapy, cytotoxic and intensive post remission treatment with high dose of Cytarabine (Ara-C) which causes reduced quality of life of the patients (Byrd et al. 2004). Due to the rising incidence of AML with aging, most eldery patients not only can not endure intensive chemotherapy but also it is associated with very poor survival (Dores et al. 2011). So, novel therapeutic strategies with less intensity but a higher rate of cure are needed urgently (Hasserjian et al. 2001; Mohammadiasl et al. 2016; Tenen 2003). In this heterogeneous failure, accumulation of genetic and epigenetic alterations in progenitor cells, alter the total expression or function of key genes (Hasserjian et al. 2001). Unlike most of the cancers, AML does not usually form tumors and it is in all of the bone marrow in the body. Subsequently, it is not staged like most of other cancers but it is classified from M0 to M7 according to the French-American-British (FAB) classification based on the type of cell from which leukemia developed and the level of maturity of the cells. Knowing the subtype of AML can be very important, as it can affect both the outlook of a patient and the best treatment. AML- M6 is called acute erythroid leukemia that originates in very early forms of red blood cells and is defined as neoplasia of myeloid and erythroid precursors in bone marrow (Acevedo-Olvera et al. 2015; Kowal-Vern et al. 2000; Santos et al. 2009).
As miRNAs signatures have been sought in AML, it has been demonstrated that they can be up and downregulated (Yuan et al. 2012). Since hsa-let-7c-5p is a tumor suppressor miRNA which is downregulated in AML, miRNA replacement therapy may affect cancerous cells. As loss of a tumor suppressor miRNA leads to activation of oncogenic pathways and tumorigenesis in related cancers. In this approach (miRNA replacement) mimic of the tumor suppressor miRNA is reintroduced in the cancer cells to restore the loss of function of a tumor suppressor miRNA which results in suppressing of oncogenic pathways and cancer cell growth (Gambari et al. 2016).
MicroRNA mimics are designed to simulate naturally occurring mature microRNAs. Also, they have the same sequence as the natural miRNAs have, so it is expected to target the same set of miRNAs that are also regulated by natural miRNAs. Off-target effect of miRNAs mimics is unlikely as they are expected to act like the natural counterpart for which the proper miRNA-mRNA interactions have evolved. These properties suggest miRNA mimics as suitable candidates for therapeutic applications (Bader et al. 2010).
In this survey, we evaluate overexpression of hsa-let-7c-5p by introducing its mimic to acute erythroleukemia cell line (KG-1). Moreover, its effect on cell proliferation, apoptosis, late apoptosis and the expression of PBX2 is assessed to evaluate the effectivity of this approach on erythroleukemia cancer cells.
Materials and methods
Cell culture
Acute erythroleukemia (AML-M6) cell line (KG-1) was purchased from the National Cell Bank of Iran (Pasteur Institute, Tehran, Iran) and cultured in Roswell Park Memorial Institute (RPMI) 1640 (Gibco, Paisley, UK) medium supplemented with 10% v/v fetal bovine serum (Gibco) and 1% penicillin and streptomycin (Sigma-Aldrich, St. Louis, MO, USA), in a humid atmosphere consisting of 5% CO2 at 37 °C in 75 cm2 culture flasks (Nunc, Roskilde, Denmark). The cells were passaged three times weekly to keep them in the exponential growth phase.
Cell transfection
The nucleotide sequence of hsa-let-7c-5p was obtained from www.mirbase.org as UGAGGUAGUAGGUUGUAUGGUU (Accession Number MIMAT0000064). miRCURY LNA miRNA mimic for hsa-let-7c-5p and mimic control (scrambled) oligonucleotides were purchased from Exiqon (Copenhagen, Denmark). Oligonucleotides of miRNA mimic of hsa-let-7c-5p (not mimic control) were labeled at their 5′ ends with 6-FAM (fluorescent dye). KG-1 cell transfection was performed by using PolyFect™ transfection reagent kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions . Initially, 5 × 105 KG-1 cells in the exponential growth phase were cultured in six-well culture plates (Nunc) containing 1.8 ml RPMI-1640 per well without antibiotic and FBS. 100 μl of Polyfect™ was added to 50-pmol miRCURY LNA miRNA mimic and then incubated for 15 min at room temperature in darkness. The complex was then added to the cells and the plate was shaken cautiously to ensure distribution over the entire plate surface. Following 6 h of incubation in the incubator, fetal bovine serum and antibiotics were added and then cells were incubated for 24, 48 and 72 h. The cell cultures of untreated cells and cells transfected with hsa-let-7c-5p mimic control were performed in the culture of miRNA mimic-transfected cells. Mimic of hsa-let-7c-5p was conjugated with 6-FAM so the KG-1 cells which were transfected with hsa-let-7c-5p mimic, were surveyed by using a fluorescence microscope.
Reverse transcriptase (RT) miRNA real-time PCR
Reverse transcriptase (RT) miRNA real-time PCR was performed to evaluate the efficiency of hsa-let-7c-5p overexpression by the hsa-let-7c-5p miRNA mimic. In short, the total cellular RNA was extracted from the three groups (24, 48 and 72 h post-transfection) with a miRCURY RNA Isolation Kit™ (Exiqon) and concentration of total RNA was measured by 260/280 absorbance, then cDNA was synthesized using the universal cDNA synthesis Kit™ (Exiqon). Real-time PCR was performed by using the SYBR® Green master mix Kit™ (Exiqon) and specific hsa-let-7c-5p primers (Exiqon). Synthetic RNA spike-in templates and its primers (Exiqon) were used as internal controls of RT-PCR. ABI_StepOnePlus (Applied Biosystems, Foster City, CA, USA) instrument was used for real-time PCR experiments and the ΔΔCt method was chosen for data calculation.
Cell viability assay
To assess the cell viability, MTT assay was used that is based on the reduction of MTT (3-[4,5dimethylthiazol-2-yl]-2,5- diphenyl tetrazolium bromide) by the mitochondrial dehydrogenases of living cells to a purple formazan product. The product of purple formazan is directly related to the number of living cells. MTT assay was carried out at intervals of 24, 48 and 72 h post transfection. Two hundred microliters of MTT at a concentration of 5 mg/ml was added to 3 × 105 KG-1 cells that were suspended in 2 ml RPMI 1640 medium and incubated for 2 h at 37 °C in the darkness. Then, 200 μl of dimethylsulfoxide (Sigma-Aldrich, Leicester, England) was added to each well and was shaken until the crystals vanished. The blank sample was prepared by using the medium corporation solely. Absorbance was measured by using a spectrophotometer (PG Instrument T80, PG Instrument, Leicestershire, England) at 570 nm. Reading was converted to the percentage of the controls.
Apoptosis and necrosis assay
The FITC Annexin-V apoptosis detection kit with propidium iodide (PI) (BioLegend, San Diego, CA, USA) was used for identification of apoptotic and necrotic KG-1 cells. Annexin-V was used to detect the phosphatidylserine on the external leaflet of apoptotic cells. To distinguish between the necrotic and apoptotic cells, PI staining was performed (untreated cells were used for control). The process was done 24, 48 and 72 h posttransfection according to the manufacturer’s instruction), and the cells were assessed using the FACSCalibur flow cytometer (BD, San Jose, CA, USA) with 488-nm excitation, 515-nm band-pass filter for fluorescein-conjugated Annexin-V detection and a 600-nm filter (for PI detection).
Reverse transcriptase PBX2 gene Real-time PCR
The expression level of PBX2 was determined by real-time qPCR. The miRCURY RNA Isolation Kit and Universal cDNA Synthesis Kit from Exiqon were used for total RNA extraction and cDNA synthesis. The SYBR Green Master Mix Kit (Exiqon) was used for PBX2 gene real-time PCR. GAPDH gene was used as the internal control. Primers used were: PBX2 F: 5′ TGCCATTTGCCCTCATTTC 3′, R: 5′ TCAACGGAGATTCCTATTCTGC 3′ and GAPDH F: 5′ TGCACCACCAACTGCTTAGC 3′ R: 5′ GGCATGGACTGTGGTCATGAG 3′. The Step One PlusTM (Applied Biosystems, Foster City, CA, USA) instrument and ΔΔCt method were used for data analysis in real-time PCR tests.
Statistical analysis
Experiments were performed in triplicate, and the results were investigated with the SPSS 22 software (IBM, New York, NY, USA). The Kolmogorov–Smirnov test was used to normalize the data and Levine’s test was used to assess homogeneity of variances. Two-way analysis of variance (ANOVA) was used to evaluate differences between the group. Data were presented as mean ± SD and p < 0.01.
Results
miRCURY LNA™ microRNA Mimic increase hsa-let-7c-5p expression
Transfection of hsa-let-7c-5p mimic into KG-1 cells was performed with PolyFect™ transfection reagent. Since the transfected oligonucleotides were FAM-conjugated, transfection efficiency was evaluated by fluorescence microscopy. The efficiency of transfection was about 87% (Fig. 1) at the optimized level.
Fig. 1.
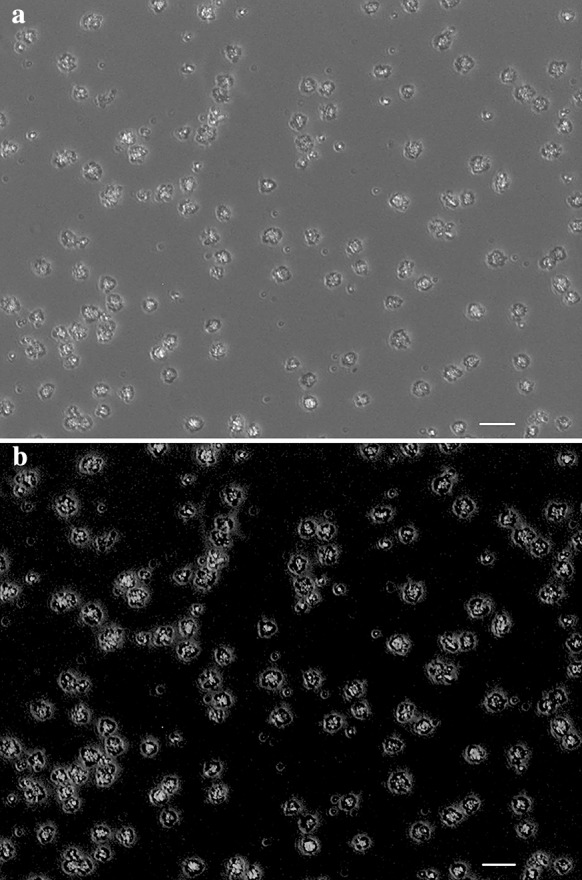
Acute erythroleukemia cells (KG-1) were transfected with 6-carboxyfluorescein-conjugated miRNA mimic oligonucleotides and then evaluation of transfection efficiency was assessed by fluorescent microscope. Phase contrast (a) and fluorescent (b) images of the same strand of KG-1 cells show that the majority of the cells were transfected. Scale bars 50 μm
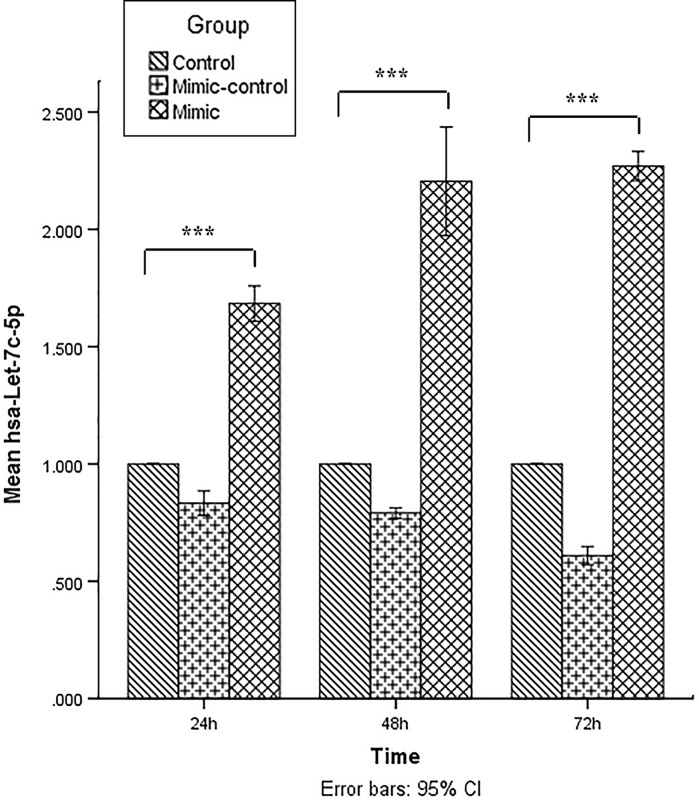
Expression of hsa-let-7c-5p was assessed by reverse transcriptase miRNA real-time PCR in KG-1 cells transfected with miRCURY LNA miRNA mimic, transfected with miRNA mimic control oligonucleotides (scrambled) or untreated KG-1 cells (untreated groups) at 24, 48 and 72 h post transfection. In all three groups, the expression of hsa-let-7c-5p was significantly increased in the miRNA mimic transfect group compared with the mimic control and untreated group (Fig. 2; p < 0.001). The hsa-let-7c-5p expression was a little lower in scrambled LNA transfected cells compared with untreated cells. The reason was presumably the transfection process that injured (affected) KG-1 cells. The expression of hsa-let-7c-5p was gradually increased over time as the expression was at the highest level 72 h post-transfection (p < 0.001; Fig. 2).
Fig. 2.
Evaluation of the hsa-let-7c-5p level by real-time PCR at 24, 48, and 72 h post transfection. The ΔΔCt method was used for data analysis and the untreated group (control) was considered as reference for each time point. Data are presented as mean ± SD of three independent experiments (***p < 0.001)
Increased expression of hsa-let-7c-5p decreased viable acute erythroleukemia cells
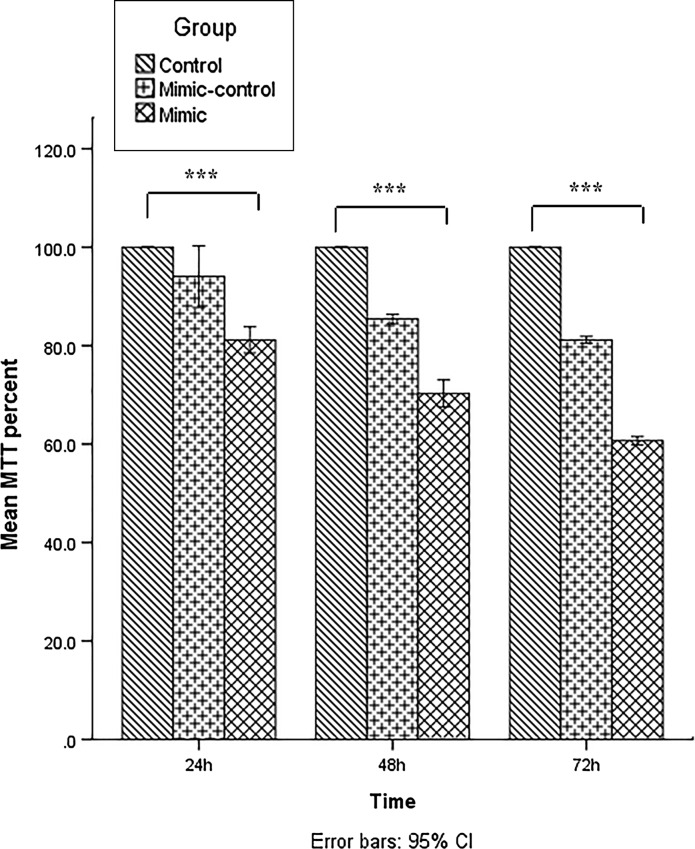
To assess the effect of hsa-let-7c-5p overexpression on cell viability, the MTT assay was performed at 24, 48 and 72 h post-transfection. In all three groups, cell viability was lower in the mimic control group compared with the untreated group. Cell viability of KG-1 cells was decreased considerably after hsa-let-7c-5p mimic transfection. The effect of miRNA mimic on the cell viability was gradually increased over time as the viability of mimic-transfected cells was at the lowest level 72 h post-transfection. The difference in cell viability at all three times points was significant in the miRNA mimic group, scrambled group and the untreated group, p < 0.001 (Fig. 3).
Fig. 3.
Evaluation of cell viability by 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT) assay performed 24, 48 and 72 h after transfection. The viability of untreated cells (control) at each time point was considered as 100% and the viability of the other groups is reported as a percentage of untreated cells at the same time point. Data are presented as mean ± SD of three independent experiments (***p < 0.001)
Increased expression of hsa-let-7c-5p increased apoptosis and necrosis in acute erythroleukemia cells
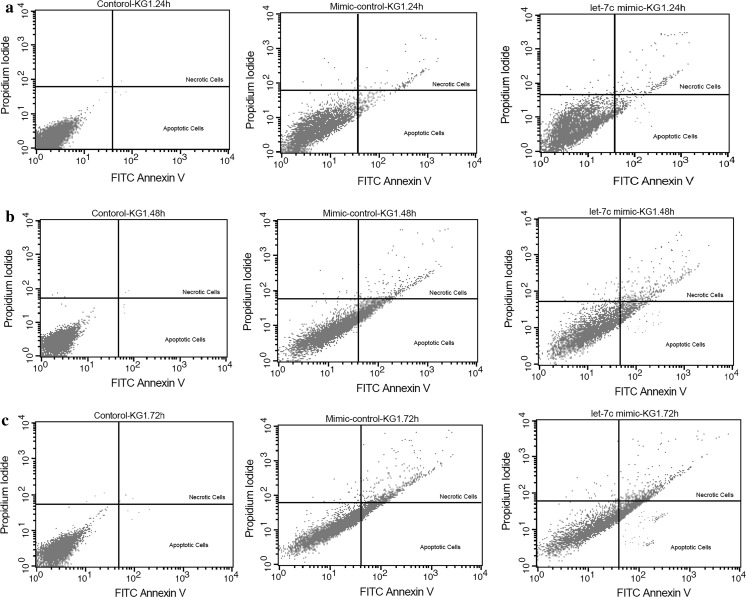
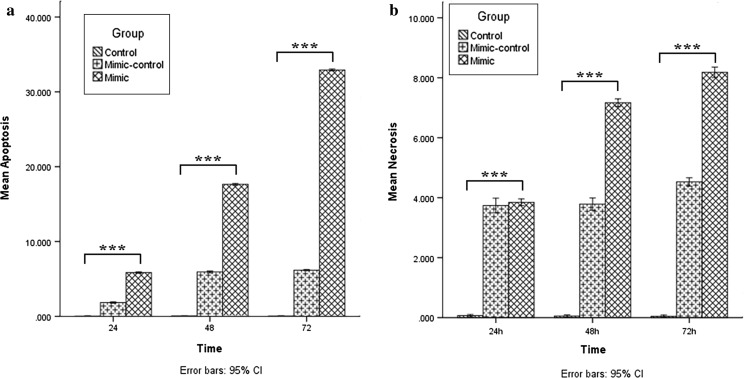
By using annexin-V and propidium iodide staining, the effect of hsa-let-7c-5p overexpression on apoptosis and necrosis in KG-1 cells at three different times was assessed. Apoptotic cells were undetectable in untreated cells whereas their numbers were gradually increased after transfection in both scrambled and miRNA mimic group at the three-time points. Apoptosis was much more elevated than that in miRNA mimic group compared to the scrambled group showing that hsa-let-7c-5p overexpression is associated with increment of apoptosis (increased apoptosis) in KG-1 cells (p < 0.001; Figs. 4, 5a).
Fig. 4.
Evaluation of apoptosis and necrosis in the untreated cells, mimic control and hsa-let-7c mimic groups by Annexin-V and propidium iodide (PI) staining performed 24, 48 and 72 h after transfection (a–c). Flow cytometry analysis was performed using 488-nm excitation and a 515-nm band-pass filter for fluorescein detection and a 600-nm filter for PI detection
Fig. 5.
Assessment of apoptosis and necrosis using the Annexin-V/PI assay performed 24, 48 and 72 h after transfection. Based on the analysis presented in Fig. 4, apoptosis of the untreated cells at each time point was considered as ~ 0%, and apoptosis of other groups is presented as the percentage of the untreated cells at the same time point (a). Data are presented as mean ± SD of three independent experiments (***p < 0.001). Necrosis of the untreated cells at each time point was considered as ~ 0% and necrosis of other groups is presented as the percentage of the untreated cells at the same time point (b). Data are presented as mean ± SD of three independent experiments (***p < 0.001)
In line with the data of apoptosis assay, necrosis was almost undetectable in untreated cells whereas the ratio of necrotic cells increased gradually after transfection in both scrambled and miRNA mimic group at the three-time points. Necrosis in the miRNA mimic group was higher than in the other groups at the three-time points showing that overexpression of hsa-let-7c-5p is associated with an increment of necrosis in KG-1 cells (p < 0.001; Figs. 4, 5b).
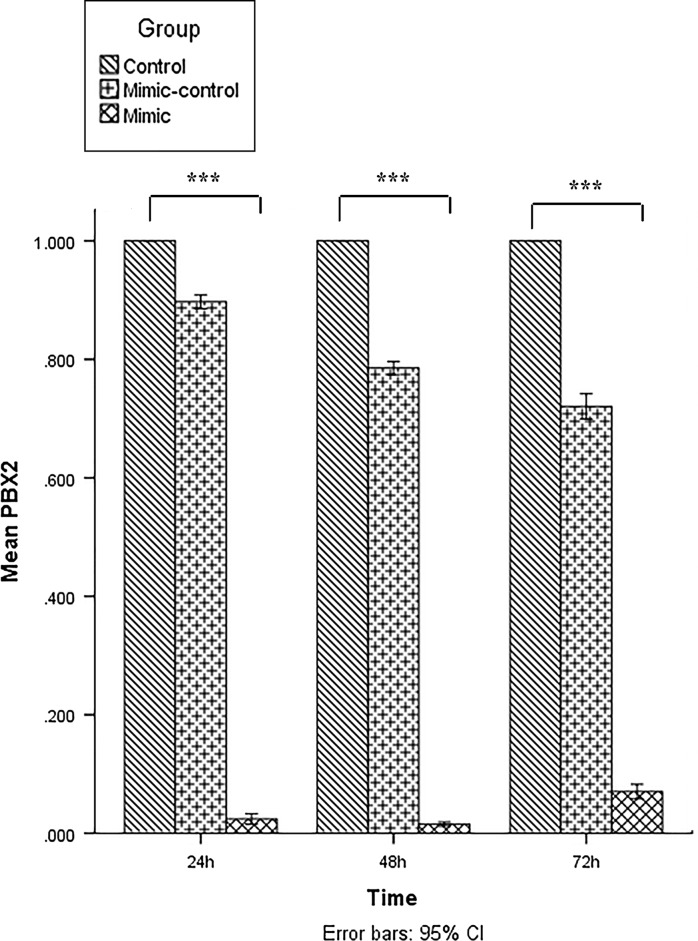
Overexpression of hsa-let-7c-5p targets PBX2 gene in acute erythroleukemia cells
The expression level of PBX2 gene was determined by RT-qPCR in miRNA mimic, scrambled and untreated groups of KG-1 cells at three-time points (24, 48 and 72 h post-transfection). The PBX2 expression was lower in the miRNA mimic group at the three-time points after transfection than in the scrambled and untreated groups (p < 0.001). The lower-level expression of PBX2 was at 48 h after transfection in KG-1 cells (Fig. 6).
Fig. 6.
Evaluation of the PBX2 level by real-time PCR at 24, 48, and 72 h post transfection. The ΔΔCt method was used for data analysis and the untreated group (control) was considered as reference for each time point. Data are presented as mean ± SD of three independent experiments (***p < 0.001)
Discussion
Acute myeloid leukemia (AML) is the most common acute leukemia cancer affecting adults and AML-M6 (acute erythroleukemia) which accounts for 3–5% of denovo AMLs and 20–30% of secondary leukemias. The incidence increases progressively with age especially in people older than 65 years (Dores et al. 2011). Also, it has been reported in children. Patients with AML-M6 have a poor prognosis and challenges in the treatment of acute erythroleukemia include induction failure, relapsed chemotherapeutic toxicity, and drug resistance (Acevedo-Olvera et al. 2015; Mazzella et al. 2000).
MicroRNAs are small, noncoding regulatory RNAs that have definitive roles in biological processes and act as the regulator of gene expression post-transcriptionally (Faller and Guo 2008; Sharifi et al. 2014). Dysregulation of miRNA expression is confirmed in multiple human cancers (Reddy et al. 2010). Some are oncogenes like miR-183 which is increased in synovial sarcoma and colon cancer, targeting the EGR1 and leads to cell migration in these tumor types (Sarver et al. 2010). Some other miRNAs are tumor suppressor genes such as miR-29c which acts as tumor suppressor gene by targeting VEGFA in lung adenocarcinoma (Liu et al. 2017).
One other example of tumor suppressor microRNA is hsa-let-7c-5p which acts as a tumor suppressor in different ways, such as cancer progression prevention by repressing of HMGA2 expression (Park et al. 2007), migration, and invasion inhibition of lung and colorectal cancer cells (Han et al. 2012; Zhao et al. 2014). Also, it shows apoptosis-inducing effect in hepatocellular carcinoma (Zhu et al. 2015). As for AML, downregulation of hsa-let-7c-5p in tissues and cells has been reported as a cause of increasing proliferation of AML. Therapeutic strategies to overexpress hsa-let-7c-5p may be useful for AML-M6 patients. Different studies suggest that reintroduction of let-7c (and other tumor suppressive miRNAs) in tumor cells has therapeutic value in different cancers including prostate cancers, colorectal adenocarcinoma, lung adenocarcinoma (Gambari et al. 2016; Rothschild 2014). One method of the reestablishment of a tumor suppressor miRNA is use of miRNA mimic that has the same sequence as the natural miRNA has, therefore, it is expected to target the same mRNAs that are regulated by the natural miRNA (Bader et al. 2010; Gambari et al. 2016). Decreasing cell proliferation, increasing apoptosis/late apoptosis and targeting of cancer-related genes (oncogenes) is the aim of cancer treatment. Therefore, miRNA mimic may be used to upregulate hsa-let-7c-5p in order to treat AML. Some efforts were made to treat some cancers by overexpression of tumor-supressor miRNAs. For example, a study on miRNA replacement, presented that, miR-34a, which is confirmed to be downregulated in Non-Small Cell Lung Cancer (NSCLC), was upregulated and as a result, it had led to inhibition of growth of NSCLC (Shi et al. 2014). In non-small lung cancer, let-7 is downregulated which is related to increased expression of RAS (a key cancer gene activated in NSCLC) (Bader et al. 2010). Also, reintroduction of let-7 mimics in cultured lung cancer cells and mouse models of lung cancer, decreases the proliferation of cancer cells and reduces growth of the tumors by targeting the RAS oncogene (Johnson et al. 2005, 2007).
We suggest miRNA replacement in AML-M6 by using hsa-let-7c-5p mimic. According to our findings, upregulated hsa-let-7c-5p has led to decreased cell proliferation, induced apoptosis, and late apoptosis. However, this perturbation of let-7c expression decreased PBX2 (an oncogenic gene of AML and target of hsa-let-7c-5p) expression level, suggesting that reestablishment of hsa-let-7c-5p in cancer cells can be potentially used for the treatment of acute leukemia. It can be used alone or in combination with conventional therapies. Furthermore, in vivo studies are essential to assess the feasibility of this strategy. However, in vivo delivery of miRNA mimic is the main challenge for successful translation into the clinic which will be the focus of therapeutic development efforts. In this study, we have used hsa-let-7c-5p miRNA mimic to increase the expression level of hsa-let-7c-5p in the KG-1 cell line. Real-time qPCR data confirmed that hsa-let-7c-5p was upregulated after hsa-let-7c-5p miRNA mimic transfection. MTT assay showed that overexpression of hsa-let-7c-5p is associated with decreased cell viability. These data were confirmed further by apoptosis/late apoptosis assay (Annexin-V and PI), showing hsa-let-7c-5p miRNA mimic transfection significantly increases apoptosis/late apoptosis. In addition, we realized that the Annexin-V and PI staining method which we used, cannot discriminate between apoptosis and late apoptosis.
Our data show that overexpression of hsa-let-7c-5p can decrease the viability of KG-1 cells that is at least partially due to increased apoptosis and late apoptosis. According to different studies on acute myeloid leukemia, PBX2, a homeodomain protein, whose aberrant expression enhances HOXA9-dependant leukemogenesis, contributes to the AML phenotype. HOXA9 shows a strong connection to AML and it has been found to be upregulated in AML. Also, the MEIS1 transcription factor is found to be upregulated in this disease. Overexpression of both HOXA9 and MEIS1 is only weakly oncogenic. Additionally, it has been observed that the PBX2 family of proteins are required for more efficient transformation in AML (Moskow et al. 1995; Shen et al. 1999). As it is shown in a study by Pelosi et al. (2013), PBX2 was determined as a novel target of hsa-let-7c-5p (Pelosi et al. 2013). Therefore, we selected the PBX2 gene not only as an oncogene in AML-M6 but also as a target of hsa-let-7c-5p in order to evaluate the effectivity of this approach on the treatment of erythroleukemia cancer cells. We used Real-time qPCR to determine the expression level of PBX2. The results show that the gene expression was at a lower level in the miRNA mimic group at the three-time points than in the two other groups showing that overexpression of hsa-let-7c-5p (a tumor suppressor miRNA) decreases the PBX2 oncogene which strengthens the effectivity of our approach to have therapeutic value in AML-M6. Our results can be used in translational medicine for future investigation in AML treatment and also, producing drugs, based on miRNA replacement therapy.
Acknowledgments
This study was conducted with the financial support of Isfahan University of Medical Sciences (IRAN) with Grant Number 395207.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the local ethics committee of Isfahan University of Medical Sciences (IRAN) and the studies have been approved by the appropriate institutional and/or national research ethics committee and have been performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Contributor Information
Deniz Mortazavi, Email: denizmortazavi_mui@yahoo.com.
Mohammadreza Sharifi, Phone: +983137929037, Email: mo_sharifi@med.mui.ac.ir.
References
- Acevedo-Olvera LF, Diaz-Garcia H, Parra-Barrera A, Caceres-Perez AA, Gutierrez-Iglesias G, Rangel-Corona R, Caceres-Cortes JR. Inhibition of the Na +/H + antiporter induces cell death in TF-1 erythroleukemia cells stimulated by the stem cell factor. Cytokine. 2015;75:142–150. doi: 10.1016/j.cyto.2015.06.020. [DOI] [PubMed] [Google Scholar]
- Amini-Farsani Z, Sangtarash MH, Shamsara M, Teimori H. MiR-221/222 promote chemoresistance to cisplatin in ovarian cancer cells by targeting PTEN/PI3K/AKT signaling pathway. Cytotechnology. 2018;70:203–213. doi: 10.1007/s10616-017-0134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Byrd JC, et al. Repetitive cycles of high-dose cytarabine benefit patients with acute myeloid leukemia and inv (16)(p13q22) or t (16; 16)(p13; q22): results from CALGB 8461. J Clin Oncol. 2004;22:1087–1094. doi: 10.1200/JCO.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Döhner H, Estey EH, Amadori S, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- Döhner H, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Ji M, Ji C. Micro-RNAs and their potential target genes in leukemia pathogenesis. Cancer Biol Ther. 2009;8:200–205. doi: 10.4161/cbt.8.3.7333. [DOI] [PubMed] [Google Scholar]
- Dores GM, Devesa SS, Curtis RE, Linet MS, Morton LM. Acute leukemia incidence and patient survival among children and adults in the United States, 2001–2007. Blood. 2011;119:34–43. doi: 10.1182/blood-2011-04-347872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakaki A, Iliopoulos D. MicroRNA gene networks in oncogenesis. Curr Genom. 2009;10:35–41. doi: 10.2174/138920209787581299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faller M, Guo F. MicroRNA biogenesis: there’s more than one way to skin a cat. Biochim Biophys Acta BBA Gene Regul Mech. 2008;1779:663–667. doi: 10.1016/j.bbagrm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Νew trends in the development of miRNA therapeutic strategies in oncology. Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammell P. MicroRNAs: recently discovered key regulators of proliferation and apoptosis in animal cells. Cytotechnology. 2007;53:55–63. doi: 10.1007/s10616-007-9049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George G, Mittal RD. MicroRNAs: potential biomarkers in cancer. Indian J Clin Biochem. 2010;25:4–14. doi: 10.1007/s12291-010-0008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, et al. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127–19137. doi: 10.1074/jbc.M110.216887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Gu J, Zuo H, et al. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J Pathol. 2012;226:544–555. doi: 10.1002/path.3014. [DOI] [PubMed] [Google Scholar]
- Hasserjian R, Howard J, Wood A, Henry K, Bain B. Acute erythremic myelosis (true erythroleukaemia): a variant of AML FAB-M6. J Clin Pathol. 2001;54:205–209. doi: 10.1136/jcp.54.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel J, Bartschat S, Wintsche A, Otto C, of the Bioinformatics Computer Lab TS, Stadler PF Evolution of the let-7 microRNA family. RNA Biol. 2012;9:231–241. doi: 10.4161/rna.18974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson CD, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Kavianpour M, Ahmadzadeh A, Shahrabi S, Saki N. Significance of oncogenes and tumor suppressor genes in AML prognosis. Tumor Biol. 2016;37:10041–10052. doi: 10.1007/s13277-016-5067-1. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Kowal-Vern A, Mazzella FM, Cotelingam JD, Shrit MA, Rector JT, Schumacher HR. Diagnosis and characterization of acute erythroleukemia subsets by determining the percentages of myeloblasts and proerythroblasts in 69 cases. Am J Hematol. 2000;65:5–13. doi: 10.1002/1096-8652(200009)65:1<5::AID-AJH2>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Liu L, et al. MicroRNA-29c functions as a tumor suppressor by targeting VEGFA in lung adenocarcinoma. Mol Cancer. 2017;16:50. doi: 10.1186/s12943-017-0620-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Mrózek K, Bloomfield CD. Molecular heterogeneity and prognostic biomarkers in adults with acute myeloid leukemia and normal cytogenetics. Curr Opin Hematol. 2005;12:68–75. doi: 10.1097/01.moh.0000149608.29685.d1. [DOI] [PubMed] [Google Scholar]
- Mazzella FM, Alvares C, Kowal-Vern A, Schumacher HR. The acute erythroleukemias. Clin Lab Med. 2000;20:119–137. doi: 10.1016/S0272-2712(18)30080-5. [DOI] [PubMed] [Google Scholar]
- Mohammadiasl J, Khosravi A, Shahjahani M, Azizidoost S, Saki N. Molecular and cellular aspects of extramedullary manifestations of acute myeloid leukemia. J Cancer Metastasis Treat. 2016;2:44–50. [Google Scholar]
- Monica K, Galili N, Nourse J, Saltman D, Cleary ML. PBX2 and PBX3, new homeobox genes with extensive homology to the human proto-oncogene PBX1. Mol Cell Biol. 1991;11:6149–6157. doi: 10.1128/MCB.11.12.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/MCB.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadiminty N, et al. MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PLoS ONE. 2012;7:e32832. doi: 10.1371/journal.pone.0032832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-M, et al. Let-7 prevents early cancer progression by suppressing expression of the embryonic gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- Pelosi A, et al. miRNA let-7c promotes granulocytic differentiation in acute myeloid leukemia. Oncogene. 2013;32:3648–3654. doi: 10.1038/onc.2012.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SDN, Gajula RP, Pakala SB, Kumar R. MicroRNAs and cancer therapy: the next wave or here to stay? Cancer Biol Ther. 2010;9:479–482. doi: 10.4161/cbt.9.7.11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild SI. microRNA therapies in cancer. Mol Cell Ther. 2014;2:7. doi: 10.1186/2052-8426-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. Chromosomal translocations: revisited yet again. Blood. 2008;112:2183–2189. doi: 10.1182/blood-2008-04-097931. [DOI] [PubMed] [Google Scholar]
- Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- Santos F, et al. Adult acute erythroleukemia: an analysis of 91 patients treated at a single institution. Leukemia. 2009;23:2275–2280. doi: 10.1038/leu.2009.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- Sharifi M, Salehi R, Gheisari Y, Kazemi M. Inhibition of microRNA miR-92a induces apoptosis and inhibits cell proliferation in human acute promyelocytic leukemia through modulation of p63 expression. Mol Biol Rep. 2014;41:2799–2808. doi: 10.1007/s11033-014-3134-5. [DOI] [PubMed] [Google Scholar]
- Shen W-F, Rozenfeld S, Kwong A, Kömüves LG, Lawrence HJ, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/MCB.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Liu C, Liu X, Tang DG, Wang J. The microRNA miR-34a inhibits non-small cell lung cancer (NSCLC) growth and the CD44hi stem-like NSCLC cells. PLoS ONE. 2014;9:e90022. doi: 10.1371/journal.pone.0090022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoicea N, Du A, Lakis DC, Tipton C, Arias-Morales CE, Bergese SD. The MiRNA journey from theory to practice as a CNS biomarker. Front Genet. 2016;7:11. doi: 10.3389/fgene.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Kasar S, Underbayev C, Prakash S, Raveche E. MicroRNAs in acute myeloid leukemia and other blood disorders. Leuk Res Treat. 2012;2012:603830. doi: 10.1155/2012/603830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- Zhu X, Wu L, Yao J, Jiang H, Wang Q, Yang Z, Wu F. MicroRNA let-7c inhibits cell proliferation and induces cell cycle arrest by targeting CDC25A in human hepatocellular carcinoma. PLoS ONE. 2015;10:e0124266. doi: 10.1371/journal.pone.0124266. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]