Abstract
Enterotoxigenic Escherichia coli (ETEC) is a leading cause of infectious diarrhea in children and postweaning piglets. ETEC infection results in induced pro-inflammatory responses in intestinal epithelial cells and dysbiosis of intestinal microbiota. Here, a Lactobacillus reuteri strain, HCM2, isolated from a healthy piglet showed a high survival rate in the harsh gastrointestinal tract environment and inhibited the growth of ETEC and its adherence to intestinal epithelial cells. Pre-supplementation with L. reuteri HCM2 for 14 days reduced the ETEC load in the jejunum of ETEC-infected mice and prevented the disruption of intestinal morphology by ETEC. The colonic microbiota of mice with or without HCM2 pre-supplementation were analyzed, and this analysis revealed that HCM2 could prevent dysbiosis caused by ETEC infection by stabilizing the relative abundance of dominant bacteria. These results indicate that L. reuteri HCM2 has the potential to attenuate the effect of ETEC on the colonic microbiota in infected mice.
Introduction
Enterotoxigenic Escherichia coli (ETEC) is a leading cause of infectious diarrhea in children from developing nations and is responsible for an estimated 3–5 million deaths annually in children under the age of five1. ETEC is also the main infectious agent of postweaning diarrhea in piglets and is responsible for 50% of piglet deaths worldwide annually2. The K88 (F4) fimbrial adhesin, heat-stable and heat-labile enterotoxins have been identified as important virulence factors leading to diarrheal diseases in piglets, and account for 93% of ETEC infections in piglets3–5. ETEC is known to adhere to the small intestinal epithelium and to secrete enterotoxins that alter the functions of enterocytes by increasing secretion, which leads to severe secretory diarrhea in pigs3. ETEC results in the loss of microvilli in the jejunum and promotes inflammation in a mouse model6. Ren et al. found that ETEC infection promotes the expression of pro-inflammatory cytokines through the activation of the NF-kB and MAPK pathways6. ETEC infection also induces the expression of intestinal IL-17 and causes dysbiosis of intestinal microbiota, increasing the abundance of Lactococcus lactis subsp. lactis in infected mice7.
Antimicrobial growth promoters and therapeutic antibiotics are widely used in animal farming to prevent neonatal and postweaning diarrhea8–10. But they also potentially contribute to the accumulation of antimicrobial drug resistance genes in both pathogenic and non-pathogenic human bacteria, which will lead to serious public health problems11. As a result, the use of all antibiotics as growth promoters has been progressively banned from European agriculture since 2006 under Regulation 1831/2003/EEC12. Thus, there is an urgent need to develop “green antibiotics” that have a minimal ecological impact on the animal commensal and environmental microbiomes11. Among the non-antibiotic alternatives, probiotics seem to have the highest potential as they are efficient against pathogenic strains in animals2. A probiotic is defined as “a live microorganism that, when administered in adequate amounts, confers a health benefit on the host”13. Several reports have elucidated the role of probiotic bacteria in preventing and treating gastrointestinal diseases5,14,15. One example is lactobacillus whose potential benefits for human and animal health include the improvement of lactose intolerance, prevention of intestinal infection, modulation of the intestinal microbiota, reduction of serum cholesterol, stimulation of the immune system, anticarcinogenic action, and antioxidative effects16–19. For instance, L. rhamnosus plus L. acidophilus protected against Shigella infection by increasing antioxidant levels16. Gao et al. also found that L. plantarum could induce a high level of immune response, stimulate the growth of many intestinal Lactobacillus spp. and accelerate intestinal microbiota maturation20.
L. reuteri is a common species that inhabits the gastrointestinal tract of human, pig, hamster, mouse, rat, dog, sheep, cattle, and different birds8,21,22. Numerous studies have demonstrated that L. reuteri is resistant to gastric acid and bile, positively improves the performance of pigs, prevents diarrhea, alleviates stress, alters gastrointestinal microbiota, reduces the abundance of colibacillus and regulates the immune system8,23–25. L. reuteri can produce a variety of antimicrobial substances, such as lactic acid and bacteriocin reuterin26. L. reuteri also has the capacity to adhere to mucin and colonize on intestinal epithelial cells through cell surface proteins27. However, whether L. reuteri protects against ETEC by altering gut microbiota in mice is unknown.
In this study, an L. reuteri strain, HCM2, was isolated from the cecum content of a 6-week-old healthy piglet and showed good probiotic characteristics in vitro. Using an ETEC-infected specific-pathogen-free mouse model, we demonstrated that L. reuteri could protect against ETEC colonization and intestinal disruption caused by ETEC by positively affecting the intestinal microbiota.
Results
L. reuteri HCM2 inhibits the growth of ETEC and its ability to adhere to intestinal epithelial cells
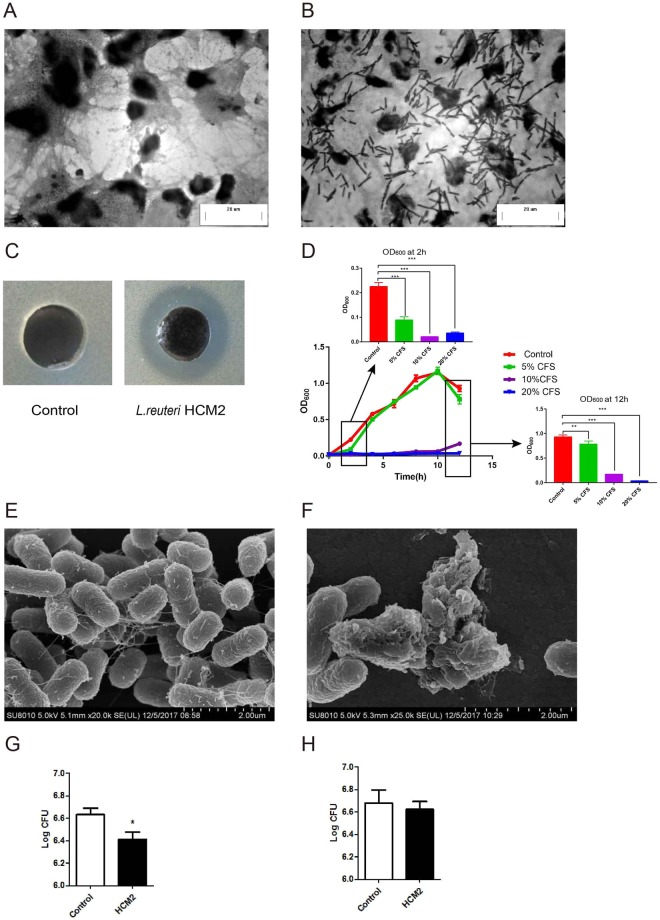
L. reuteri HCM2 showed a high rate of survival in artificial gastric juice (88.35%) and artificial small intestine fluid (73.93%) (Table 1). It could survive in bile salt concentrations of up to 0.3%, and the survival rates of L. reuteri HCM2 in 0.1% and 0.2% bile salt were 30.12% and 10.2%, respectively (data not shown). It also appeared to adhere to Caco-2 cells in chains when observed under a microscope (Fig. 1A,B); (16.1 ± 6.5) ×106 colony forming units (CFUs) L. reuteri HCM2 cells were found to adhere to one square centimeter of Caco-2 cells. These results suggest that L. reuteri HCM2 is tolerant to the environment of the gastrointestinal tract and may adhere to the intestinal epithelial cells.
Table 1.
Survival of L. reuteri HCM2 in artificial gastric juice (pH 2.5) and artificial small intestine fluid after incubation for 0 min and 2 h.
| L. reuteri HCM2 | Log CFU/ml (% survival) | |
|---|---|---|
| 0 min | 2 h | |
| Artificial gastric juice | 7.54 ± 0.08 (100) | 7.48 ± 0.10 (88.35) |
| Artificial small intestine fluid | 8.10 ± 0.02 (100) | 7.97 ± 0.05 (73.93) |
Figure 1.
The probiotic properties of L. reuteri HCM2 (A) Caco-2 cells observed using light microscopy. (B) Adhesion of L. reuteri HCM2 to Caco-2 cells observed using light microscopy after Gram-staining. (C) An agar diffusion assay showing the antibacterial activity of L. reuteri HCM2 against ETEC. (D) The inhibition of ETEC growth by L. reuteri HCM2 was examined by adding different concentrations (5%, 10% and 20%) of L. reuteri HCM2 cell free supernatant (CFS) to ETEC inoculations and then measuring growth over 12 h. OD600 values of the inoculations at 2 h and 12 h are shown as bar graphs. The data are presented as mean ± SD with n = 5. (E) An SEM image of ETEC cells after 12 h of cultivation at 37 °C. (F): An SEM image of ETEC cultivated in medium containing 20% L. reuteri HCM2-CFS for 12 h at 37 °C. The scale bar is 2 μm in (E,F). (G) Number of ETEC cells bound to Caco-2 cells after ETEC (108 CFUs) and L. reuteri HCM2 (108 CFUs) cells were incubated with Caco-2 cells at 37 °C for 90 min. H: Number of ETEC cells bound to Caco-2 cells after ETEC (108 CFUs) cells were incubated with Caco-2 cells at 37 °C for 45 min, and then with L. reuteri HCM2 (108 CFUs) and Caco-2 cells for another 45 min. These results include data from three independent experiments. *Significant difference at p < 0.05.
We next tested the effect of L. reuteri HCM2 on ETEC growth. We found that the CFS (cell free supernatant) of L. reuteri HCM2 produced a clear zone with a diameter ≥14 mm in ETEC lawns (Fig. 1C), indicating that L. reuteri HCM2 may be capable of inhibiting the growth of ETEC. To test this, different concentrations (5%, 10% and 20%) of the CFS of L. reuteri HCM2 were added to the ETEC culture medium, and ETEC growth was monitored. ETEC was obviously inhibited by 10% and 20% L. reuteri HCM2 CFS (Fig. 1D). Observations under an SEM (scanning electron microscope) revealed that the CFS destroyed the rod-shaped structures of the ETEC cells (Fig. 1E,F). These results indicate that the CFS of L. reuteri HCM2 might inhibit ETEC growth by damaging the cell wall. ETEC cells had the ability to adhere to Caco-2 cells; however, the number of ETEC cells binding to Caco-2 cells was significantly reduced when ETEC were co-cultured with L. reuteri HCM2 (Fig. 1G). These results suggest that L. reuteri HCM2 could compete with ETEC cells and prevent them from adhering to Caco-2 cells. By contrast, no obvious difference in the number of ETEC cells bound to Caco-2 cells was observed when L. reuteri HCM2 cells were added to a pre-culture of ETEC and Caco-2 cells (Fig. 1H). Thus, it seems that L. reuteri HCM2 contributes little to replace ETEC that have already bound to Caco-2 cells. We hypothesized that L. reuteri HCM2 protects intestinal epithelial cells against ETEC.
L. reuteri HCM2 reduces ETEC load in the jejunum and preserves intestinal morphology in mice
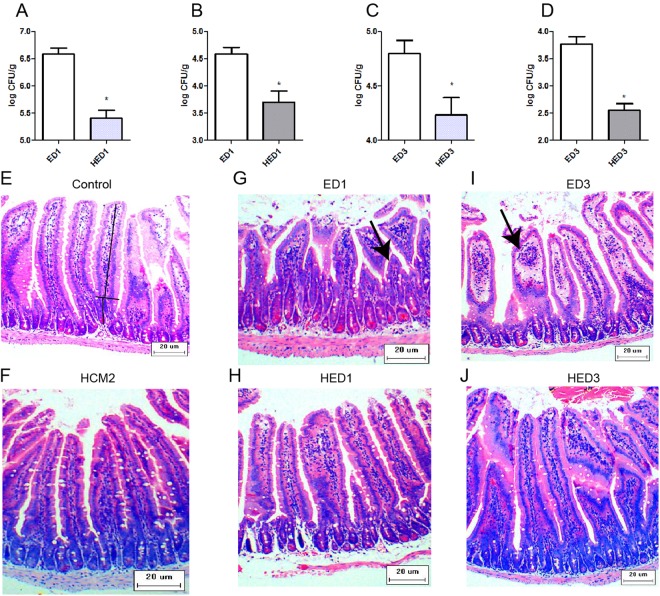
The number of ETEC CFUs in the jejunal tissues and jejunal contents were determined to estimate the ETEC load in mice. Before challenging mice with ETEC at day 15 (D15), no ETEC were detected in mice in the Control and HCM2 groups. The mice infected by ETEC all survived. At day 16 (D16), after 1 day of recovery from the ETEC challenge, the ETEC loads in the jejunal tissues (p < 0.05) and jejunal contents (p < 0.05) of mice pre-supplemented with L. reuteri HCM2 (HED1 group) were significantly lower than those in mice without L. reuteri HCM2 pre-treatment (ED1 group) (Fig. 2A,B). At day 18 (D18), after 3 days of recovery from the ETEC challenge, the ETEC loads in the jejunal contents and jejunal tissues of mice pre-supplemented with L. reuteri HCM2 for 14 days (HED3) were significantly lower (p < 0.05) than those in mice without L. reuteri HCM2 pre-treatment (ED3) (Fig. 2C,D). These results demonstrate that L. reuteri HCM2 can reduce ETEC loads in the mouse jejunum.
Figure 2.
L. reuteri HCM2 preserves intestinal morphology in ETEC infected mice. (A) The load of ETEC in the jejunal tissues at D16; (B) The load of ETEC in the jejunal contents at D16; (C) The load of ETEC in the jejunal tissues at D18; (D) The load of ETEC in the jejunal contents at D18; E to J: Representative images of HE staining of the jejunum of weanling mice are shown (×100; n = 5). The villus length and crypt depth were measured as indicated in the image in (E). The black arrows in G and I indicate jejunal tissues that were damaged by ETEC. Control: mice received a basal diet. HCM2: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks. ED1: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 1 day. HED1: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15, and then received a basal diet for 1 day. ED3: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 3 days. HED3: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15, and then received a basal diet for 3 days.
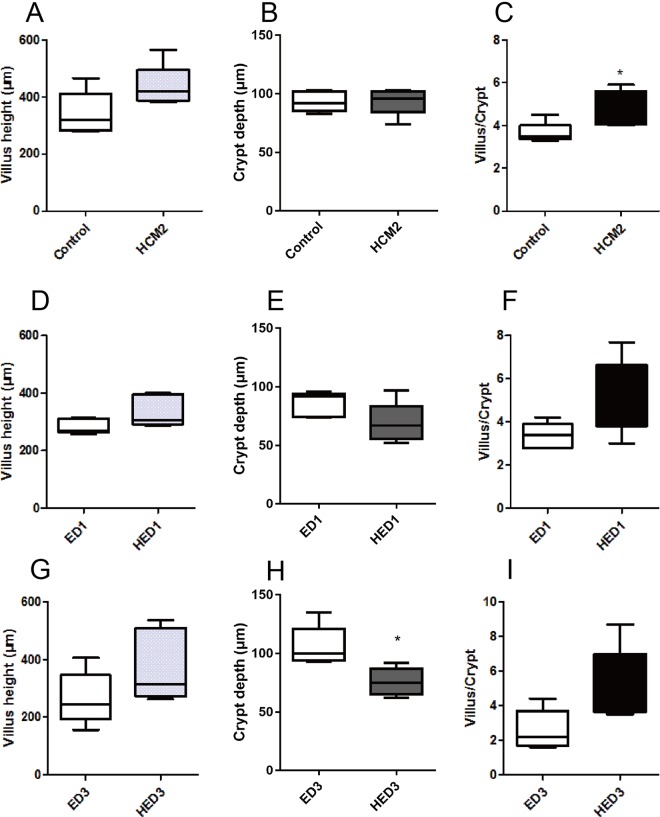
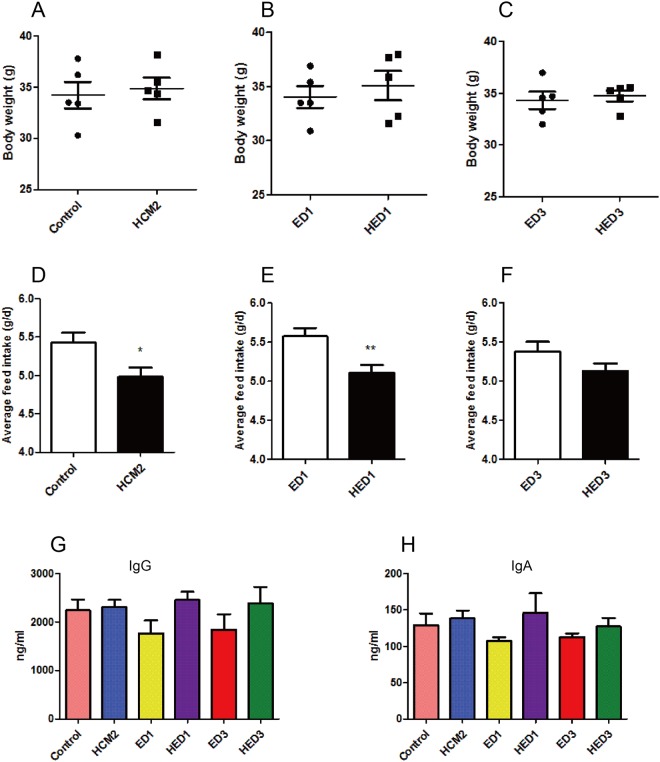
Morphological analyses revealed that ETEC infection led to inflammatory infiltration and loss of microvilli in the jejunum tissues of mice in the ED1 and ED3 groups (Fig. 2G,I), while inflammatory infiltration was not observed in mice in the HED1 and HED3 groups (Fig. 2H,J). These results indicate that L. reuteri HCM2 could protect the intestine from disruption by ETEC. At D15, mice pre-treated with L. reuteri HCM2 had a significantly higher villus/crypt ratio than mice in the control group (p < 0.05, Fig. 3C). At D18, three days after the ETEC challenge, the crypt depth was significantly lower in HED3 mice than in ED3 mice (p < 0.05, Fig. 3H). Although the feed intake at D15 (Fig. 4E) and D16 (Fig. 4F) was lower in mice receiving L. reuteri HCM2 supplementation, there was no obvious difference in body weight between groups (Fig. 4A–C). Levels of serum IgG and IgA, which reflect the system immune state28, were also determined and compared between the different groups. At D15, the HCM2 group had 2.9% and 7.67% higher serum IgG and IgA levels, respectively than the control group (Fig. 4G,H). At D16, the HED1 group had 38.93% and 35.63% higher levels of serum IgG and IgA, respectively, than the ED1 group (Fig. 4G,H). At D18, the HED3 group had 29.41% and 13.27% higher levels of serum IgG and IgA, respectively, than the ED3 group (Fig. 4G,H).
Figure 3.
The villus length, crypt depth and the ratio of the villus to crypt in different groups of mice. (A–C) villus length (A), crypt depth (B), and the ratio of villus to crypt (C) at D15. (D–F) Villus length (D), crypt depth (E) and the villus to crypt ratio (F) at D16. (G–I) Villus length (G), crypt depth (H) and the villus to crypt ratio (I) at D18. The data are presented as mean ± SD with n = 5. *Indicates significance at p < 0.05. Control: mice received a basal diet. HCM2: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks. ED1: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received basal diet for 1 day. HED1: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15, and then received a basal diet for 1 day. ED3: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 3 days. HED3: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15, and then received a basal diet for 3 days.
Figure 4.
Body weight and feed intake for different groups of mice. (A–C) The body weight of mice in different groups. (D–F) The average feed intake for different groups. (G) The expression level of serum IgG in mice from different groups. (H) The expression level of serum IgA in mice from different groups. The data are presented as mean ± SD with n = 5. * indicates significance at p < 0.05. Control: mice received a basal diet. HCM2: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks. ED1: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 1 day. HED1: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15 and then received a basal diet for 1 day. ED3: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 3 days. HED3: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15, and then received a basal diet for 3 days.
Colonic microbiota diversity and composition analysis
The colonic microbiota of mice in the six groups were analyzed by sequencing the bacterial 16 S rRNA V3 + V4 region. High-throughput pyrosequencing of the samples (n = 5) produced a total of 2 757 344 raw reads. The effective reads were clustered into OTUs based on a similarity threshold of 97%. The 2 004 194 clean tags remaining after removing low-quality sequences were clustered into a total of 2247 OTUs, which were present in at least five samples. The rarefaction and rank abundance curves showed that the total richness of the microbial community was sampled completely (Fig. S1A,B). The Chao1, OS (Observed species), ACE, Shannon, Simpson, and PD (phylogenetic distance whole tree) indexes were calculated (Table S1). Based on these indexes there was not a big difference in microbial structure between mice receiving the L. reuteri HCM2 pre-treatment and those that did not.
The sequences across all samples were assigned to 27 phyla and 280 genera. Firmicutes and Bacteroidetes were the two predominant phyla in the mice gut microbiota, followed by Proteobacteria, Actinobacteria, and Saccharibacteria (Table 2). To investigate the gut bacteria in each group of mice, namely Control, HCM2, ED1, HED1, ED3 and HED3, the relative abundance of bacteria in each group was estimated based on the number of representative OTUs. The dominant sequences, comprising > 1% of the total bacteria composition, belonged to 22 genera: Lactobacillus, Lachnospiraceae-_NK4A136_group, Candidatus Arthomitus, Ruminococcaceae_UCG-014, unidentified_Lachnospiraceae, Roseburia, Anaerotruncus, unidentified_Ruminococcaceae, Lachnospiraceae_UCG-006, Lachnoclostridium and Ruminiclostridium_9, which belong to Firmicutes; Alloprevotella, Alistipes, Bacteroides, Odoribacter, Parabacteroides, Rikenella and Rikenellaceae_RC9_gut_group which belong to Bacteroidete; Helicobacter and Desulfovibrio which belong to Proteobacteria; Bifidobacterium, which belong to Actinobacteria; and Candidatus Saccharimonas, which belong to Saccharibacteria. We observed significant differences in the composition of microbiota between groups. Major changes in composition were mainly observed for Firmicutes, Bacteroidete, and Proteobacteria. Changes in the abundance of Firmicutes were mainly attributable to Lactobacillus. The relative abundances of all genera belonging to Bacteroides changed at D16, especially in the HED1 group. Changes in the abundance of Proteobacteria were mainly attributable to Helicobacter (Table 2).
Table 2.
Relative abundance (%) of bacterial genera in the colonic microbiota of mice during ETEC treatment, determined by Illumina sequencing of 16S rRNA tags.
| Genus | D15 | D16 | D18 | |||
|---|---|---|---|---|---|---|
| Control | HCM2 | ED1 | HED1 | ED3 | HED3 | |
| Firmicutes | ||||||
| Lactobacillus | 75.08 ± 10.5ab | 86.06 ± 3.75a | 62.89 ± 10.1bc | 44.63 ± 23.13c | 52.76 ± 25.68c | 86.27 ± 3.57a |
| Lachnospiraceae_NK4A136_group | 0.75 ± 1.02 | 0.35 ± 0.29 | 1.13 ± 0.56 | 1.68 ± 2.26 | 2.4 ± 2.93 | 0.35 ± 0.21 |
| Candidatus_Arthromitus | 1.98 ± 0.69 | 2.19 ± 1.88 | 1.45 ± 0.69 | 1.85 ± 0.49 | 1.94 ± 2.77 | 1.4 ± 0.52 |
| Ruminococcaceae_UCG-014 | 1.3 ± 0.6bc | 0.64 ± 0.61c | 1.02 ± 0.23c | 2.68 ± 1.82a | 1 ± 0.67c | 1.18 ± 0.97c |
| unidentified_Lachnospiraceae | 0.03 ± 0.01b | 0.04 ± 0.04b | 0.17 ± 0.05b | 0.17 ± 0.26b | 1.19 ± 1.66a | 0.08 ± 0.05b |
| Roseburia | 0.13 ± 0.12ab | 0.07 ± 0.05b | 0.19 ± 0.09ab | 0.19 ± 0.27ab | 0.81 ± 1.25a | 0.09 ± 0.08b |
| Anaerotruncus | 0.14 ± 0.06b | 0.04 ± 0.01b | 0.18 ± 0.07ab | 0.3 ± 0.4ab | 0.7 ± 0.95a | 0.08 ± 0.06ab |
| unidentified_Ruminococcaceae | 0.05 ± 0.03b | 0.03 ± 0.01b | 0.11 ± 0.06ab | 0.03 ± 0.02b | 0.57 ± 0.95a | 0.02 ± 0.01b |
| Lachnospiraceae_UCG-006 | 0.06 ± 0.02 | 0.03 ± 0.01 | 0.11 ± 0.04 | 0.52 ± 0.92 | 0.11 ± 0.04 | 0.11 ± 0.09 |
| Lachnoclostridium | 0.16 ± 0.15 | 0.11 ± 0.06 | 0.14 ± 0.06 | 0.49 ± 0.86 | 0.27 ± 0.25 | 0.09 ± 0.07 |
| Ruminiclostridium_9 | 0.15 ± 0.08b | 0.06 ± 0.05b | 0.2 ± 0.06ab | 0.19 ± 0.13ab | 0.47 ± 0.56a | 0.07 ± 0.03b |
| Coprococcus_1 | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.13 ± 0.11 | 0.1 ± 0.11 | 0.3 ± 0.55 | 0.01 ± 0.01 |
| Ruminiclostridium | 0.03 ± 0.01b | 0.01 ± 0.01b | 0.08 ± 0.02ab | 0.03 ± 0.03b | 0.32 ± 0.49a | 0.01 ± 0.08b |
| Oscillibacter | 0.03 ± 0.02b | 0.01 ± 0.01b | 0.05 ± 0.02ab | 0.04 ± 0.05ab | 0.24 ± 0.38a | 0.002 ± 0.004b |
| Clostridium_sensu_stricto_1 | 0.22 ± 0.35 | 0.04 ± 0.02 | 0.05 ± 0.02 | 0.05 ± 0.02 | 0.04 ± 0.03 | 0.04 ± 0.02 |
| Staphylococcus | 0.04 ± 0.02c | 0.03 ± 0.02c | 0.09 ± 0.01bc | 0.02 ± 0.01c | 0.27 ± 0.32a | 0.24 ± 0.01ab |
| Erysipelatoclostridium | 0.08 ± 0.04 | 0.06 ± 0.08 | 0.56 ± 1.01 | 0.29 ± 0.31 | 0.19 ± 0.27 | 0.07 ± 0.08 |
| Allobaculum | ND | 0.02 ± 0.42 | ND | ND | ND | 0.01 ± 0.01 |
| Other (P: Firmicutes) | 3.44 ± 0.79abc | 2.36 ± 0.81c | 3.76 ± 1.15ab | 2.94 ± 0.55bc | 4.59 ± 1.34a | 3.65 ± 0.95ab |
| Bacteroidetes | ||||||
| Alloprevotella | 0.21 ± 0.16 | 0.1 ± 0.1 | 1.79 ± 2.44 | 6.61 ± 12.04 | 0.73 ± 0.64 | 0.06 ± 0.04 |
| Alistipes | 1.33 ± 1.06b | 0.23 ± 0.22b | 1.77 ± 1.27b | 5.79 ± 5.72a | 1.44 ± 1.4b | 0.24 ± 0.12b |
| Bacteroides | 0.05 ± 0.03b | 0.05 ± 0.04b | 1.67 ± 0.98ab | 2.89 ± 3.49a | 0.28 ± 0.19b | 0.07 ± 0.05b |
| Odoribacter | 0.64 ± 0.45ab | 0.26 ± 0.36b | 0.98 ± 0.62ab | 2.22 ± 2.55a | 1.49 ± 2.01ab | 0.11 ± 0.05b |
| Parabacteroides | 0.09 ± 0.09b | 0.02 ± 0.03b | 0.41 ± 0.32ab | 1.11 ± 1.36a | 0.09 ± 0.09b | 0.02 ± 0.01b |
| Rikenella | 0.13 ± 0.12b | 0.02 ± 0.02b | 0.37 ± 0.29ab | 0.66 ± 0.86a | 0.14 ± 0.12b | 0.03 ± 0.01b |
| Rikenellaceae_RC9_gut_group | 0.34 ± 0.11bc | 0.24 ± 0.09c | 1.02 ± 0.22a | 0.97 ± 0.28a | 0.55 ± 0.19b | 0.38 ± 0.01bc |
| Other (P: Bacteroidetes) | 0.17 ± 0.04c | 0.27 ± 0.29bc | 0.86 ± 0.31a | 0.61 ± 0.09ab | 0.53 ± 0.5abc | 0.2 ± 0.07c |
| Proteobacteria | ||||||
| Helicobacter | 4.34 ± 4.31b | 0.14 ± 0.09b | 6.93 ± 4.27b | 1.8 ± 1.79b | 13.59 ± 15.7a | 0.42 ± 0.14b |
| Desulfovibrio | 1.63 ± 1.97a | 0.24 ± 0.17b | 0.31 ± 0.14b | 0.41 ± 0.36b | 0.9 ± 0.97ab | 0.37 ± 0.23b |
| Acinetobacter | 0.02 ± 0.01 | 0.02 ± 0.01 | 0.03 ± 0.01 | 0.01 ± 0.01 | 0.23 ± 0.46 | 0.21 ± 0.01 |
| [F:Enterobacteriaceae] | 0.003 ± 0.00b1 | 0.001 ± 0.00c1 | 0.039 ± 0.05aa | 0.008 ± 0.009abb | 0.042 ± 0.075aa | 0.003 ± 0.003bc |
| Other (P: Proteobacteria) | 0.15 ± 0.04 | 0.12 ± 0.04 | 0.19 ± 0.1 | 0.13 ± 0.08 | 0.16 ± 0.05 | 0.12 ± 0.04 |
| Actinobacteria | ||||||
| Bifidobacterium | 0.06 ± 0.02 | 0.87 ± 1.76 | ND | ND | 0.04 ± 0.04 | 0.05 ± 0.02 |
| Saccharibacteria | ||||||
| Candidatus_Saccharimonas | 1.43 ± 0.38ab | 0.14 ± 0.06b | 0.98 ± 0.61ab | 2.08 ± 3.29a | 1.48 ± 0.88ab | 0.4 ± 0.5ab |
| Tenericutes | ||||||
| [C:Mollicutes] | 0.17 ± 0.16 | 0.07 ± 0.08 | 0.12 ± 0.09 | 0.1 ± 0.07 | 0.05 ± 0.03 | 0.03 ± 0.02 |
| Spirochaetes | ||||||
| [F:Spirochaetaceae] | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.02 | 0.01 ± 0.01 |
| Others | 5.5 ± 2.65b | 4.84 ± 2.63b | 10.22 ± 4.82b | 18.38 ± 8.68a | 10.09 ± 7.11b | 3.48 ± 1.02b |
| Total | 99.98 | 100.00 | 99.96 | 99.99 | 99.96 | 99.99 |
Data are presented as means ± standard deviation of values at D15 (Control and HCM2), D16 (ED1 and HED1), and D18 (ED3 and HED3). Data in the same row that do not share a common superscript are significantly different (p < 0.05).
ND, not detected.
“Others” means the assignment is ambiguous.
L. reuteri HCM2 stabilized the gut microbiota of mice challenged with ETEC
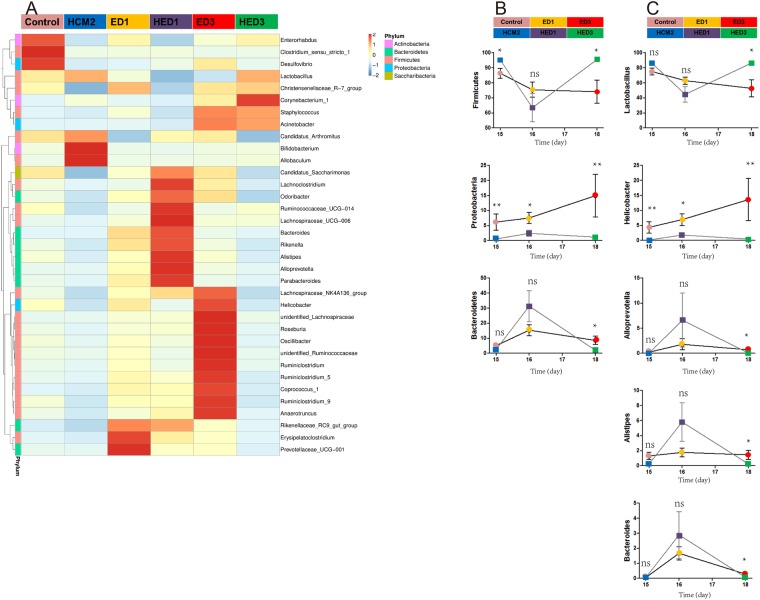
The colonic microbiome was characterized by analyzing the relative abundance of bacterial taxa (Fig. 5A). The dynamic changes in several dominant phyla in each group of mice are shown in Fig. 5B. After ETEC challenge, the proportion of Firmicutes in mice without HCM2 supplementation (Control group) decreased from 86.4% to 75.4% after 1 day of recovery (ED1 group) and then to 74.2% after 3 days of recovery (ED3 group). This result indicates that ETEC challenge could reduce the proportion of Firmicutes in the mouse colon. The proportion of Firmicutes in mice supplemented with HCM2 decreased from 95.2% to 63.4% 1 day after ETEC challenge (HED1 group), but increased to the original level of 95.4% 3 days after ETEC challenge (HED3 group). This result demonstrates that although ETEC infection leads to a decreased proportion of Firmicutes in the short term, pre-treatment with L. reuteri HCM2 could help Firmicutes recover to the pre-infection level in the gut. Supplementation with L. reuteri HCM2 in advance also contributed to the recovery of Bacteroidetes to the pre-infection level in the gut 3 days after ETEC challenge. ETEC challenge led to an increase in the proportion of Proteobacteria from 6.2% in the control group to 7.4% in the ED1 group, and this proportion increased to 15% in the ED3 group. The proportion of Proteobacteria was different in the L. reuteri HCM2 supplementation groups. It increased from 0.4% to 2.4% in the HED1 group, and then decreased to 1.2% in the HED3 group (Fig. 5B). As shown in Table 2, after ETEC challenge, the proportion of Enterobacteriaceae in mice without HCM2 supplementation increased from 0.003% in the Control group to 0.039% in the ED1 group and then to 0.042% in the ED3 group. This result demonstrates that ETEC challenge could increase the proportion of Enterobacteriaceae in the mouse colon. The proportion of Enterobacteriaceae in mice supplemented with HCM2 increased from 0.001% in the Control to 0.008% 1 day after ETEC challenge (HED1 group), but decreased to pre-infection proportion of 0.003% 3 days after ETEC challenge (HED3 group). This result demonstrates that although ETEC infection leads to an increased proportion of Enterobacteriaceae in the short term, pre-treatment with L. reuteri HCM2 could help Enterobacteriaceae recover to the pre-infection level in the gut. Taken together, these results indicate that pretreatment of mice with L. reuteri HCM2 for 14 days before ETEC challenge could alleviate the disruption of the bacterial community caused by ETEC infection and help the gut microbiota to recover to pre-infection levels.
Figure 5.
A heatmap of bacterial genera and dynamic changes in the dominant genera in each group. (A) A heatmap showing the abundances of bacterial genera in different groups. Relative abundance is scaled by the relative abundance within a genus. The color indicates the relative abundance as shown in the legend provided in the top right. (B) The relative abundances (mean ± SD) of the phyla Firmicutes, Bacteroidetes and Proteobacteria were plotted against time for each treatment. (C) The relative abundances (mean ± SD) of the genera Lactobacillus, Allprevotella, Alistipes, Bacteroides and Helicobacter were plotted against time for each treatment. * indicates significance at p < 0.05 for a single time point. Control: mice received a basal diet. HCM2: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks. ED1: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 1 day. HED1: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15 and then received a basal diet for 1 day. ED3: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 3 days. HED3: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15, and then received a basal diet for 3 days.
The dynamic changes in the dominant genera in each group of mice were also analyzed. Lactobacillus was the predominant genera in the mouse gut microbiota and showed a similar dynamic trend as Firmicutes after ETEC challenge (Fig. 5C). This result suggests that Lactobacillus might be the main genus contributing to the changes in Firmicutes after ETEC challenge. The proportion of bacteria in the Helicobacter genus also increased with increasing time after infection in groups without L. reuteri HCM2 supplementation and fluctuated in groups with L. reuteri HCM2 supplementation, a trend similar to that observed for Proteobacteria. This result suggests that Helicobacter might be the main genus contributing to the changes in Proteobacteria (Fig. 5C). The relative abundance of Alloprevotella, which belongs to the phylum Bacteroidetes, increased from 0.1% in the HCM2 group to 6.6% in the HED1 group, and then decreased to 0.1% in the HED3 group (Fig. 5C). Two other dominant genera, Alistipes and Bacteroides, which also belong to the phylum Bacteroidetes, showed similar dynamic trends as the phylum Bacteroidetes (Fig. 5C).
We next compared the colonic microbiota of mice in groups ED3 and HED3 with those of mice in the Control group using the linear discriminant analysis (LDA) effect size (LEfSe) method. A cladogram representing the structure of the colonic microbiota and the predominant bacteria taxa is shown in Fig. S2. Compared with the Control group, mice in the HED3 group had a significantly higher abundance of Lactobacillus, while mice in the ED3 group had an increased abundance of Bacteroidia, Lachnospiraceae, Campylobacterales and Helicobacter (Fig. S2A,B). These results demonstrate that L. reuteri HCM2 could protect mice from ETEC challenge by modulating the gut microbiota, restoring the pre-infection structure and promoting high levels of probiotic bacteria.
Discussion
L. reuteri is a common species that inhabits the gastrointestinal tract of humans and many animals. Here we demonstrated that an L. reuteri HCM2 strain isolated from a healthy piglet could protect against ETEC infection in a mouse model by altering gut microbiota. The growth of ETEC was obviously inhibited by L. reuteri HCM2, and this might be caused by damage to the ETEC cell wall. As we did not observe any antimicrobial peptide production by L. reuteri HCM2, it seems that lactic acid might function as the antimicrobial agent inhibiting ETEC growth (data not shown). Competition for adhesion sites is one of the mechanisms by which probiotics fight against pathogen infection29,30. We found that L. reuteri HCM2 could survive in the harsh gastrointestinal tract environment and had the capacity to adhere to Caco-2 cells. Furthermore, a competition assay revealed that L. reuteri HCM2 was able to reduce the number of ETEC cells. These results demonstrate that L. reuteri HCM2 potentially protects against ETEC. We further observed that the ETEC load in both the jejunum tissues and contents of the mouse model was significantly decreased after L. reuteri HCM2 pre-supplementation. A similar phenomenon was also observed by Yang et al., who found that L. reuteri TMW1.656 could decrease the abundance of E. coli and K88 fimbriae in ETEC-challenged weanling pigs31.
Probiotics are important because they protect the host gastrointestinal micro-environment from invading pathogens2. In this study, ETEC infection led to inflammatory infiltration and loss of microvilli in jejunum tissues in mice not receiving L. reuteri HCM2 pre-supplementation (ED1 and ED3 groups). This is consistent with the finding of a previous study that ETEC infection promotes the expression of pro-inflammatory cytokines by activating the NF-kB and MAPK pathways, leading to the loss of microvilli in the jejunum6. However, inflammatory infiltration was not found in mice of the HED1 and HED3 groups, which were pre-supplemented with L. reuteri HCM2. The expression levels of IgA and IgG did not obviously increase in mice pre-supplemented with L. reuteri HCM2 compared with control mice. Gao et al. also reported that supplementation with L. plantarum did not obviously increase the expression levels of IgG in broiler chickens20, which supports our finding.
Several recent studies have noted that supplementation with L. reuteri improves the growth and feed efficiency of neonatal and growing pigs8. For example, Liu et al. reported that L. reuteri I5007 supplementation increased the average daily weight gain in formula-fed piglets18. Wang et al. found that pigs supplemented with L. fermentum I5007 had higher weight gain and feed intake than pigs without L. fermentum I5007 treatment32. In our study, mice supplemented with L. reuteri HCM2 took in less food, however, no significant difference in body weight was detected between control mice and mice supplemented with L. reuteri HCM2. L. reuteri HCM2 slightly decreased the feed conversion in HCM2 and HED1 mice (Fig. S3), which is consistent with the report that L. fermentum I5007 could slightly decrease feed conversion in piglets32. The decreased feed conversion in HCM2 and HED1 mice may due to the increased ratio of Firmicutes to Bacteroidetes (Fig. S4), which has been reported to be associated with improved energy-harvesting capacity33. Furthermore, L. reuteri HCM2 significantly increased the villus/crypt ratio in mice, which indicates that the absorption of nutrients might be increased by feeding mice L. reuteri HCM2.
The healthy colon is usually dominated by obligate anaerobes, while dysbiosis is often associated with a sustained increase in the abundance of facultative anaerobic Proteobacteria, which results in the disruption of anaerobiosis34. For example, Zhang et al. found that ETEC infection could increase the relative abundance of Proteobacteria in newly weaned pigs compared with the uninfected control35. In our study, we analyzed the dynamics of gut microbiota and found that the abundance of Proteobacteria gradually increased after ETEC infection (ED1 and ED3). By contrast, not much difference in Proteobacteria abundance was observed in the L. reuteri HCM2 pre-supplementation groups (HED1 and HED3). Higher levels of Enterobacteriaceae (belonging to Proteobacteria) were detected in mice without L. reuteri HCM2 pre-supplementation, and this may contribute to a thinner and more penetrable mucus layer, which increases the risk of chronic colitis36. ETEC are members of Enterobacteriaceae, and there was a significant difference in the relative abundance of Enterobacteriaceae between the Control and HCM2 groups. It is interesting that there was no big difference in the relative abundance of Enterobacteriaceae between the ED1 and HED1 and groups (p > 0.05). This might be due to the amount of ETEC challenge. However, after 3 days of recovery, the relative abundance of Enterobacteriaceae in ED3 was significantly higher than that in the group HED3 (p = 0.0397). It is clear that HCM2 pre-supplementation decreases the relative abundance of Enterobacteriaceae and may prevent the dysbiosis of gut microbiota caused by ETEC.
It is noteworthy that the relative abundance of Helicobacter was increased by ETEC infection, while there was no obvious difference in the abundance of Helicobacter in the L. reuteri HCM2 pre-supplementation groups (HED1 and HED3) after ETEC infection. It has been reported that many cases of non-H.pylori Helicobacter (NHPH) infection in humans occur in immunocompromised patients, because NHPH can induce high levels of inducible nitric oxide synthase and the development of DNA double-stranded breaks, but the properties of Lactobacillus can prevent Helicobacter infection or its related pathologies37. In our study, L. reuteri HCM2 supplementation not only maintained the abundance of Helicobacter at a relatively low level, but also increased the relative abundance of Lactobacilli. It has been reported that the abundance of Helicobacteraceae in a rat model of colon cancer was decreased by administering a probiotic cocktail (Lactobacillus acidophilus, Bifidobacteria bifidum, and Bifidobacteria infantum), which promoted the growth of Lactobacilli and thus altered the gut microbiota38. We conclude that pre-supplementation with L. reuteri HCM2 may alter the gut microbiota and protect against ETEC and the increase in abundance of detrimental bacteria caused by ETEC.
We monitored the dynamic changes in dominant genera in each group of mice and intriguingly found that ETEC could reduce the relative abundance of Lactobacillus in ED1 and HED1 mice. The relative abundance of Lactobacillus in the ED3 group was lower than that of both the Control and ED1 groups, while in the HED3 group, the abundance of Lactobacillus was the same as that in the control, suggesting that L. reuteri HCM2 pre-supplementation prevented the reduction in Lactobacillus abundance. It was reported that lactobacilli abundance in the colon could be reduced by ETEC, while pre-administration of probiotics could increase the abundance of lactobacilli and enhance goblet cell function to ameliorate enteritis35. Lactobacilli, which are considered health promoting probiotics39, might promote defense against detrimental bacteria in the gut by creating an acidic environment (e.g., pH 4.5), synthesizing exopolysaccharides5, competitively excluding intestinal pathogens40, improving antioxidant activity19, or activating and enhancing local cell-mediated immunity against certain enteric pathogens41. So, stabilizing the relative abundance of Lactobacillus is important for maintaining the balance of gut microbes. The relative abundances of the dominant genera Alistipes, Bacteroides and Alloprevotella, which belong to the Bacteroidetes, were also higher after ETEC infection, but the abundances returned back to the pre-infections after 3 days of recovery (HED3). This also indicates that L. reuteri HCM2 pre-supplementation could stabilize the relative abundance of Bacteroidetes. In conclusion, L. reuteri HCM2 pre-supplementation may modulate the gut microbiota, preventing the dysbiosis of intestinal microbiota caused by ETEC.
Materials and Methods
Bacterial strains and cell lines
E. coli F4-producing strain W25K (ETEC; O149:K91, K88ac; LT, STb, EAST)4,42 was used and cultured in LB medium. Lactobacillus reuteri HCM2 was isolated from a 6-week-old healthy piglet and was anaerobically cultured in MRS medium. All strains were sub-cultured twice prior to being used for experiments. The Caco-2 cell line was cultivated in complete Dulbecco’s modified Eagle’s minimal essential medium (DMEM, C12430500BT, purchased from Thermo Fisher Scientific (China) Co., Ltd.) supplemented with heat-inactivated fetal bovine serum (10% v/v) and 100 U/ml penicillin-Streptomycin. The medium was replaced every 2 days. Cells were grown at 37 °C, 5% CO2/95% air in T25 flasks.
Animals, feeding procedures, and infection
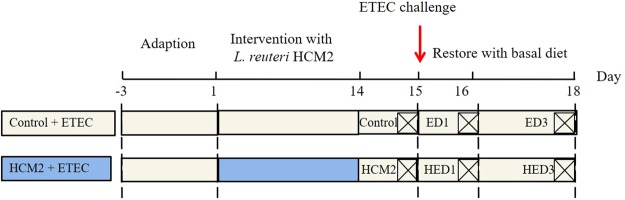
ICR6,7,42 male mice (5 weeks of age) were purchased from SLACCAS (Shanghai Laboratory Animal Center). Only male mice were used to avoid differences in microbiota composition resulting from sex and maternal factors43. Animal experiments were approved by the Laboratory Animal Ethical Commission of the Chinese Academy of Sciences and performed according to its guidelines. In order to test the protective effect of L. reuteri HCM2 against ETEC in vivo, we developed a mouse model as described in a previous study6,35 with minor modifications. The mice were housed in a pathogen-free mouse colony (temperature, 25 ± 2 °C; relative humidity, 45–60%; lighting cycle, 12 h/d). After acclimatization for three days, the mice were randomly divided into six groups (n = 5 for each group) as shown in Fig. 6. The Control group received a basal diet44; the HCM2 group received a basal diet and a 200 μL suspension of 5 × 109 CFU/ml L.reuteri HCM2 daily45 for two consecutive weeks; the ED1 (basal diet + ETEC + Day 1) group and the ED3 (basal diet + ETEC + Day 3) group received a basal diet, were challenged with 100 μL of 109 CFU/ml ETEC6,7 by intragastric administration at day 15, then received a basal diet for 1 and 3 days, respectively; the HED1 (HCM2 + ETEC + Day 1) group and the HED3 (HCM2+ ETEC+ Day 3) group received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 1 and 3 days, respectively. Mice were intragastrically administered L. reuteri HCM2 or ETEC via 12-gauge gavage needles (Bio-Medical Needles, Beijing Solarbio Science & Technology Co., Ltd.). Control and HCM2 mice were sacrificed at D15, ED1 and HED1 mice were sacrificed at D16, and ED3 and HED3 mice were sacrificed at D18. All mice were sacrificed at 10:00 am and samples were collected. Briefly, after cervical dislocation, blood was collected by removing the eyeball, then the jejunum, the contents of the jejunum and the contents of colon were collected. The body weights and food intake of the mice were regularly monitored during the experiment.
Figure 6.
A schematic of the experimental design. The gray boxes indicate that mice received a common diet, and blue boxes indicate that mice were treated with L. reuteri HCM2. The red arrow indicates when mice were challenged with ETEC. Control: mice received a basal diet. HCM2: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks. ED1: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 1 day. HED1: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15 and then received a basal diet for 1 day. ED3: mice received a basal diet, were challenged with 108 CFUs ETEC by intragastric administration at day 15, then received a basal diet for 3 days. HED3: mice received a basal diet and 109 CFUs L. reuteri HCM2 daily for two consecutive weeks, were challenged with 108 CFUs ETEC by intragastric administration at day 15, and then received a basal diet for 3 days.
Antimicrobial activity assay
ETEC were cultured overnight in Shake-tubes and were standardized to OD600 = 2. The supernatants of the L. reuteri HCM2 overnight cultures were filtered through a 0.22 μm filter, and 100 μl of CFS was placed into each well of an ETEC indicator strain agar plate. MRS medium was tested as a negative control. After incubation at 37 °C for 24 h, the diameters of the inhibition zones were measured. To further validate the capacity of L. reuteri HCM2 to antagonize ETEC, ETEC cells at a density of 1 × 106 CFUs/ml were seeded into the wells of a 96-well microtiter plate, and 0%, 5%, 10% or 20% (v/v) CSF of L. reuteri HCM2 was added to each well. The absorbance of ETEC at 600 nm was monitored every 2 h.
Scanning electron microscopy
SEM experiments were performed as described by Aiba et al. with slight modifications46. Briefly, a 108 CFUs/ml inoculum of ETEC was treated with 20% L. reuteri HCM2 CFS for 12 h at 37 °C, and a non-treated ETEC culture served as a control. After washing with fresh PBS, cells were fixed with 2.5% glutaraldehyde for 12 h. The bacteria were then washed with demineralized water 3 times, 6 min each time. After dehydration with ethanol (stepwise gradient of 50%, 70%, 85% 95% and 100%), the specimens were freeze-dried in a critical-point dryer (Czech FEI company) and coated with gold. The morphology of the cells was visualized by examining the specimens with a Hitachi cold field emission scanning electron microscope.
Characterization of the tolerance of L. reuteri HCM2
L. reuteri HCM2 cells from an overnight culture were washed twice with PBS (pH 7.2), and the concentration was adjusted to about 108 to 109 CFUs/ml. One hundred microliters of cells were transferred to 900 μl of artificial gastric juice (pH 2.5)47 or artificial small intestine fluid (pH 6.8) and incubated anaerobically for 2 h at 37 °C. After recovery for 48 h, viable bacterial cells were counted by plating serial dilutions of the culture in PBS on MRS medium.
In vitro adhesion assay
Caco-2 cells were prepared on a Millicell EZ SLIDE 4-well and seeded at a concentration of 105 cells per well. Caco-2 cells were cultured for 15 days in cell culture medium to obtain confluence, and the medium was changed on alternate days. Caco-2 cells were washed with D-Hanks buffer solution and fresh culture medium without antibiotic solution was added 48 h before the adhesion assay40. The overnight L. reuteri HCM2 culture was washed with D-Hanks buffer solution and then re-suspended in fresh DMEM. The L. reuteri HCM2 suspension was diluted to 108 CFUs/ml and 1 ml was added into the Millicell EZ SLIDE 4-well. After co-culturing for 2 h, the Caco-2 monolayers were washed three times and fixed for 20 min at room temperature with 4% paraformaldehyde (w/v) fix solution. The Caco-2 monolayers were then washed five times with D-Hanks buffer solution, air-dried and Gram stained48. The images of L. reuteri HCM2 binding to Caco-2 cells were captured under microscope.
The amount of L. reuteri HCM2 binding to Caco-2 cells was further quantified as described by Todoriki et al. with slight modifications49. Briefly, the Caco-2 cells were cultivated in 6-well plates until confluence was reached and then used for adhesion experiments. Caco-2 monolayers were washed twice with PBS, and 3 ml bacterial suspension (5 × 108 cells/ml) was added to each well of the tissue culture plate. The plates were incubated at 37 °C in 5% CO2/95% air. After 2 h of incubation, the monolayers were washed three times with PBS. Following the last wash, Caco-2 cell monolayers were covered with 3 ml distilled water and mechanically agitated by vigorous pipetting to suspend the Caco-2 cells and bacteria. Adherent bacteria were serial diluted by 10-fold and plated on MRS media. The number of L. reuteri HCM2 colonies were counted after anaerobic incubation for 48 h.
Competition and displacement assays
Caco-2 cells were cultivated in 6-well plates to 80% confluence and then used for adhesion experiments40,50. Suspensions of L. reuteri HCM2 and ETEC were prepared with DMEM medium. For the competition assays, bacterial cells were washed twice with PBS, and of 100 μl of L. reuteri HCM2 suspension (108 CFUs) and 100 μl of ETEC suspension (108 CFUs) were added to each well simultaneously. The cultures were incubated at 37 °C for 90 min, then the monolayers were washed twice with PBS and digested with 100 μl trypsin (0.25%) for 2 min. For displacement assays, 100 μl ETEC suspension (108 CFUs) was added to each well, and the samples were incubated for 45 min. Then 100 μl of L. reuteri HCM2 suspension (108 CFUs) was added, and the samples were incubated for another 45 min. The plates were incubated and digested as described above. For both assays, serial dilutions of the adherent bacteria were plated on MacConkey Agar containing 50 μg/ml streptomycin and incubated at 37 °C for 16 h. The number of colonies were then counted. The ETEC strain used in this study is resistant to streptomycin.
Morphological analyses
Mouse jejunums were fixed with 4% paraformaldehyde-PBS overnight and then dehydrated and embedded in paraffin blocks. Sections (5 μm) from these blocks were deparaffinized, hydrated, and then stained with hematoxylin and eosin (H&E). At least three villus lengths and crypt depths per slide were measured using Image-Pro Plus software 6. Five mice were studied from each group. The data collectors were unaware of the treatment status of the examined slides.
Detection of serum IgG and IgA
Blood (500 μl) was collected from each mouse. Serum was collected after centrifugation at 4500 rpm for 10 min at 4 °C and stored at −20 °C until IgG and IgA were quantified. Total IgG and IgA in the serum were measured by enzyme-linked immunosorbent assay (Mouse IgG ELISA Kit and Mouse IgG ELISA Kit, AMEKO). The concentrations were then calculated from standard curves.
Illumina HiSeq sequencing and data processing
Total genomic DNA was extracted from the colonic contents of mice from the six groups using the Qiagen QIAamp DNA Stool Mini Kit. The V3-V4 hypervariable region of the bacteria 16 S rRNA gene was amplified using the primers 338 F 5′-ACTCCTACGGGAGGCAGCA-3′ and 806 R 5′-GGACTACHVGGGTWTCTAAT-3′. PCR products were mixed in equidensity ratios and purified with the Qiagen Gel Extraction Kit (Qiagen, Germany). Sequencing libraries were generated using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA), and index codes were added. The library quality was assessed using the Qubit@ 2.0 Fluorometer (Thermo Scientific) and the Agilent Bioanalyzer 2100 system. Finally, the library was sequenced on the Illumina HiSeq2500 platform, and 250 bp paired-end reads were generated, assigned to samples based on their unique barcode and truncated by cutting off the barcode and primer sequences. Paired-end reads were merged using FLASH51, which was designed to merge paired-end reads when at least some of the reads overlap with the read generated from the opposite end of the same DNA fragment, and the trimmed sequences were called raw tags. The raw tags were filtered using the QIIME quality control process to obtain high-quality clean tags51. The tags were compared with the reference database (Gold database) using the UCHIME algorithm to detect chimeric sequences, which were later removed52,53. The effective tags were finally obtained.
Bioinformatics analysis
Sequence analysis was performed using Uparse software (Uparse v7.0.1001). Sequences sharing greater than 97% similarity were assigned to the same OTUs54. A representative sequence for each OTU was selected for further annotation. For each representative sequence, the GreenGene Database was used to obtain taxonomic information using the RDP classifier algorithm55,56. OTU abundance was normalized to the number of sequences in the sample with the fewest sequences. Subsequent analysis of alpha diversity and beta diversity were performed using the normalized data. Species diversity complexity of a sample was analyzed using six indices of alpha diversity, including Observed-species, Chao1, Shannon, Simpson, ACE and Good-coverage. All indices were calculated with QIIME (Version 1.7.0) and visualized using R software (Version 2.15.3). The differences in dominant bacterial communities between groups were determined based on LDA Effect Size.
Statistical analysis
Data shown are the means ± SD or SEM. Differences in the means between two groups were analyzed by performing an unpaired t test (Prism 7.0) if the data were normally distributed and the samples had equal variance, or by performing a non-parametric test (Mann–Whitney U test, Prism 7.0) if the data were not normally distributed. Means of more than two groups were analyzed by performing one-way ANOVA followed by the Dunnett multiple comparisons test (Prism 7.0) if the data were followed a Gaussian distribution and had equal variance, or by performing the Kruskal-Wallis test followed by Dunn’s multiple comparisons test (Prism 7.0) if the data were not normally distributed. The Kolmogorov-Smirnov test (Prism 7.0) was used to determine if the data followed a Gaussian distribution. The homogeneity of variance test (SPSS 22.0) or the Brown-Forsythe test (Prism 7.0) was used to test for equal variance. Differences with p < 0.05 were considered significant.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31570114 and 31772642) and the Special Fund for Agro-scientific Research in the Public Interest (201503134). The authors thank professor Shiyan Qiao from China Agricultural University for providing the Caco-2 cell line.
Author Contributions
J.Z., G.L. and K.L.T. designed the study; T.W.W. and K.L.T. wrote the manuscript; T.W.W., Y.Y.L., J.Z., X.Z. and M.Z. performed the experiments; Y.T. and J.Z. edited the manuscript. All authors have discussed the results and reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Tianwei Wang and Kunling Teng contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35702-y.
References
- 1.Fleckenstein JM, et al. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes and infection. 2010;12:89–98. doi: 10.1016/j.micinf.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gresse R, et al. Gut Microbiota Dysbiosis in Postweaning Piglets: Understanding the Keys to Health. Trends in microbiology. 2017 doi: 10.1016/j.tim.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Nagy B, Fekete PZ. Enterotoxigenic Escherichia coli in veterinary medicine. International journal of medical microbiology: IJMM. 2005;295:443–454. doi: 10.1016/j.ijmm.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Ren W, et al. Draft genome sequence of enterotoxigenic Escherichia coli strain W25K. Genome announcements. 2014;2:e00593–00514. doi: 10.1128/genomeA.00593-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen XY, Woodward A, Zijlstra RT, Gänzle MG. Exopolysaccharides synthesized by Lactobacillus reuteri protect against enterotoxigenic Escherichia coli in piglets. Applied and environmental microbiology. 2014;80:5752–5760. doi: 10.1128/AEM.01782-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren W, et al. Mouse intestinal innate immune responses altered by enterotoxigenic Escherichia coli (ETEC) infection. Microbes and infection. 2014;16:954–961. doi: 10.1016/j.micinf.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Ren W, et al. Intestinal Microbiota-Derived GABA Mediates Interleukin-17 Expression during Enterotoxigenic Escherichia coli Infection. Frontiers in immunology. 2016;7:685. doi: 10.3389/fimmu.2016.00685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou C, Zeng X, Yang F, Liu H, Qiao S. Study and use of the probiotic Lactobacillus reuteri in pigs: a review. Journal of animal science and biotechnology. 2015;6:14. doi: 10.1186/s40104-015-0014-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubreuil JD. Enterotoxigenic Escherichia coli and probiotics in swine: what the bleep do we know? Bioscience of Microbiota. Food and Health. 2017;36:75–90. doi: 10.12938/bmfh.16-030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Docic M, Bilkei G. Differences in Antibiotic Resistance in Escherichia coli, Isolated from East-European Swine Herds With or Without Prophylactic Use of Antibiotics. Journal of Veterinary Medicine, Series B. 2003;50:27–30. doi: 10.1046/j.1439-0450.2003.00609.x. [DOI] [PubMed] [Google Scholar]
- 11.Toutain PL, Ferran AA, Bousquet-Melou A, Pelligand L, Lees P. Veterinary Medicine Needs New Green Antimicrobial Drugs. Frontiers in microbiology. 2016;7:1196. doi: 10.3389/fmicb.2016.01196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samanidou VF, Evaggelopoulou EN. Chromatographic analysis of banned antibacterial growth promoters in animal feed. J Sep Sci. 2008;31:2091–2112. doi: 10.1002/jssc.200800075. [DOI] [PubMed] [Google Scholar]
- 13.Joint, F. WHO working group report on drafting guidelines for the evaluation of probiotics in food. London, Ontario, Canada 30 (2002).
- 14.Lebeer S., Vanderleyden J., De Keersmaecker S. C. J. Genes and Molecules of Lactobacilli Supporting Probiotic Action. Microbiology and Molecular Biology Reviews. 2008;72(4):728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaton KA, Honkala A, Auchtung TA, Britton RA. Probiotic Lactobacillus reuteri ameliorates disease due to enterohemorrhagic Escherichia coli in germfree mice. Infection and immunity. 2011;79:185–191. doi: 10.1128/IAI.00880-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moorthy G, Murali MR, Devaraj SN. Protective role of lactobacilli in Shigella dysenteriae 1-induced diarrhea in rats. Nutrition. 2007;23:424–433. doi: 10.1016/j.nut.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Seegers JF. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends in biotechnology. 2002;20:508–515. doi: 10.1016/S0167-7799(02)02075-9. [DOI] [PubMed] [Google Scholar]
- 18.Liu H, et al. Oral administration of Lactobacillus fermentum I5007 favors intestinal development and alters the intestinal microbiota in formula-fed piglets. Journal of agricultural and food chemistry. 2014;62:860–866. doi: 10.1021/jf403288r. [DOI] [PubMed] [Google Scholar]
- 19.Lin M-Y, Chang F-J. Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Digestive diseases and sciences. 2000;45:1617–1622. doi: 10.1023/A:1005577330695. [DOI] [PubMed] [Google Scholar]
- 20.Gao P, et al. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome. 2017;5:91. doi: 10.1186/s40168-017-0315-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh PL, et al. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. The ISME journal. 2010;4:377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- 22.Walter J, Britton RA, Roos S. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4645–4652. doi: 10.1073/pnas.1000099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Hongbin, Hou Chengli, Wang Gang, Jia Hongmin, Yu Haitao, Zeng Xiangfang, Thacker Philip A, Zhang Guolong, Qiao Shiyan. Lactobacillus reuteri I5007 Modulates Intestinal Host Defense Peptide Expression in the Model of IPEC-J2 Cells and Neonatal Piglets. Nutrients. 2017;9(6):559. doi: 10.3390/nu9060559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang F, et al. Lactobacillus reuteri I5007 modulates tight junction protein expression in IPEC-J2 cells with LPS stimulation and in newborn piglets under normal conditions. BMC microbiology. 2015;15:32. doi: 10.1186/s12866-015-0372-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Y, Zhao X, Le MH, Zijlstra RT, Ganzle MG. Reutericyclin producing Lactobacillus reuteri modulates development of fecal microbiota in weanling pigs. Frontiers in microbiology. 2015;6:762. doi: 10.3389/fmicb.2015.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chung TC, Axelsson L, Lindgren SE, Dobrogosz WJ. In VitroStudies on Reuterin Synthesis by Lactobacillus reuteri. Microbial Ecology in Health and Disease. 1989;2:137–144. doi: 10.3109/08910608909140211. [DOI] [Google Scholar]
- 27.Wang B, et al. Identification of a surface protein from Lactobacillus reuteri JCM1081 that adheres to porcine gastric mucin and human enterocyte-like HT-29 cells. Current microbiology. 2008;57:33–38. doi: 10.1007/s00284-008-9148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo Q, et al. Intestinal IgA(+) cell numbers as well as IgA, IgG, and IgM contents correlate with mucosal humoral immunity of broilers during supplementation with high fluorine in the diets. Biological trace element research. 2013;154:62–72. doi: 10.1007/s12011-013-9713-9. [DOI] [PubMed] [Google Scholar]
- 29.Kim PI, et al. Probiotic properties of Lactobacillus and Bifidobacterium strains isolated from porcine gastrointestinal tract. Applied microbiology and biotechnology. 2007;74:1103–1111. doi: 10.1007/s00253-006-0741-7. [DOI] [PubMed] [Google Scholar]
- 30.Valeriano VD. M., P. B. & Kang, D. K. Probiotic Roles of Lactobacillus spp. in Swine: Insights from Gut Microbiota. Journal of applied microbiology. 2016 doi: 10.1111/jam.13364. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, Galle S, Le MH, Zijlstra RT, Ganzle MG. Feed Fermentation with Reuteran- and Levan-Producing Lactobacillus reuteri Reduces Colonization of Weanling Pigs by Enterotoxigenic Escherichia coli. Applied and environmental microbiology. 2015;81:5743–5752. doi: 10.1128/AEM.01525-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang A, Yu H, Gao X, Li X, Qiao S. Influence of Lactobacillus fermentum I5007 on the intestinal and systemic immune responses of healthy and E. coli challenged piglets. Antonie Van Leeuwenhoek. 2009;96:89–98. doi: 10.1007/s10482-009-9339-2. [DOI] [PubMed] [Google Scholar]
- 33.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 34.Litvak Y, Byndloss MX, Tsolis RM, Baumler AJ. Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Current opinion in microbiology. 2017;39:1–6. doi: 10.1016/j.mib.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, W. et al. Oral Administration of a Select Mixture of Bacillus Probiotics Affects the Gut Microbiota and Goblet Cell Function following Escherichia coli Challenge in Newly Weaned Pigs of Genotype MUC4 That Are Supposed To Be Enterotoxigenic E. coli F4ab/ac Receptor Negative. Applied and environmental microbiology 83, 10.1128/AEM.02747-16 (2017). [DOI] [PMC free article] [PubMed]
- 36.Carvalho FA, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell host & microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Péré-Védrenne Christelle, Flahou Bram, Loke Mun Fai, Ménard Armelle, Vadivelu Jamuna. Other Helicobacters, gastric and gut microbiota. Helicobacter. 2017;22:e12407. doi: 10.1111/hel.12407. [DOI] [PubMed] [Google Scholar]
- 38.Kuugbee ED, et al. Structural Change in Microbiota by a Probiotic Cocktail Enhances the Gut Barrier and Reduces Cancer via TLR2 Signaling in a Rat Model of Colon Cancer. Digestive diseases and sciences. 2016;61:2908–2920. doi: 10.1007/s10620-016-4238-7. [DOI] [PubMed] [Google Scholar]
- 39.Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Applied and environmental microbiology. 2008;74:4985–4996. doi: 10.1128/AEM.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quilodran-Vega SR, et al. Isolation of lactic acid bacteria from swine milk and characterization of potential probiotic strains with antagonistic effects against swine-associated gastrointestinal pathogens. Canadian journal of microbiology. 2016;62:514–524. doi: 10.1139/cjm-2015-0811. [DOI] [PubMed] [Google Scholar]
- 41.Hardy H, Harris J, Lyon E, Beal J, Foey AD. Probiotics, prebiotics and immunomodulation of gut mucosal defences: homeostasis and immunopathology. Nutrients. 2013;5:1869–1912. doi: 10.3390/nu5061869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren W, et al. Melatonin alleviates weanling stress in mice: involvement of intestinal microbiota. Journal of pineal research. 2017 doi: 10.1111/jpi.12448. [DOI] [PubMed] [Google Scholar]
- 43.Xiao L, et al. A catalog of the mouse gut metagenome. Nature biotechnology. 2015;33:1103–1108. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 44.Ren W, et al. Dietary arginine supplementation of mice alters the microbial population and activates intestinal innate immunity. The Journal of nutrition. 2014;144:988–995. doi: 10.3945/jn.114.192120. [DOI] [PubMed] [Google Scholar]
- 45.Wu CM, Chung TC. Mice protected by oral immunization with Lactobacillus reuteri secreting fusion protein of Escherichia coli enterotoxin subunit protein. FEMS Immunol Med Microbiol. 2007;50:354–365. doi: 10.1111/j.1574-695X.2007.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aiba, Y., Ishikawa, H., Tokunaga, M. & Komatsu, Y. Anti-Helicobacter pylori activity of non-living, heat-killed form of lactobacilli including Lactobacillus johnsonii No.1088. FEMS microbiology letters 364, 10.1093/femsle/fnx102 (2017). [DOI] [PubMed]
- 47.Casey PG, et al. Isolation and characterization of anti-Salmonella lactic acid bacteria from the porcine gastrointestinal tract. Letters in applied microbiology. 2004;39:431–438. doi: 10.1111/j.1472-765X.2004.01603.x. [DOI] [PubMed] [Google Scholar]
- 48.Tuomola EM, Salminen SJ. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. International journal of food microbiology. 1998;41:45–51. doi: 10.1016/S0168-1605(98)00033-6. [DOI] [PubMed] [Google Scholar]
- 49.Todoriki K, Mukai T, Sato S, Toba T. Inhibition of adhesion of food-borne pathogens to Caco-2 cells by Lactobacillus strains. Journal of applied microbiology. 2001;91:154–159. doi: 10.1046/j.1365-2672.2001.01371.x. [DOI] [PubMed] [Google Scholar]
- 50.Gopal PK, Prasad J, Smart J, Gill HS. In vitro adherence properties of Lactobacillus rhamnosus DR20 and Bifidobacterium lactis DR10 strains and their antagonistic activity against an enterotoxigenic Escherichia coli. International journal of food microbiology. 2001;67:207–216. doi: 10.1016/S0168-1605(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 51.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haas BJ, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome research. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 55.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and environmental microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.