Abstract
To investigate the functions of fructokinase (FRK) in apple (Malus domestica) carbohydrate metabolism, we cloned the coding sequences of MdFRK1 and MdFRK2 from the ‘Royal Gala’ apple. The results showed that MdFRK2 expression was extremely high in shoot tips and young fruit. Analyses of heterologously expressed proteins revealed that MdFRK2 had a higher affinity for fructose than did MdFRK1, with Km values of 0.1 and 0.62 mM for MdFRK2 and MdFRK1, respectively. The two proteins, however, exhibited similar Vmax values when their activities were significantly inhibited by high concentrations of fructose. MdFRK2 ectopic expression was associated with a general decrease in fructose concentration in transgenic lines. In leaves, increased FRK activity similarly resulted in reduced concentrations of glucose and sucrose but no alterations in sorbitol concentration. When compared with those in the untransformed control, genes involved in sorbitol synthesis (A6PR) and the degradation pathway (SDH1/2) were significantly upregulated in transgenic lines, whereas those involved in sucrose synthesis (SPS1) and other degradation processes (SUSY4, NINV1/2, and HxK2) were downregulated. The activity of enzymes participating in carbohydrate metabolism was proportional to the level of gene expression. However, the growth performance and photosynthetic efficiency did not differ between the transgenic and wild-type plants. These results provide new genetic evidence to support the view that FRK plays roles in regulating sugar and sorbitol metabolism in Rosaceae plants.
Sugar metabolism: a master regulator
An international study led by Mingjun Li at Northwest A&F University in Shaanxi, China, reveals that an enzyme regulating the utilization of fructose in apple plants, fructokinase 2 (FRK2), has a key role in sugar metabolism. Overexpressing the gene encoding FRK2 in apple plants did not affect their growth or levels of sorbitol, the major product of photosynthesis in these plants, but it significantly lowered the levels of fructose, glucose and sucrose in mature leaves and increased the levels of starch. Accordingly, the activity of enzymes involved in sucrose and glucose metabolism was downregulated, whereas the activity of those involved in sorbitol metabolism was upregulated. Future analysis of the fruit and flowers will shed further light on the role of FRK in controlling sugar levels and the distribution of carbon in apple plants.
Introduction
Photosynthetically active leaves produce various forms of carbohydrates, which comprise primarily sucrose and starch in most plant species. These compounds serve as energy and carbon sources for metabolism, including the synthesis of cell wall polymers. Soluble sugars—primarily sucrose, glucose, and fructose—can also act as signal molecules to regulate the expression of various genes involved in defense, development, and metabolism1–4. Therefore, understanding the mechanisms of carbohydrate metabolism is important for exploiting sugar metabolism to improve plant growth and crop quality.
In the mature green leaves of most plants, sucrose is the main end product of photosynthesis in mesophyll cells4. One key enzyme for sucrose synthesis is sucrose-phosphate synthase (SPS; EC 2.4.1.14)5,6. Sucrose can be converted to fructose and glucose by invertases (EC 3.2.1.26), including cell wall invertase (CWINV), neutral invertase (NINV), and vacuolar invertase (VINV) or to fructose and UDP-glucose (UDPG) by sucrose synthase (SUSY; EC 2.4.1.13)4. The resulting fructose can be phosphorylated by fructokinase (FRK; EC 2.7.1.4) to fructose-6-phosphate (F6P), whereas the resulting glucose can be phosphorylated by hexokinase (HxK; EC 2.7.1.1) to glucose-6-phosphate (G6P)7–9. HxK and FRK are central in sugar metabolism and homeostasis.
As outlined above, FRK is the gateway to fructose metabolism, regulating fructose flux in cells10. Genes encoding FRK with distinct kinetic properties have been identified from multiple tissues and species, including tomato (Lycopersicon esculentum)11–13, potato (Solanum tuberosum)14, maize (Zea mays)15, rice (Oryza sativa)16, and Arabidopsis thaliana17. Generally, FRK2 orthologs have much higher affinity for fructose than do other FRKs9,17,18 and play important roles in controlling fructose utilization and carbon flux repartition19. In plants that have sucrose as the end product of photosynthesis, scientists have explored the roles of FRK2 orthologs in regulating carbon partition and plant growth via transgenic experiments. The inhibition of the expression of FRK2 orthologs in transgenic lines of tomato and potato have been found to have negative effects on growth, potato yield, and development of tomato active xylem20,21, whereas sugar levels in leaves are largely unaltered20–22. However, in transgenic aspen, the reduction of FRK activity by using an RNAi-FRK2 approach increased the leaf concentrations of fructose, glucose, and sucrose but decreased cell wall fiber thickness and the proportion of cellulose in the cell wall, implying that FRK2 is required for carbon partitioning to cellulose in the wood19. Additionally, overexpression of tomato low-affinity LeFRK1 in cotton (Gossypium hirsutum) results in a decreased level of sucrose in young leaves but increases in seed and fiber yields per plant due to the increased area per leaf and leaf number23. Although the physiological roles of FRK in plant development have been partly elucidated in these plant species, the underlying function of FRK in regulating carbohydrate concentrations remains obscure, and no studies have investigated the impacts of enhanced high-affinity FRK activity on sugar metabolism and plant growth.
Unlike most higher plant species, in which sucrose is the end product of photosynthesis, in apple and many other Rosaceae fruit trees, sorbitol is the primary end product of photosynthesis. It is catalyzed via aldose-6-phosphate reductase (A6PR; EC 1.1.1.200) and accounts for 60 to 80% of the newly fixed carbon in source leaves of these trees24,25. In addition to being a key metabolite in carbohydrate metabolism, sorbitol acts as a signal that regulates stamen development, pollen tube growth and resistance to Alternaria alternata in apple26,27. In apple sink cells, almost all the sorbitol available is converted to fructose by sorbitol dehydrogenase (SDH; EC 1.1.1.14), whereas half of the sucrose is converted to fructose28,29. As a result, in apple sink cells, more than 80% of the total carbon flux passes through fructose30, whereas in other model plant systems that transport and utilize only sucrose (e.g., Arabidopsis, tomato, and Populus), 50% of the total carbon flux is via fructose4. The utility of such high levels of fructose in apple sink cells must require high FRK activity. MdFRK expression is thought to be involved in regulating fructose utilization and accumulation in apple30,31. In the fruit of transgenic apple with decreased sorbitol synthesis, MdFRK2 was suggested as a main regulating factor of fructose homeostasis32. We hypothesized that apple FRK plays direct roles in metabolizing fructose and controlling its accumulation in apple. Based on analyses of transcript, enzyme activity and metabolite data in different varieties, it was suggested that FRK might play roles in regulating sugar metabolism and concentrations in peach fruits33. However, there is no genetic evidence available to clarify whether FRK plays roles in regulating sugar and sorbitol metabolism in Rosaceae plants.
Here, we examined the role of MdFRK2 in regulating carbohydrate metabolism in apple leaves. Its expression patterns and enzymatic properties suggest that MdFRK2 is the major fructose-phosphorylating gene and that its expression is closely correlated, both temporally and spatially, with fructose metabolism in apple. To investigate its roles in plant growth and sugar metabolism, we generated MdFRK2-overexpression (OE) transgenic plants in which the expression and activity of MdFRK2 was increased significantly relative to that of untransformed wild-type (WT) plants. In the transgenic plants, all fructose, glucose, and sucrose concentrations were decreased; furthermore, the expression of genes and the activities of enzymes involved in sorbitol metabolism were greatly enhanced, whereas those of sucrose metabolism were decreased. The results demonstrate that overexpression of MdFRK2 in apple alters the allocation of photosynthetic carbon flux from sucrose to sorbitol, thereby changing sugar concentrations and homeostasis in apple leaves.
Results
Cloning and expression characterization of MdFRK2
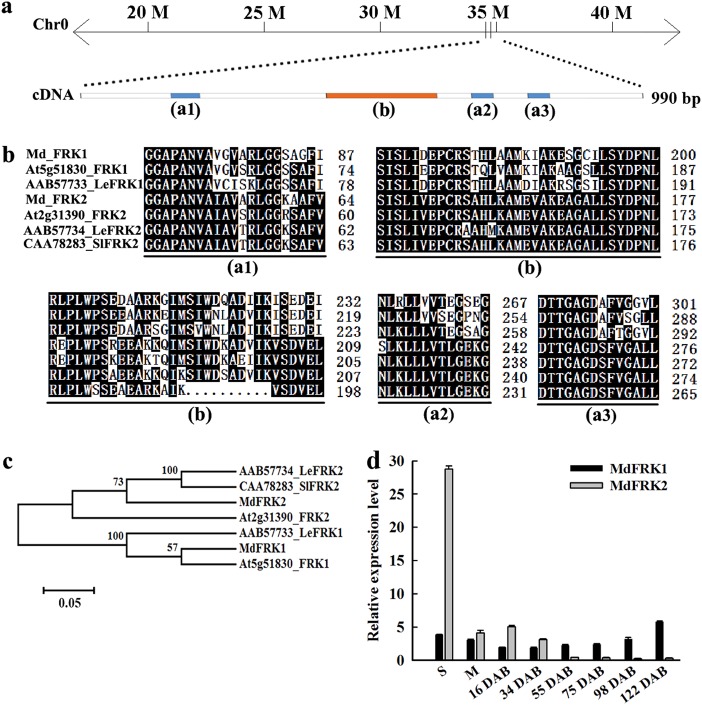
In apple, four MdFRK genes are mainly expressed and are homologous to LeFRK-1 to -4 from tomato30. Our previous studies indicated that MdFRK2 might be a key gene in determining FRK activity and fructose homeostasis30–32. In the present study, to investigate the function of the MdFRK2 gene, we cloned the coding sequence of MdFRK2 from ‘Royal Gala’ apple plants, and the MdFRK1 coding sequence was also cloned as a reference gene. Both genes showed similar sequences as the predicted protein from ‘Golden Delicious’ apple (Fig. 1a). Furthermore, MdFRK2 showed high amino acid sequence identity to LeFRK2 (82.4%) and StFRK2 (79.3%) while MdFRK1 was homologous to AtFRK1 and LeFRK1 (Fig. 1c). Both of the deduced amino acid sequences contained highly conserved motifs and domains of characterized FRK genes (Fig. 1b)10. In Arabidopsis mesophyll protoplasts and onion epidermal cells, we observed that, analogous to tomato SlFRK1 and SlFRK2, both MdFRK1::GFP and MdFRK2::GFP occurred in plasma membrane and cytoplasm, whereas MdFRK2::GFP was also detected in the nucleus (Fig. S1).
Fig. 1. Sequence analysis of MdFRK1/2.
a Schematic diagram of gene positions and structure of MdFRK2. Conserved regions are indicated with (a1), (a2), (a3) and (b). b Alignment of conserved regions in deduced amino acid sequences of MdFRK1/2 with other plant fructokinases. (a1–a3), 3 signature patterns for phosphofructokinase B (PfkB) family; (b), region specific to fructokinase. Black boxes indicate proteins with >50% identical amino acids. c Maximum likelihood phylogeny of MdFRK1/2 deduced amino acid sequences and those from Arabidopsis, Lycopersicon esculentum, and Solanum tuberosum. d Patterns of mRNA relative expression for MdFRK1 and MdFRK2 in developing fruit and different tissues. S, shoot tips; M, mature leaves; DAB, days after bloom
We further investigated the expression patterns of MdFRK1 and MdFRK2 in different tissues from the ‘Royal Gala’ apple. Our results revealed that MdFRK1 mRNA was expressed at an approximately constant level in shoot tips (shoot apex meristem with primordia and early leaves including young stem) and mature leaves, whereas MdFRK2 mRNA levels were approximately six-fold higher in shoot tips than those in mature leaves (Fig. 1d). While the fruits were developing, the transcript levels of MdFRK1 remained unchanged until just before fruit ripening, when they increased markedly. In contrast, the level of MdFRK2 mRNA was highest in the earlier stages of fruit development (Fig. 1d).
Enzymatic properties of MdFRK2
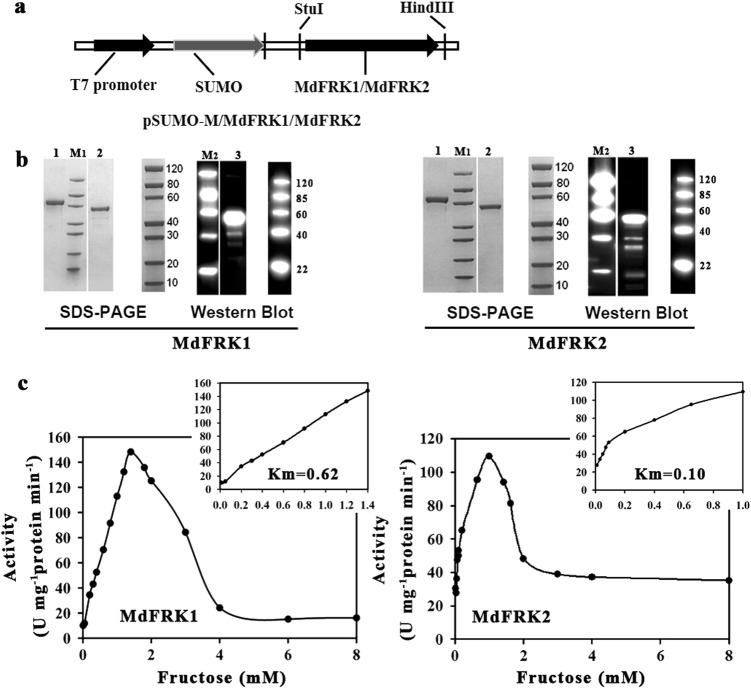
To examine the catalytic characteristics of the proteins encoded by MdFRK1 and MdFRK2, we constructed pSUMO-M vectors of MdFRK1 (predicated to be a low-affinity FRK, similar to that in tomato) and MdFRK2 (Fig. 2a). The two plasmids were individually expressed in E. coli with polyhistidine (His) tags and purified using Ni-agarose resin. Analysis using SDS polyacrylamide gel electrophoresis (SDS-PAGE) showed that the molecular weight for each isoform was approximately 60 KD (Fig. 2b). Both of the heterologous proteins had high specificity for fructose but no affinity for glucose (data not shown). Their enzyme activities were significantly inhibited by fructose when the concentration exceeded 2 mM (Fig. 2c). Furthermore, the maximum activity was measured at 1.0 and 1.4 mM fructose for MdFRK2 and MdFRK1, respectively, with Km values of 0.10 mM for MdFRK2 and 0.62 mM for MdFRK1 (Fig. 2c). The results indicate that MdFRK2 has much higher affinity for fructose than does MdFRK1 and a lower Vmax for fructose.
Fig. 2. Kinetics analysis of 2 fructokinase isoform proteins from E. coli expressing MdFRKs.
a Schematic gene structure of pSUMO-M/MdFRK1 and pSUMO-M/MdFRK2 constructs. b SDS-PAGE and western blot results. For MdFRK1 (left): Lanes M1, M2, protein markers; 1, MdFRK1 (2.00 μg); 2, BSA (2.00 μg); 3, FRK1 (His antibody). For MdFRK2 (right): Lanes M1, M2, protein markers; 1, MdFRK2 (2.00 μg); 2, BSA (2.00 μg); 3, FRK2 (His antibody). c Effect of fructose concentration on MdFRK1 (left) and MdFRK2 (right) activity in extracts prepared from E. coli. Inset shows the level at which fructokinase activity inhibition begins
Overexpression of MdFRK2 increased FRK enzyme activity in leaves
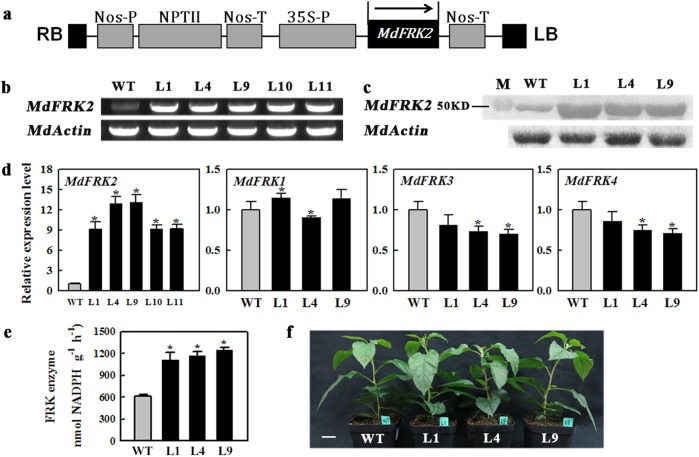
In an attempt to determine the potential physiological functions of MdFRK2 in apple, we tried to generate MdFRK2-inhibited lines using RNAi technology. However, the buds from the transgenic seedlings did not grow well and eventually died in the selection medium, possibly because the gene is indispensable for the growth of the shoot apical meristem, as indicated by its high expression in the shoot tip (Fig. 1d). As an alternative, we constructed an overexpression cassette with its open reading frame (ORF) inserted under the control of a 35 S promoter and transformed it into apple (Fig. 3a). We identified five overexpression transgenic lines through RT-PCR (Fig. 3b) and qRT-PCR (Fig. 3d), which exhibited evident increases in MdFRK2 transcript levels in mature leaves relative to the levels in WT controls (Fig. 3b, d). Among these lines, L1, L4, and L9 displayed 9.2, 13.0 and 13.2-fold increases, respectively, in leaf mRNA levels, respectively, when compared with that of relative to the levels of the untransformed WT controls (Fig. 3d). All three lines also exhibited protein abundance using specific antibody (Fig. 3c) and significantly increased enzyme activity (Fig. 3e). For the other FRK members, the expression of MdFRK1 did not differ from that of WT controls, whereas the transcript levels of MdFRK3 and MdFRK4 were slightly decreased, indicating that any phenotype observed in the transgenic plants would be largely due to the overexpression of MdFRK2.
Fig. 3. Levels of transcripts, enzyme activities, and MdFRK2 protein in leaves from transgenic lines.
a Schematic diagram of vector construct. b Semiquantitative RT-PCR. WT, wild-type; L1, L4, and L9, MdFRK2-OE lines. c Western blotting using the specific monoclonal antibody generated from rabbit. Amount of protein extract loading was referenced by immunoblot analysis with anti-Actin antibody. M: protein marker. d, e Quantitative RT-PCR of MdFRKs expression and MdFRK activity in mature leaves from wild-type (WT) and transgenic lines (L1, L4, and L9), respectively. f Phenotype of wild-type (WT) and transgenic lines (L1, L4, and L9). In (b, d), the MdActin gene was used as internal control. Error bars represent SD based on 3 independent replicates. Statistically significant differences among values (at P < 0.05) were detected with independent t-tests and SPSS software. Scale bar = 2 cm
Overexpression of MdFRK2 altered carbohydrate levels in leaves
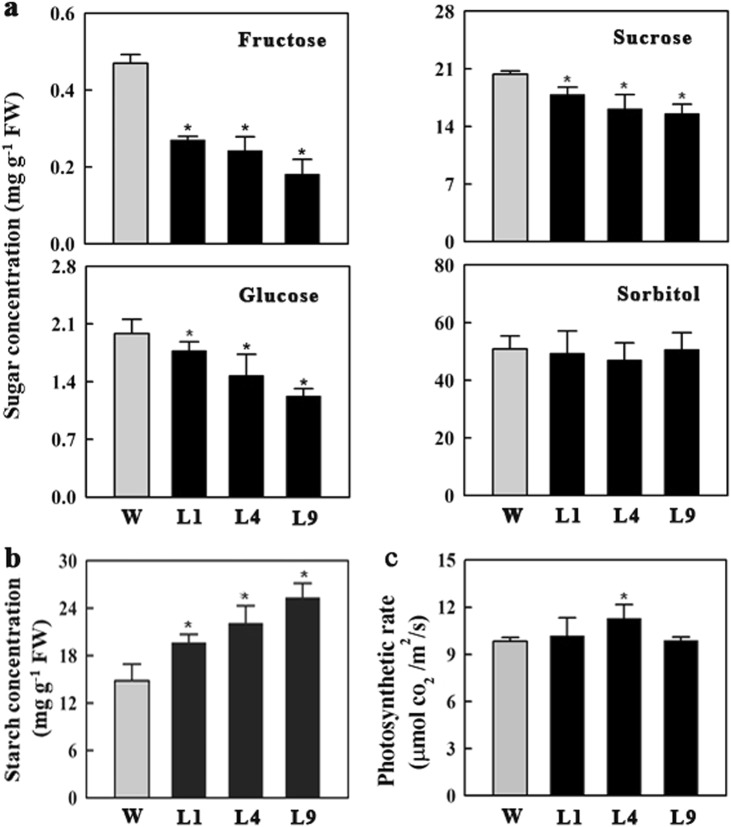
Increased MdFRK activity in the leaves resulted in significantly lower concentrations of fructose in the transgenic lines, with levels reduced to 52% of the control level for L1, 50% for L4, and 41% for L9 (Fig. 4a). In the transgenic lines, the concentrations of sucrose and glucose were also decreased (Fig. 4a). By contrast, the level of starch was increased greatly in all three OE lines relative to the level in WT (Fig. 4b). However, the level of sorbitol did not differ between the transgenic and wild-type plants. These data demonstrated that MdFRK2 modulates sugar concentrations in the leaves; not only those of fructose but also those of glucose and sucrose. In addition, regardless of these observed differences, the transgenic plants showed no obvious alterations in their growth performance or photosynthetic efficiency (Fig. 3f; Fig. 4c; Table S1)
Fig. 4. Changes in carbohydrate concentrations and photosynthetic rates in mature leaves of wild-type (WT) and transgenic lines (L1, L4, and L9).
a Concentrations of sorbitol, sucrose, glucose, and fructose. b Starch concentrations. c Photosynthetic rates. Error bars represent SD based on 3 independent replicates. Statistically significant differences among values (at P < 0.05) were detected with independent t-tests and SPSS software
Overexpression of MdFRK2 altered the activities of enzymes involved in carbohydrate metabolism
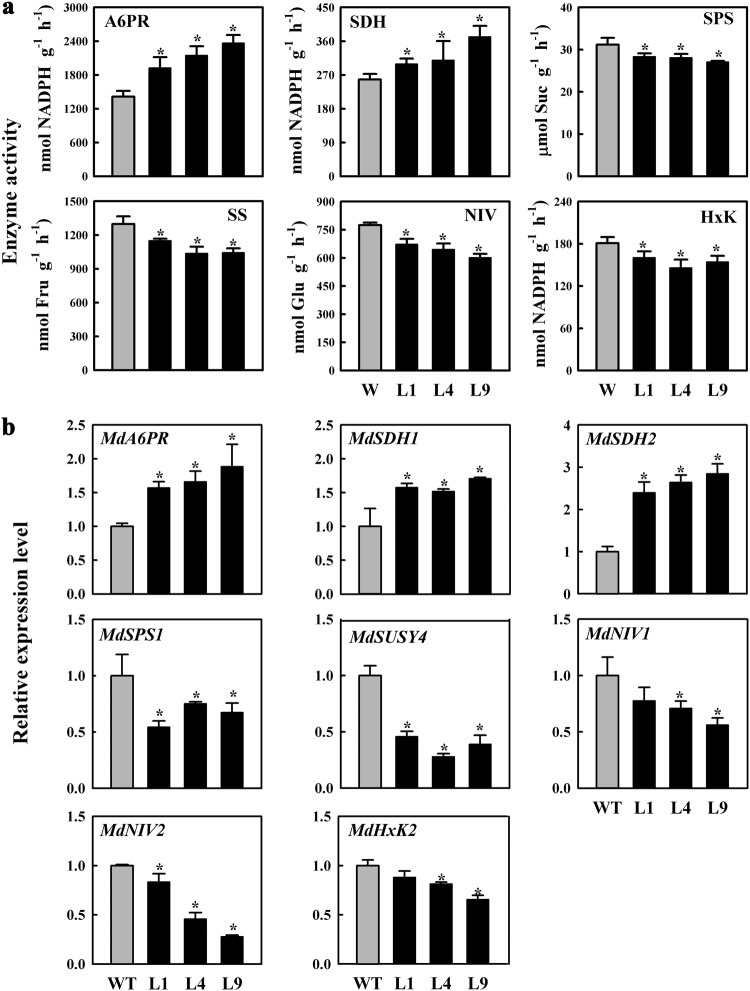
To determine why glucose and sucrose concentrations were reduced in the leaves of MdFRK2-overexpression transgenic plants, we analyzed the key enzymes involved in apple soluble sugar metabolism (Fig. 5a). The activities of A6PR and SDH were markedly increased in the OE lines (Fig. 5a). However, key enzymes involved in sucrose metabolism were suppressed to varying degrees. For example, SPS activity was decreased to 77 and 75% of the WT level for L4 and L9, respectively. A similar pattern was observed for SUSY, NIV, and HxK activities. Changes in the status of sucrose and sorbitol (Fig. 4a) agreed with the patterns of enzyme activities involved in sucrose and sorbitol metabolism (Fig. 5a). We also detected the expression levels of main genes involved in sugar metabolism (Fig. 5b, S3). Notably, genes involved in sorbitol synthesis (A6PR) and the degradation pathway (SDH1 and SDH2) were significantly upregulated in the transgenic lines (Fig. 5b). In contrast, the expression of several genes responsible for sucrose synthesis (SPS1) and degradation (SUSY4, NINV1, and NINV2) was downregulated in these lines. Corresponding to the reduced glucose concentrations (Fig. 4a), HxK members such as HxK2 were also slightly suppressed. However, the expression levels of the genes MdSPS6, MdSUSY1/2/3, MdNIV3, MdHxK1/3/6 were unchanged relative to control levels in transgenic apple leaves (Fig. S3).
Fig. 5. Changes in enzyme activity and gene expression in mature leaves.
Activities of enzymes (a) and relative mRNA expression of genes (b) related to sucrose and sorbitol metabolism in mature leaves of wild-type (WT) and transgenic lines (L1, L4, and L9). Quantitative RT-PCR was performed with gene-specific primers using MdActin as an internal control. Error bars represent SD based on 3 independent replicates. Statistically significant differences among values (at P < 0.05) were detected with independent t-tests and SPSS software
Discussion
MdFRK2 is a major player in fructose metabolism in apple
Fructose metabolism in apple exhibits a well-defined developmental pattern. Total FRK activity, as well as fructose levels in different tissues and at various fruit development stages, has been reported previously30. The early stage of fruit development was characterized by high FRK activity and low levels of fructose accumulation. FRK expression and activity decrease significantly as the concentrations of fructose peaks in mature fruit30. Orthologs of FRK2 that are mainly located in cytoplasm have been confirmed as the major fructose-phosphorylating enzymes in several species, and they show high expression/abundance in sink tissues11,15,16,20,34. Both MdFRK1 and MdFRK2 were mainly located in cytoplasm (Fig. S1). The results indicate that the two encoding proteins are involved in fructose phosphorylation in cytoplasm. MdFRK2 was also expressed in the nucleus, and this expression implies that it plays a role in sugar phosphorylation in the nucleus or has other unknown functions. MdFRK2 had significantly high expression levels in the shoot tips and in young developing fruit, whereas the levels were substantially lower in mature fruit (Fig. 1d). The expression pattern of MdFRK2 closely matches the activity of FRK (Fig. 1d)30. In contrast, MdFRK1 showed significantly different tissue-specific and developmental expression patterns from those of MdFRK2 (Fig. 1d). Additionally, the MdFRK2-OE lines exhibited significantly increased enzyme activities and decreased relative fructose levels in mature leaves. (Figs. 3e, 4a). These findings suggest that MdFRK2 is a major contributor to fructose metabolism in apple; as suggested in peach, high-affinity FRK could be responsible for the low fructose-to-glucose ratio phenotype33.
Sorbitol accounts for approximately 60–80% of all photosynthates produced in apple leaves25. Almost all of the sorbitol and half of the sucrose are converted to fructose in sink organs. Therefore, at least 80% of the total carbon flux is estimated to pass through fructose in apple sink organs30. We expect those organs that do not accumulate significant amounts of fructose (i.e., shoot tips and young fruit) to have stronger FRK activity for fructose utilization than those organs that actively accumulate fructose (e.g., in fruit during cell expansion). Two characteristics of MdFRK support this idea. First, MdFRK2 was more highly expressed in tissues without fructose accumulation, such as shoot tips and young fruit, than in those of mature fruit (Fig. 1d). Second, and more importantly, our results indicated that apple MdFRK2 purified from E. coli had a Km value for fructose of ~ 100 µm, whereas the corresponding value for MdFRK1 was ~ 620 µm (Fig. 2c). The Km value indicated that MdFRK2 has a much higher affinity for fructose than does MdFRK1. However, the maximum fructokinase activity was lower for MdFRK2 (~ 110 U) than for MdFRK1 (~ 150 U) (Fig. 2c). This finding is in contrast to findings for tomato, in which the maximum enzyme activity for LeFRK2 protein was less than 10% of that measured for LeFRK111. The high catalytic capability of MdFRK2 is related to its great capacity for fructose phosphorylation in apple sink cells due to high fructose flux. These results further support our conclusion that MdFRK2 plays a key role in regulating fructose metabolism in apple.
Similar to the LeFRK2 enzymes11, the MdFRK1 and MdFRK2 enzymes in vitro showed substrate inhibition by fructose at pH 8.0. This MdFRK substrate inhibition implied that FRK plays an important gateway role in sugar metabolism and in determining fructose concentrations in apple. This inhibition can effectively control carbon flux into glycolysis and other pathways via F6P once excess fructose exists in the cytosol (especially in sink cells) given that excess F6P can lead to a stress reaction because the cell requirement is limited for carbon flux4. In apple, this substrate inhibition is particularly important because the sink cells have much greater fructose carbon flux. This substrate inhibition induces serial accumulations of fructose in the cytosol10, and these accumulations then accelerate the storage of fructose in the vacuoles. This outcome suggests that FRK and SUSY work in tandem to control the utilization of sucrose, as is also evidenced by the elevated fructose levels in tomato fruit9.
Increased fructokinase activity alters sugar metabolism in apple leaves
The significant increase in FRK enzyme activity in apple plants that overexpressed MdFRK2 further confirmed that the MdFRK2 gene has a decisive role in controlling fructose phosphorylation. As we had expected, elevated FRK activity significantly decreased the fructose concentrations in mature leaves, similar to the connection between diminished FRK expression and higher fructose levels in potato21 and aspen19. Transgenic cotton plants that overexpressed tomato LeFRK1 have significantly increased FRK enzyme activity; however, fructose concentrations are unchanged in the leaves of homozygous T3 plants23. Km values are higher for LeFRK1 (1300 µm) than those for LeFRK2 (54–220 µm), and almost no enzyme activity occurs under low-fructose conditions in yeast systems11. Given the enzymatic property of LeFRK1, the lack of change in fructose levels when tomato LeFRK1 is overexpressed in cotton can be attributed to its low affinity23. We also confirmed, based on the reduced fructose concentrations, that MdFRK2 protein has higher affinity for fructose than does MdFRK1 protein.
In plant cells, fructose is a signal molecule for regulating growth and development35. A system that controls fructose levels might help avoid the negative effects associated with fluctuations in fructose concentration. In transgenic lines of tomato and potato, fructose levels are largely unaltered after FRK activities are modulated21,22,36. Because they produce sorbitol, apple plants have at least two alternative approaches for compensating for reduced fructose concentrations in cells. These approaches include the conversion of sorbitol to fructose by SDH and a sucrose-cleavage pathway via SUSY or invertase catalysis. We found that leaves from our transgenic apple had significantly increased SDH expression and activity but reduced levels of SUSY and invertase relative to controls (Fig. 5). MdFRK2 transcription was also strongly increased by exogenous sorbitol feeding than that by sucrose feeding (Fig. S4), confirming that sorbitol is a more versatile substrate than sucrose for fructose metabolism, as reported in peach by Desnoues et al. (2018)33. Our results indicated that the decreased fructose concentrations in the leaves were partly compensated by increased sorbitol dehydrogenization via SDH in the MdFRK2-OE lines.
When sorbitol synthesis is downregulated in the leaves of transgenic lines, the decreased content of sorbitol leads to a decline in SDH activity and expression level in the shoot tips29 and fruit37. Sorbitol availability is also thought to affect SDH activity and fructose concentrations in apple fruit38. Accordingly, although the ratio of sorbitol/sucrose was increased (Fig. 4a) along with a significant rise in transcript levels and A6PR activity in the transgenics (Fig. 5), sorbitol levels did not differ from control levels because of increased SDH activity in our MdFRK2-OE lines.
Although the capacity to fix CO2 in the leaves was unchanged in OE lines relative to controls (Fig. 4c, Table S1), sugar metabolism and accumulation were significantly changed from control levels in OE lines. SPS1 is thought to be the main gene responsible for SPS activity30,39. Accordingly, decreased SPS1 expression led to reduced sucrose concentrations. In apple shoot tips, the expression levels and enzyme activities of SUSY and neutral invertase are positively regulated by sucrose concentration29, and SUSY activity is correlated with the sink strength of the storage organs40. We speculate that the decreases in sucrose synthesis and concentrations reduced SUSY and NIVN activities in MdFRK2-OE leaves and downregulated the expression levels of MdSUSY4 and MdNINV1 (Fig. 5), both of which are expressed in apple leaves30. The decreases in HxK activity and transcript levels of MdHxK2 (Fig. 5) in the transgenics indicate that MdHxK2 contributes to HXK activity7. Based on these changes in sugar metabolism, we conclude that via a regulated pathway, overexpression of MdFRK2 leads to declines in the concentrations of fructose, sucrose, and glucose (Fig. 6).
Fig. 6. Alteration of pathways for sucrose and sorbitol metabolism in leaves from transgenic apple.
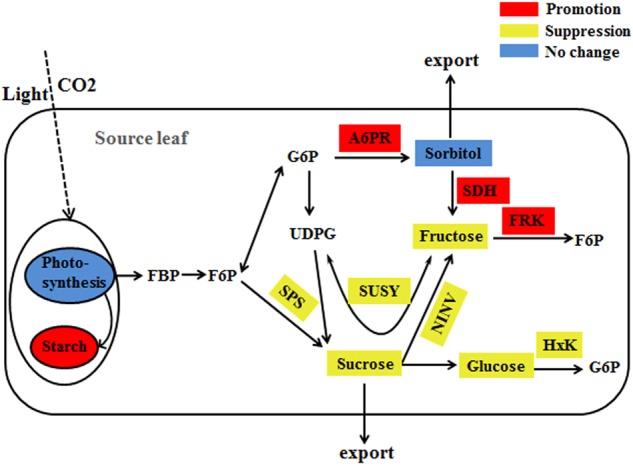
Increased FRK activity contributes to the upregulation of sorbitol metabolism (synthesis and dehydrogenation) and downregulation of sucrose metabolism (synthesis and disassembly). Red boxes indicate increases in sugar concentrations or enzyme activity; yellow boxes, reduction in sugar concentrations or enzyme activity; blue boxes, no change in photosynthetic rate or sugar concentrations. Bold lines indicate that metabolism is accelerated
Although we did not detect the level of F6P or G6P, we expected to see an increase in F6P flux because of elevated fructose phosphorylation in the MdFRK2-OE lines. Such an increase would mean more F6P being reversibly converted to G6P or fructose-1,6-bisphosphatase41. As part of a feedback response, such changes in carbon flux may contribute to greater fixing of CO2 for starch synthesis in the daytime; however, the transgenic samples contained significantly more starch in mature leaves (Fig. 4b). It appears that in sink tissues, FRK regulates starch metabolism based on the concomitant activities of fructokinase, SUSY, and starch-related genes42. However, silencing of LeFRK1 and LeFRK2 in tomato and StFRK1 in potato does not alter the patterns of starch accumulation in those species21,36. Therefore, the mechanism that links FRK with regulated starch concentrations might be associated with its capacity to control sugar metabolism and carbon distribution.
In summary, MdFRK2 encodes a high-affinity fructokinase, and its overexpression increases fructose phosphorylation and alters carbohydrate metabolism in apple leaves. In apple sink cells, at least 80% of the total carbon goes primarily through fructose into metabolism flux because of sorbitol30. Accordingly, apple contains high-affinity FRK that utilizes and maintains fructose at a low level in the cytosol. Here, we confirmed that compared with other plant FRK2s, MdFRK2 is highly expressed in apple sink tissues, and its protein not only shows high affinity for fructose but also has high maximum enzymatic activity. As a main gateway to carbon metabolism, MdFRK activity is strongly inhibited by high fructose concentrations. As in the cases of tomato20 and aspen19, MdFRK2 might be required for apple plant development and growth. This outcome might explain why we were unable to obtain MdFRK2-silencing lines under the 35S promoter. As an alternative approach, it may be feasible to use an inducible promoter with MdFRK2-silencing for apple cultivars that are difficult to transform. For MdFRK2-OE plants, the decrease in fructose level was partly compensated by an increase in sorbitol synthesis and a reduction in sucrose synthesis in the leaves. This outcome caused the concentrations of fructose, sucrose, and glucose to decline in the leaves of our transgenics. The molecular mechanism by which sugar metabolism is regulated through a fructose signal is unknown; it is unknown how the signal is perceived and how its homeostasis is regulated via altered metabolism and other pathways. There have been previous reports of fructose homeostasis in apple fruits under different crop loadings24, fruits from transgenic apple plants with inhibited sorbitol synthesis37, and aspen leaves that show increased CWINV activity43. The lack of change in sorbitol level in our transgenic apple showed that this sugar has primary roles in the vegetative growth of Rosaceae fruit trees37,44. Furthermore, we did not see any phenotypic differences among genotypes, wild or transformed, even though these plants had been grown in the greenhouse for two years. We expect the transgenic apple trees to flower and fruit after four to six years, at which time we will further investigate fruit traits.
Materials and methods
Plant materials and growth conditions
Plants of Arabidopsis thaliana (‘Columbia’, ecotype Col-0) were maintained in a controlled culture room at 22 °C under conditions of 100 μmol photons m−2 s−1, 70% relative humidity, and a 16-h, long-day photoperiod. Tobacco plants (Nicotiana tabacum L., ‘Samsun NN’) were grown at 25 °C under a 16-h photoperiod and lamp-supplemented light at 120 μmol photons m −2 s −1.
Tissue-cultured plants of WT and MdFRK2-transformed ‘Royal Gala’ apples were initially grown on MS medium supplemented with 0.2 mg L−1 IAA and 0.3 mg L−1 6-BA for four weeks. They were then transferred to rooting medium (MS + 0.5 mg IBA and 0.5 mg IAA). After rooting, plants of both genotypes were transferred to a culture room maintained at 23 °C under a 14-h photoperiod supplemented with fluorescent light (60 μmol m−2 s−1)45. After these plants had grown for two months, the fifth to eighth leaves from the base of the stem (fully mature leaves) were removed from healthy, uniformly sized stocks. The samples were immediately frozen in liquid nitrogen and stored at –80 °C for further analysis.
Gene cloning, phylogenetic analysis, and subcellular localization
The predicted sequences of MdFRK1/2 (MDP0000173131 and MDP0000323311) in the apple genome were retrieved from the Malus Genome Database (http://www.rosaceae.org), and primers were designed for gene cloning (Table S2). Total RNA was extracted from apple shoot tips by the CTAB method46, and the full cDNAs of MdFRK1/2 were cloned by RT-PCR.
Amino acid alignments were aligned using ClustalW and Lasergene software (DNASTAR, USA). We then constructed maximum likelihood (ML) phylogenetic trees with 100 bootstrap replicates with MEGA version 5 (http://www.megasoftware.net/). The evolutionary history was inferred according to the ML method based on the JTT matrix-based model30.
For determining subcellular localizations, the full-length ORFs of MdFRK1 and MdFRK2 without the stop codons were cloned into PBI121-GFP vectors under the control of the CaMV35S promoter. To examine transient expression, we transformed the plasmids into onion epidermal cells with a biolistic helium gun device (PDS-1000; BioRad, Hercules, CA, USA). Cell culturing and fluorescence observations were conducted as previously described47. These constructs were also transiently expressed in Arabidopsis protoplasts via polyethylene glycol-mediated transformation based on a protocol previously described48. After 12 h of transformation, protoplasts were observed with confocal microscopy (LSM 510 META, Carl Zeiss, Germany).
Expression of MdFRKs in E. coli, protein purification, and kinetics analysis
The expression of MdFRKs was monitored in E. coli, and proteins were purified according to published protocols49. After cloning, MdFRK1 and MdFRK2 were inserted into pSUMO-M vector. The two recombinant plasmids, pSUMO-M/MdFRK1, and pSUMO-M/MdFRK2, were then transferred into E. coli BL21 (DE3), and the recombinant strains were induced by 1 mM isopropyl–D thiogalactopyranoside (IPTG) to produce fusion proteins tagged with His at 15 °C or 37 °C for different durations. The expressed fusion proteins were detected by SDS-PAGE and western blotting (Fig. S2). After determining that expression peaked at 4 h at 37 °C, we obtained the protein from the supernatant and purified it on 5 × 5 mL HisTrap HP columns prepacked with Ni Sepharose (GE healthcare, Beijing, China) to obtain soluble protein.
For the Km and substrate-specific analyses, fructose phosphorylation activity was measured in a final volume of 0.5 mL that contained 50 mM Tri-HCl (pH 8.0), 4 mM MgCl2, 2.5 mM ATP, 0.33 mM NAD+, 1 U of G6P dehydrogenase, and 1 U of phosphoglucoisomerase30. Fructose concentrations ranged from 1 to 5 mM, whereas glucose concentrations ranged from 0 to 20 mM. Both sugars were measured using a continuous assay that coupled fructose phosphorylation to NADP+ reduction at 340 nm.
Vector constructs and apple transformation
To construct the vector for the MdFRK2-overexpression lines, we introduced the coding region of MdFRK2 into the pGWB401 binary vector, which is driven by the CaMV 35S promoter and carries the kanamycin (Kan) selectable marker. The recombinant plasmid was introduced into Agrobacterium tumefaciens strain EHA105. Leaf fragments of the ‘Royal Gala’ apple were transformed based on a protocol previously described50. The regenerated Kan-resistant buds were screened on an MS medium containing 25 mg L−1 Kan as a selectable marker. Afterward, plants displaying normal growth were evaluated by PCR analysis of extracted DNA. Overexpression of MdFRK2 was confirmed by quantitative real-time PCR and western blotting. From the five independently transformed lines obtained here, we selected three for further analysis. Untransformed ‘Royal Gala’ plants were cultured in the same way and served as the control plants.
Western blotting
Total protein samples were extracted from the mature leaves of OE-lines and WT plants as described previously45, and the total concentrations were determined with protein assay kits (Bio-Rad), using bovine serum albumin as a standard. Specific monoclonal antibodies to a peptide (CNPSADMLLKPDELN) from a highly conserved region in MdFRK2 (Genscript, Nanjing, China) were generated in rabbit. MdActin was monitored with a monoclonal antibody (CWBIO, Beijing, China). The procedures for western blotting followed Sun et al. (2017)45. The antigen–antibody complexes were detected using ClarityTM Western ECL Substrate (Bio-Rad) according to the manufacturer’s instructions.
Measurements of soluble sugars and starch
As we described previously30, soluble sugars were extracted and derivatized sequentially with methoxyamine hydrochloride and N-methyl-N-trimethylsilyl-trifluoroacetamide. Then, the metabolites were analyzed with a Shimadzu GCMS-2010 SE (Shimadzu Corporation, Kyoto, Japan). The tissue residue that remained after 75% methanol extraction for GC‒MS analysis was re-extracted three times with 80% (v/v) ethanol at 80 °C, and the pellet was retained for starch determinations30. The assimilation of CO2 in seedling leaves was monitored between 9:30 and 11:30 A.M. using a LI-COR 6400 portable photosynthesis system (LI-COR, Huntington Beach, CA, USA).
Assays of enzyme activities
We applied methods previously reported methods30 to assay the activities of FRK, SDH, NINV, SUSY, HK, and SPS in leaf samples from five biological replications30. The A6PR enzyme was extracted and assayed according to the method of Cheng et al. (2005)25.
Analysis of mRNA expression
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to analyze the expression of genes involved in sugar metabolism. All of the genes, primers, and procedures used here were described previously30. qRT-PCR was performed on an ABI7300 Real-Time PCR System (Thermo Fisher Scientific). Transcripts of Actin (CN938023) served to standardize the cDNA from our test genes. For each sample, total RNA was extracted from three biological replicates before the qRT-PCR experiments were conducted. All data were examined according to the ddCT method.
Statistical analysis
All data were analyzed via IBM SPSS Statistics 21 and graphed with Sigma Plot 10.0 software. Data were analyzed using independent t-tests with significance evaluated at P < 0.05. Values were presented as the means ± standard deviation of 6 plants, with triplicate observations for each measurement.
Electronic supplementary material
Acknowledgements
This work was supported by the Program for the National Natural Science Foundation of China (No. 31372038) and by the earmarked fund for the China Agriculture Research System (CARS-28). The authors thank Mr. Xuanchang Fu and Xiaowei Ma for maintaining the plants.
Author contributions
J.Y. performed the majority of the experiments. L.C.Z., W.F.C., C.Z., and D.X.L. performed experiments. J.Y. and L.C.Z. analyzed and discussed the data. M.J.L. supervised the work. J.Y. prepared a draft of the manuscript. All authors contributed to the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41438-018-0099-x).
References
- 1.Koch KE. Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Smeekens S. Sugar-induced signal transduction in plants. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 2000;51:49–81. doi: 10.1146/annurev.arplant.51.1.49. [DOI] [PubMed] [Google Scholar]
- 3.Cirilli M, Bassi D, Ciacciulli A. Sugars in peach fruit: a breeding perspective. Hortic. Res. 2016;3:15067. doi: 10.1038/hortres.2015.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruan YL. Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant. Biol. 2014;65:33–67. doi: 10.1146/annurev-arplant-050213-040251. [DOI] [PubMed] [Google Scholar]
- 5.Huber SC, Huber JL. Role of sucrose-phosphate synthase in sucrose metabolism in leaves. Plant Physiol. 1992;99:1275–1278. doi: 10.1104/pp.99.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solis-Guzman MG, et al. Arabidopsis thaliana sucrose phosphate synthase (sps) genes are expressed differentially in organs and tissues, and their transcription is regulated by osmotic stress. Gene Expr. Patterns. 2017;92:25–26. doi: 10.1016/j.gep.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Roessner-Tunali U, et al. Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 2003;133:84–99. doi: 10.1104/pp.103.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandel-Kfir M, et al. Two newly identified membrane-associated and plastidic tomato HXKs: characteristics, predicted structure and intracellular localization. Planta. 2006;224:1341–1352. doi: 10.1007/s00425-006-0318-9. [DOI] [PubMed] [Google Scholar]
- 9.Granot D. Role of tomato hexose kinases. Funct. Plant. Biol. 2007;34:564–570. doi: 10.1071/FP06207. [DOI] [PubMed] [Google Scholar]
- 10.Pego JV, Smeekens SC. Plant fructokinases: a sweet family get-together. Trends Plant Sci. 2000;5:531–536. doi: 10.1016/S1360-1385(00)01783-0. [DOI] [PubMed] [Google Scholar]
- 11.Kanayama Y, et al. Divergent fructokinase genes are differentially expressed in tomato. Plant Physiol. 1997;113:1379–1384. doi: 10.1104/pp.113.4.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez-Barajas E, Luethy MH, Randall DD. Molecular cloning and analysis of fructokinase expression in tomato (Lycopersicon esculentum Mill.) Plant Sci. 1997;125:13–20. doi: 10.1016/S0168-9452(97)04620-7. [DOI] [Google Scholar]
- 13.German MA, et al. Cloning, expression and characterization of LeFRK3, the fourth tomato (Lycopersicon esculentum Mill.) gene encoding fructokinase. Plant Sci. 2004;166:285–291. doi: 10.1016/j.plantsci.2003.09.017. [DOI] [Google Scholar]
- 14.Taylor MA, Ross HA, Gardner A, Davies HV. Characterisation of a cDNA encoding fructokinase from potato (Solanum tuberosum) J. Plant Physiol. 1995;145:253–256. doi: 10.1016/S0176-1617(11)81885-7. [DOI] [Google Scholar]
- 15.Zhang S, Nichols SE, Dong JG. Cloning and characterization of two fructokinases from maize. Plant Sci. 2003;165:1051–1058. doi: 10.1016/S0168-9452(03)00298-X. [DOI] [Google Scholar]
- 16.Jiang HW, Dian WM, Liu FY, Wu P. Isolation and characterization of two fructokinase cDNA clones from rice. Phytochemistry. 2003;62:47–52. doi: 10.1016/S0031-9422(02)00428-4. [DOI] [PubMed] [Google Scholar]
- 17.Riggs JW, Cavales PC, Chapiro SM, Callis J. Identification and biochemical characterization of the fructokinase gene family in Arabidopsis thaliana. BMC Plant Biol. 2017;17:83. doi: 10.1186/s12870-017-1031-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petreikov M, Dai N, Granot D, Schaffer AA. Characterization of native and yeast-expressed tomato fruit fructokinase enzymes. Phytochemistry. 2001;58:841–847. doi: 10.1016/S0031-9422(01)00331-4. [DOI] [PubMed] [Google Scholar]
- 19.Roach M, et al. Fructokinase is required for carbon partitioning to cellulose in aspen wood. Plant J. 2012;70:967–977. doi: 10.1111/j.1365-313X.2012.04929.x. [DOI] [PubMed] [Google Scholar]
- 20.German MA, et al. Suppression of fructokinase encoded by LeFRK2 in tomato stem inhibits growth and causes wilting of young leaves. Plant J. 2003;34:837–846. doi: 10.1046/j.1365-313X.2003.01765.x. [DOI] [PubMed] [Google Scholar]
- 21.Davies HV, et al. Modulation of fructokinase activity of potato (Solanum tuberosum) results in substantial shifts in tuber metabolism. Plant Cell Physiol. 2005;46:1103–1115. doi: 10.1093/pcp/pci123. [DOI] [PubMed] [Google Scholar]
- 22.Damari-Weissler H, et al. LeFRK2 is required for phloem and xylem differentiation and the transport of both sugar and water. Planta. 2009;230:795–805. doi: 10.1007/s00425-009-0985-4. [DOI] [PubMed] [Google Scholar]
- 23.Mukherjee T, et al. Constitutively overexpressing a tomato fructokinase gene (LeFRK1) in cotton (Gossypium hirsutum L. cv. Coker 312) positively affects plant vegetative growth, boll number and seed cotton yield. Funct. Plant. Biol. 2015;42:899–908. doi: 10.1071/FP15035. [DOI] [PubMed] [Google Scholar]
- 24.Klages K, Donnison H, Wunsche J, Boldingh H. Diurnal changes in non-structural carbohydrates in leaves, phloem exudate and fruit in ‘Braeburn’ apple. Aust. J. Plant. Physiol. 2001;28:131–139. [Google Scholar]
- 25.Cheng L, Zhou R, Reidel EJ, Sharkey TD, Dandekar AM. Antisense inhibition of sorbitol synthesis leads to up-regulation of starch synthesis without altering CO2 assimilation in apple leaves. Planta. 2005;220:767–776. doi: 10.1007/s00425-004-1384-5. [DOI] [PubMed] [Google Scholar]
- 26.Meng D, et al. Decreased sorbitol synthesis leads to abnormal stamen development and reduced pollen tube growth via a MYB transcription factor, MdMYB39L, in apple (Malus domestica) New Phytol. 2018;217:641–656. doi: 10.1111/nph.14824. [DOI] [PubMed] [Google Scholar]
- 27.Meng D, et al. Sorbitol modulates resistance to Alternaria alternata by regulating the expression of an NLR resistance gene in apple. Plant Cell. 2018;30:1562–1581. doi: 10.1105/tpc.18.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada K, Oura Y, Mori H, Yamaki S. Cloning of NAD-dependent sorbitol dehydrogenase from apple fruit and gene expression. Plant Cell Physiol. 1998;39:1375–1379. doi: 10.1093/oxfordjournals.pcp.a029345. [DOI] [PubMed] [Google Scholar]
- 29.Zhou R, Cheng L, Dandekar AM. Down-regulation of sorbitol dehydrogenase and up-regulation of sucrose synthase in shoot tips of the transgenic apple trees with decreased sorbitol synthesis. J. Exp. Bot. 2006;57:3647–3657. doi: 10.1093/jxb/erl112. [DOI] [PubMed] [Google Scholar]
- 30.Li M, Feng F, Cheng L. Expression patterns of genes involved in sugar metabolism and accumulation during apple fruit development. PLoS One. 2012;7:e33055. doi: 10.1371/journal.pone.0033055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, et al. Proteomic analysis reveals dynamic regulation of fruit development and sugar and acid accumulation in apple. J. Exp. Bot. 2016;67:5145–5157. doi: 10.1093/jxb/erw277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, M., Li, P., Ma, F. & Cheng, L. Sugar metabolism and accumulation in the fruit of transgenic apple trees with decreased sorbitol synthesis. Hortic. Res. 5, 60 (2018). [DOI] [PMC free article] [PubMed]
- 33.Desnoues Elsa, Génard Michel, Quilot-Turion Bénédicte, Baldazzi Valentina. A kinetic model of sugar metabolism in peach fruit reveals a functional hypothesis of a markedly low fructose-to-glucose ratio phenotype. The Plant Journal. 2018;94(4):685–698. doi: 10.1111/tpj.13890. [DOI] [PubMed] [Google Scholar]
- 34.Kanayama Y, et al. Tomato fructokinases exhibit differential expression and substrate regulation. Plant Physiol. 1998;117:85. doi: 10.1104/pp.117.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho YH, Yoo SD. Signaling role of fructose mediated by FINS1/FBP in Arabidopsis thaliana. PLoS Genet. 2011;7:e1001263. doi: 10.1371/journal.pgen.1001263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odanaka S, Bennett AB, Kanayama Y. Distinct physiological roles of fructokinase isozymes revealed by gene-specific suppression of Frk1 and Frk2 expression in tomato. Plant Physiol. 2002;129:1119–1126. doi: 10.1104/pp.000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teo G, et al. Silencing leaf sorbitol synthesis alters long distance partitioning and apple fruit quality. P Natl. Acad. Sci. Usa. 2006;103:18842–18847. doi: 10.1073/pnas.0605873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Archbold DD. Carbohydrate availability modifies sorbitol dehydrogenase activity of apple fruit. Physiol. Plant. 1999;105:391–395. doi: 10.1034/j.1399-3054.1999.105301.x. [DOI] [Google Scholar]
- 39.Sun J, Zhang J, Larue CT, Huber SC. Decrease in leaf sucrose synthesis leads to increased leaf starch turnover and decreased RuBP regeneration-limited photosynthesis but not Rubisco-limited photosynthesis in Arabidopsis null mutants of SPSA1. Plant Cell Environ. 2011;34:592–604. doi: 10.1111/j.1365-3040.2010.02265.x. [DOI] [PubMed] [Google Scholar]
- 40.Chourey PS, Taliercio EW, Carlson SJ, Ruan YL. Genetic evidence that the two isozymes of sucrose synthase present in developing maize endosperm are critical, one for cell wall integrity and the other for starch biosynthesis. Mol. Gen. Genet. 1998;259:88–96. doi: 10.1007/s004380050792. [DOI] [PubMed] [Google Scholar]
- 41.Zhou R, Cheng L. Biochemical characterization of cytosolic fructose-1,6-bisphosphatase from apple (Malus domestica) leaves. Plant Cell Physiol. 2004;45:879–886. doi: 10.1093/pcp/pch096. [DOI] [PubMed] [Google Scholar]
- 42.Schaffer AA, Petreikov M. Sucrose-to-starch metabolism in tomato fruit undergoing transient starch accumulation. Plant Physiol. 1997;113:739–746. doi: 10.1104/pp.113.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C, et al. Symplastic Phloem Loading in Poplar. Plant Physiol. 2014;166:306–313. doi: 10.1104/pp.114.245845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanamaru N, et al. Transgenic apple transformed by sorbitol-6-phosphate dehydrogenase cDNA-Switch between sorbitol and sucrose supply due to its gene expression. Plant Sci. 2004;167:55–61. doi: 10.1016/j.plantsci.2004.02.024. [DOI] [Google Scholar]
- 45.Sun X, et al. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 2018;16:545–557. doi: 10.1111/pbi.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant. Mol. Biol. Rep. 1993;11:113–116. doi: 10.1007/BF02670468. [DOI] [Google Scholar]
- 47.Tan Y, Li M, Ma F. Overexpression of MpCYS2, a phytocystatin gene from Malus prunifolia (Willd.) Borkh., confers drought tolerance and protects against oxidative stress in Arabidopsis. Plant Cell Tiss. Org. 2015;123:15–27. doi: 10.1007/s11240-015-0809-0. [DOI] [Google Scholar]
- 48.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 49.Chen X, et al. The stability, and efficacy against penicillin-resistant Enterococcus faecium, of the plectasin peptide efficiently produced by Escherichia coli. J. Microbiol. & Biotechnol. 2015;25:1007–1014. doi: 10.4014/jmb.1501.01056. [DOI] [PubMed] [Google Scholar]
- 50.Dai H, et al. Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci. Hortic.-Amst. 2013;164:202–208. doi: 10.1016/j.scienta.2013.09.033. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.