Abstract
Auxin signaling, which is crucial for normal plant growth and development, mainly depends on ARF–Aux/IAA interactions. However, little is known regarding the regulatory effects of auxin signaling on anthocyanin metabolism in apple (Malus domestica). We investigated the functions of MdARF13, which contains a repression domain and is localized to the nucleus. This protein was observed to interact with the Aux/IAA repressor, MdIAA121, through its C-terminal dimerization domain. Protein degradation experiments proved that MdIAA121 is an unstable protein that is degraded by the 26S proteasome. Additionally, MdIAA121 stability is affected by the application of exogenous auxin. Furthermore, the overexpression of MdIAA121 and MdARF13 in transgenic red-fleshed apple calli weakened the inhibitory effect of MdARF13 on anthocyanin biosynthesis. These results indicate that the degradation of MdIAA121 induced by auxin treatment can release MdARF13, which acts as a negative regulator of the anthocyanin metabolic pathway. Additionally, yeast two-hybrid, bimolecular fluorescence complementation, and pull-down assays confirmed that MdMYB10 interacts with MdARF13. A subsequent electrophoretic mobility shift assay and yeast one-hybrid assay demonstrated that MdARF13 directly binds to the promoter of MdDFR, which is an anthocyanin pathway structural gene. Interestingly, chromatin immunoprecipitation–quantitative real-time PCR results indicated that the overexpression of MdIAA121 clearly inhibits the recruitment of MdARF13 to the MdDFR promoter. Our findings further characterized the mechanism underlying the regulation of anthocyanin biosynthesis via Aux/IAA–ARF signaling.
A green light for red apples
Insights into a hormonal signaling pathway controlling apple coloration could help growers to consistently produce eye-catching fruit. Auxins are signaling molecules that regulate many important metabolic pathways in plants. Research from a team led by Xue-sen Chen at Shandong Agricultural University in China now reveals how the auxin naphthalene acetic acid (NAA) controls production of anthocyanin, the molecule that gives red apples their distinctive coloration. The researchers determined that increasing NAA levels result in reduced production of anthocyanin. These effects are mediated by a subset of auxin-associated proteins that regulate the expression of genes that contribute to the synthesis of this pigment. The color of fruit can strongly influence consumer purchasing decisions, and this signaling pathway could help horticulturalists to ensure that apples retain their visual appeal even under inclement growing conditions.
Introduction
Color is an important fruit quality index that influences the purchasing behavior of consumers to some degree1. As a type of secondary metabolite, anthocyanins constitute one of the main pigments responsible for fruit color2,3. Additionally, their positive effects on human health include enhancing vascular elasticity, preventing cardiovascular disease, and protecting the liver from damage4–7.
Three main protein families (MYB, bHLH, and WD40) regulate anthocyanin biosynthesis by forming the MYB–bHLH–WD40 complex8,9. The MYB transcription factors (TFs) belong to one of the largest plant TF families, with some members associated with anthocyanin biosynthesis having been isolated and identified in many plant species, including Arabidopsis thaliana, maize, strawberry, and grape10–13. In apple, the three genes encoding MYB activators (MYB10, MYB1, and MYBA) involved in anthocyanin biosynthesis are alleles14. Two MYB TF genes (MdMYB1 and MdMYBA), which were first isolated from fruit skin, have been confirmed to be responsible for the accumulation of anthocyanin15,16. Furthermore, MdMYB10 consists of a characteristic R6 rearrangement in its promoter that is associated with increased anthocyanin biosynthesis in red-fleshed apples17.
Anthocyanin biosynthesis is influenced by environmental stimuli (e.g., light and temperature) and plant hormones (e.g., auxin and jasmonate)18–21. Moreover, auxins (naphthalene acetic acid (NAA) or 2,4-dichlorophenoxyacetic acid (2,4-D)) regulate secondary metabolic pathways, including those related to phenylpropanoid, flavonoid, and anthocyanin metabolism22–25. An earlier study involving a carrot suspension culture revealed that 2,4-D strongly inhibits anthocyanin biosynthesis20. Additionally, the application of exogenous NAA and 2,4-D can decrease the anthocyanin contents of transgenic tobacco calli24. In apple, studies involving red-fleshed apple calli revealed that increasing auxin concentrations can significantly inhibit anthocyanin biosynthesis26,27. However, the role of the auxin signaling pathway during the regulation of anthocyanin biosynthesis is unclear.
Auxin (indole-3-acetic acid (IAA)) regulates plant development by inducing rapid cellular responses and changes in gene expression. Auxin response factors (ARFs) together with auxin/IAA (Aux/IAA) proteins are TFs that function as key regulators of auxin-responsive transcription in plants28–30. Current auxin signaling models indicate that auxin responses mainly depend on the interaction between the homologous C-terminal domains of Aux/IAA and ARF proteins31,32. When cellular auxin concentrations are low, ARF activators at the promoters of auxin-responsive genes are thought to be inactive because of their association with Aux/IAA repressors29,31. When auxin concentrations increase, the Aux/IAA repressors are recruited to auxin receptors and degraded via the ubiquitin–proteasome pathway33,34. This degradation enables ARF activators to repress/activate the expression of auxin-responsive genes33.
Most ARFs consist of an N-terminal DNA-binding domain (DBD), a middle region that functions as an activation domain (AD) or a repression domain (RD), and a C-terminal dimerization domain (CTD)35. ARFs can specifically bind to the TGTCTC auxin-response element (AuxRE) in the promoters of primary/early auxin-response genes35. In the last few years, several new details regarding the role of ARFs in plant growth and development have been revealed. For example, AtARF3 and AtARF4 are important for the development of reproductive and vegetative tissues36,37. Additionally, AtARF5 is involved in embryonic and vascular tissue development38,39, while OsARF8 mediates hypocotyl elongation and influences auxin homeostasis40. Meanwhile, SlARF7 may negatively regulate fruit set, whereas the upregulated expression of SIARF10 suggests that the encoded protein positively affects fruit development41,42. Thus, ARFs exhibit dynamic functions in plants and have a central role in the hub of transcriptional networks.
In the present study, we investigated the effects of different auxin concentrations on anthocyanin biosynthesis in red-fleshed apple calli. We also characterized an ARF gene, MdARF13, and confirmed that the MdARF13-mediated auxin signaling pathway helps regulate anthocyanin biosynthesis in apple, thereby revealing a novel metabolic function for ARF proteins.
Materials and methods
Plant materials and hormone treatments
Calli induced as previously described were cultured on Murashige and Skoog (MS) medium supplemented with 2 μmol/L 6-benzylaminopurine and 5, 10, 20, or 40 μmol/L NAA43. The pH of all media was adjusted to 5.8 ± 0.1. The red-fleshed apple calli were incubated under a 16 h light/8 h dark photoperiod (light intensity: 1000–2000 lx) for 18 days before being harvested.
Anthocyanin extraction and absorbance measurements
Harvested calli (0.5 g each) were ground to a powder in liquid nitrogen and then treated with 15 mL 1% (v/v) HCl-methanol for 24 h at 4 °C in darkness. The solutions were then centrifuged for 10 min at 8000 × g. The absorbance of the supernatants was measured at 530 nm using a UV-2450 spectrophotometer (Shimadzu, Kyoto, Japan).
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was isolated using an RNAprep Pure Plant Kit (Tiangen, Beijing, China). First-strand complementary DNA (cDNA) was synthesized using the RevertAid First-Strand cDNA Synthesis Kit (Fermentas, St. Leon-Roth, Germany). Three replicates were prepared for each sample. Quantitative real-time polymerase chain reaction (qPCR) analyses were conducted using 50 ng/μL cDNA as the template, SYBR Green PCR Master Mix (TransGen Biotech, Beijing, China), and the iCycler iQ5 system (Bio-Rad, Hercules, CA, USA). Details regarding the qPCR primers are provided in Supplementary Table S1. MdActin was used as the internal control.
Amino acid sequence analysis and phylogenetic tree construction
The ARF amino acid sequences were obtained from the NCBI (National Center for Biotechnology Information) database and aligned using the DNAMAN software (Lynnon Biosoft, USA). A phylogenetic tree was constructed according to the neighbor-joining method using the MEGA5.0 program.
Red-fleshed apple callus transformation
The intact MdARF13 coding sequence (CDS) was inserted into the pRI101 vector containing the 35S promoter and a green fluorescent protein (GFP) tag sequence to generate the 35S::MdARF13-GFP construct. Additionally, the intact MdIAA121 CDS was inserted into the pCAMBIA1301 vector containing the 35S promoter and a GFP tag sequence to prepare the 35S::MdIAA121-GFP construct. The resulting recombinant vectors were transferred to Agrobacterium tumefaciens LBA4404 cells. Two-week-old calli grown in liquid medium were cocultured with A. tumefaciens LBA4404 cells carrying the 35S::MdARF13-GFP construct on MS medium containing 2 μmol/L NAA and 4 μmol/L 6-benzylaminopurine at 25 °C for 2 days in darkness. The calli were then transferred to fresh MS medium supplemented with 662 μmol/L carbenicillin and 74 μmol/L kanamycin to screen for transgene-carrying calli. To obtain calli cotransfected with MdARF13+MdIAA121, the same method was used, but calli were transferred to MS medium supplemented with 662 μmol/L carbenicillin, 74 μmol/L kanamycin, and 40 μmol/L hygromycin. GFP-overexpressing calli were used as controls.
Yeast two-hybrid assay
Yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) assays were conducted according to the manufacturer’s instructions (Clontech, Mountain View, CA, USA). The MdARF13 CDS was inserted into the pGBKT7 vector (i.e., bait), while the MdIAA7, MdIAA121, MdMYB9, and MdMYB10 CDSs were inserted into the pGADT7 vector (i.e., prey). All recombinant vectors were cotransformed into yeast strain Y2H Gold cells using the lithium acetate method. The cells were cultured on synthetic defined (SD) medium lacking leucine and tryptophan (SD/−Leu/−Trp). Putative transformants were transferred to SD medium lacking adenine, histidine, leucine, and tryptophane (SD/−Ade/−His/−Leu/−Trp; Clontech) with or without X-α-gal.
Bimolecular fluorescence complementation assay
For a bimolecular fluorescence complementation (BiFC) assay, the MdARF13 CDS was inserted into the pSPYNE-35S vector (with an NYFP tag sequence) to generate the MdARF13-NYFP recombinant vector. The MdIAA121 and MdMYB10 CDSs were inserted into the pSPYCE-35S vector (with a CYFP tag sequence) to generate the MdIAA121-CYFP and MdMYB10-CYFP recombinant vectors. A. tumefaciens LBA4404 cells were transformed with the recombinant vectors and then cultured in liquid medium until the optical density (600 nm) reached 0.6. Onion epidermal cells were treated with equal volumes of different combinations of A. tumefaciens strains. Onion cells were cultured at 23 °C for 48 h. The yellow fluorescence protein (YFP) signal was detected using a Zeiss CLSM-5 confocal laser scanning microscope (excitation wavelength of 488 nm).
Pull-down assay
The MdIAA121 and MdMYB10 open reading frames were cloned into the pET32a vector containing a poly-histidine (HIS) tag sequence. The intact MdARF13 CDS was ligated into the pGEX-4T-1 vector containing a glutathione S-transferase (GST) tag sequence. The recombinant vectors were inserted into Escherichia coli BL21 (DE3) cells (TransGen) to induce the production of fusion proteins. The resulting proteins were mixed together and then purified in columns using the HIS tag. The purified mixed proteins were subsequently analyzed by western blotting with anti-HIS or anti-GST antibodies (Abmart, Shanghai, China).
Yeast one-hybrid analysis
Yeast one-hybrid (Y1H) assays were conducted using yeast strain Y187 cells (Clontech) according to the manufacturer’s instructions. MdARF13 was cloned into the pGADT7 vector, while the MdDFR promoter was inserted into the pHIS2 vector. Different combinations of recombinant and empty vectors were cotransformed into yeast Y187 cells, and the interactions were examined on SD medium lacking Trp, Leu, and His (SD/−Trp/−Leu/−His) with an optimal concentration of 3-amino-1,2,4-triazole.
Electrophoretic mobility shift assays
Electrophoretic mobility shift assay (EMSA) experiments were completed using an EMSA kit (Pierce, Rockford, IL, USA) and biotin-labeled probes. Briefly, biotin-labeled probes were incubated in 1× binding buffer (2.5% glycerol, 10 mM EDTA, 5 mM MgCl2, and 50 mM KCl) with or without proteins at 24 °C for 25 min. An unlabeled probe was added to the reactions.
Chromatin immunoprecipitation–quantitative real-time PCR analysis
The chromatin immunoprecipitation (ChIP) experiment was completed using a modified version of a published method44. The ChIP Assay Kit (Upstate Biotechnology, Lake Placid, NY, USA) and anti-GFP antibody (Abmart) were used for cross-linking, the removal of cross-linkers, immunoprecipitation, and elution. The ChIP signal was quantified by qPCR as the percentage of the total input DNA. The experiment was repeated three times.
Subcellular localization analysis
Protoplasts isolated from apple callus cells were prepared and transformed as described by Hu et al.45. Transformed apple protoplasts were stained with the DNA-specific dye 4′,6-diamidino-2-phenylindole, which highlights the location of the nucleus. The protoplasts from transgenic calli were observed by fluorescence microscopy.
In vitro protein degradation assay
Protein degradation assays were used to detect the dynamic changes to MdIAA13 in vitro. The assay buffer contained 25 mM Tris-HCl, pH 7.5, 10 mM NaCl, 10 mM MgCl2, 5 mM dithiothreitol, 10 mM ATP, and 4 mM phenylmethylsulfonyl fluoride. Wild-type red-fleshed calli were treated with buffer, and the extract was incubated with the MdIAA13-His fusion protein for specific periods. The relative MdIAA121 abundance was detected by western blot using anti-His monoclonal antibodies.
Results
Anthocyanin content and related gene expression under different auxin concentrations
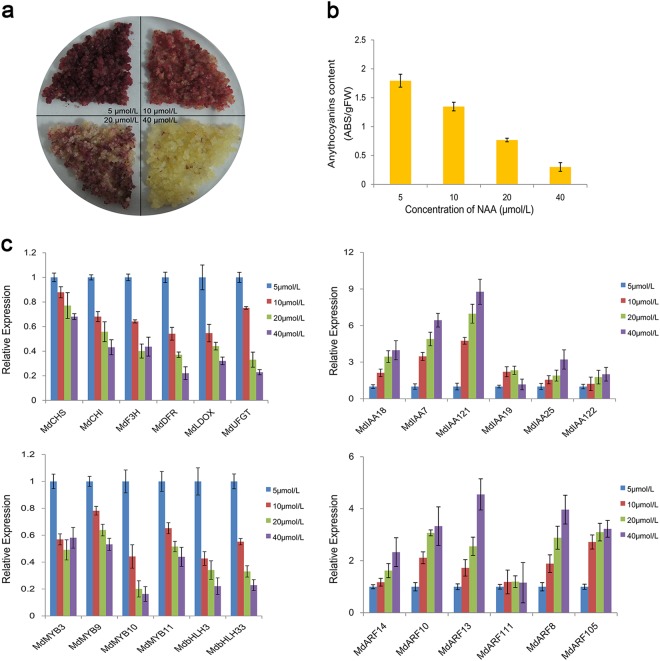
The anthocyanin contents of red-fleshed apple calli gradually decreased with increasing NAA concentrations (5–40 μmol/L), which caused the callus color to fade (Fig. 1a). Moreover, the anthocyanin content of calli cultured in 5 μmol/L NAA was 1.39-, 2.42-, and 5.98-fold higher than that of calli grown in 10, 20, and 40 μmol/L NAA, respectively (Fig. 1b). These results are consistent with the findings of Ji et al.26,27 and implied that low auxin concentrations were conducive to anthocyanin accumulation in red-fleshed apple calli.
Fig. 1. Red-fleshed apple calli on medium supplemented with different NAA concentrations (5, 10, 20, and 40 μmol/L).
a Phenotypes of calli treated with different NAA concentrations, as well as the extraction of anthocyanin. b Relative anthocyanin content at 18 days. The relative anthocyanin content was calculated as follows: absorbance (530 nm)/fresh weight (g). c Relative expression levels of anthocyanin biosynthesis-related genes in response to different NAA treatments at 18 days
The changes in the anthocyanin content of calli grown under different auxin conditions were also reflected at the transcript level. As the auxin concentration increased, the expression levels of anthocyanin biosynthesis-related genes decreased by varying degrees (Fig. 1c). However, the transcript levels for the Aux/IAA and ARF genes increased with increasing NAA concentrations (Fig. 1c). In particular, the MdARF13 expression levels in calli treated with 40 μmol/L NAA were 2–5-fold higher than in calli exposed to the other NAA concentrations. This finding suggested that MdARF13 may repress anthocyanin biosynthesis.
Bioinformatics analysis of the nuclear protein MdARF13
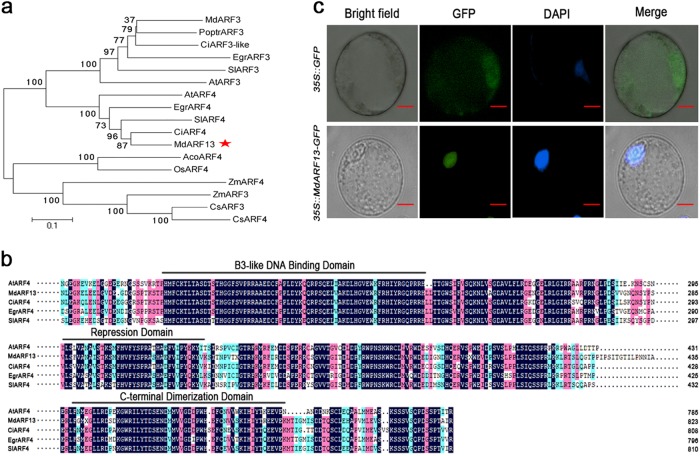
Extensive bioinformatics and biochemical analyses of plants have clarified the number of ARF genes and their potential functions. In A. thaliana, experiments have revealed that AtARF4 contains an active RD and functions as a repressor in plant protoplast transfection assays35. A phylogenetic analysis to elucidate the relationship between MdARF13 and ARF proteins from other plant species indicated that MdARF13 and AtARF4 belong to the same clade (Fig. 2a). Additionally, the aligned protein sequences revealed that MdARF13 and ARF proteins from other plant species contain a conserved B3-like DBD, an RD, and a domain homologous to the Aux/IAA proteins (CTD) (Fig. 2b).
Fig. 2. Bioinformatic analyses and MdARF13 subcellular localization.
a Phylogenetic analysis of ARF transcription factors in different species. b Alignment of MdARF13, AtARF4, CiARF4, EgrARF4, and SiARF4 proteins. c Subcellular localization of the MdARF13-GFP fusion protein in transformed apple callus protoplasts. The protoplasts producing GFP alone were used as the control. Bars = 50 μm
The subcellular localization of MdARF13 was studied by introducing a 35S::MdARF13-GFP construct into the protoplasts of red-fleshed apple calli. The transgenic calli carrying the empty vector (35S::GFP) were used as the control. In protoplasts expressing 35S::MdARF13-GFP, the GFP signal was observed only in the nucleus, while the GFP signal was detected throughout the control protoplasts (Fig. 2c), implying that MdARF13 is localized to the nucleus.
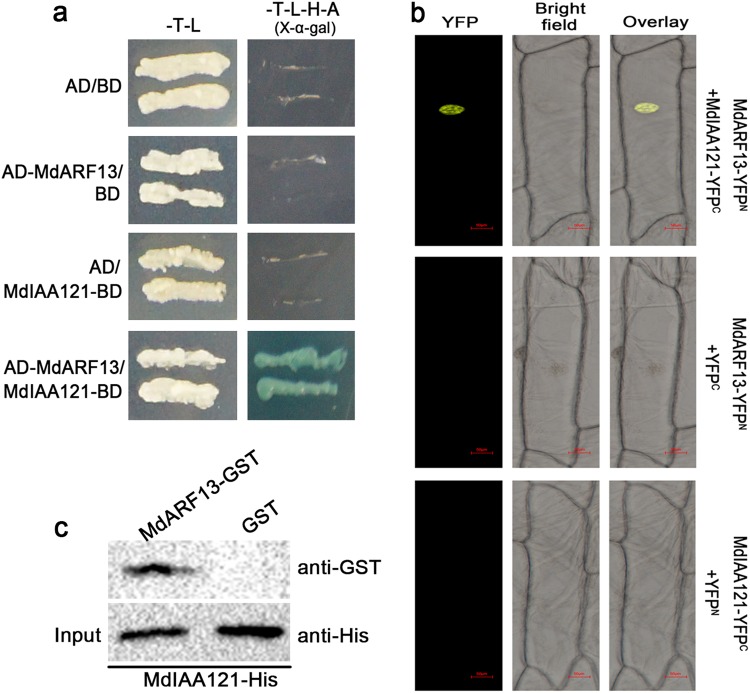
MdARF13 interacts with MdIAA121
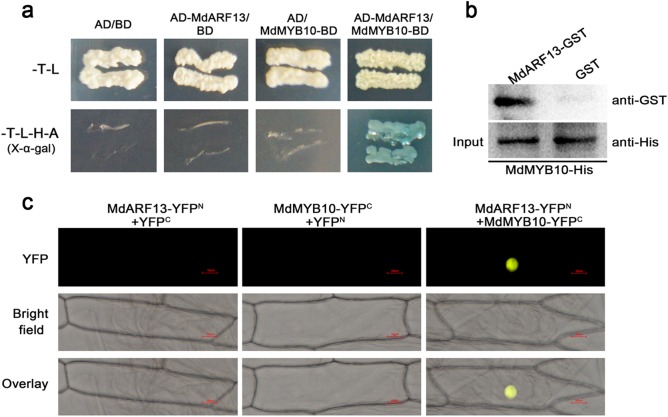
The ARFs contain a dimerization domain responsible for the formation of heterodimers with Aux/IAA proteins31. The Y2H assays, which were used to test whether MdARF13 interacts with Aux/IAA proteins, revealed that MdARF13 interacts only with MdIAA121 (Fig. 3a). The BiFC assay involving onion epidermal cells cotransformed with MdARF13-NYFP and MdIAA121-CYFP constructs confirmed that MdARF13 interacts with MdIAA121 (Fig. 3b). The nuclei of these onion epidermal cells produced a strong YFP signal according to laser scanning confocal microscopy. In contrast, the YFP signal was not detected in cells in which MdIAA121-CYFP was replaced with CYFP alone. Meanwhile, our pull-down assays revealed that the recombinant MdIAA121-HIS fusion protein could be purified with MdARF13-GST, but not with GST alone (Fig. 3c). These results were consistent with those of the Y2H and BiFC assays. Our data suggested that MdARF13 can interact with MdIAA121 under in vivo and in vitro conditions.
Fig. 3. MdARF13 is targeted by MdIAA121.
The interaction between MdARF13 and MdIAA121 was verified in a a yeast two-hybrid assay, b a bimolecular fluorescence complementation assay, and c a pull-down assay
Auxin affects MdIAA121 stability
Ubiquitination, which represents an important posttranslational modification, is a key regulator of various processes, such as hormone signaling, responses to light and sugars, plant development, and plant immunity. In the Aux/IAA–ARF signaling pathway, auxin treatments promote the degradation of Aux/IAA by the 26S proteasome to release ARF proteins46. We conducted protein degradation assays to study the posttranslational regulation of MdIAA121 in vitro. The MdIAA121-His proteins were rapidly degraded after NAA was added (Fig. 4a). Furthermore, samples were treated with MG132 and dimethyl sulfoxide (DMSO). The MG132 treatment noticeably weakened the effect of auxin on MdIAA121 stability (Fig. 4b). These findings indicated that auxin induces the degradation of MdIAA121 via the 26S proteasome.
Fig. 4. MdIAA13 is degraded by the 26S proteasome in the auxin signaling pathway.
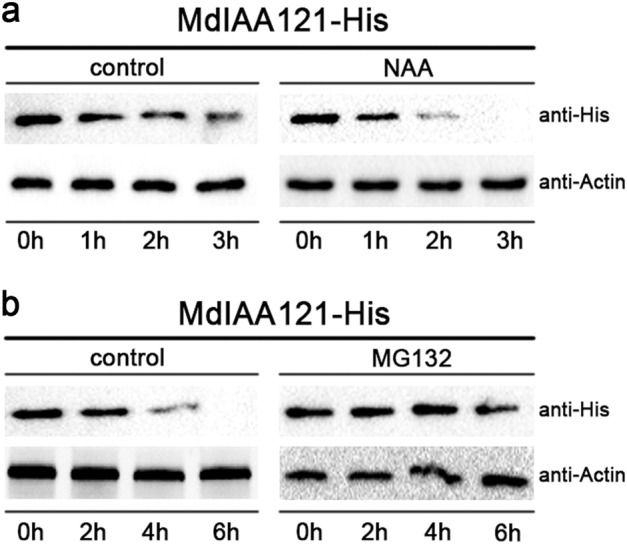
a A wild-type callus was treated with DMSO or 40 mmol NAA. Callus extracts were treated with the MdIAA121-His fusion protein and then incubated for the indicated periods. Actin was used as a loading control. b MG132 inhibited the degradation of MdIAA121. A wild-type apple callus extract was treated with DMSO or 100 mM MG132 for 0.5 h and then incubated with MdIAA121-His for the indicated periods
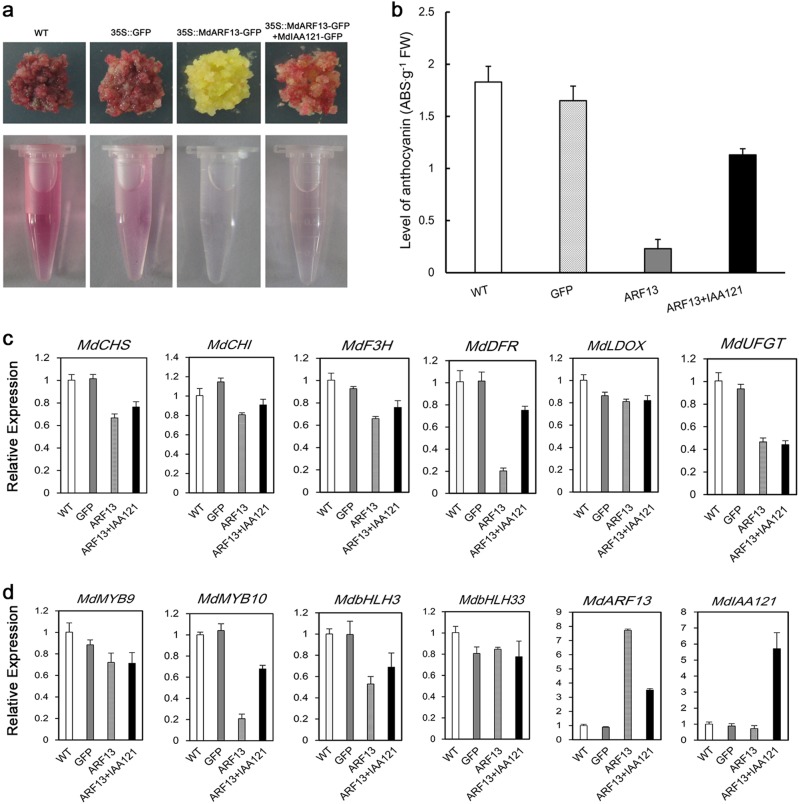
Overexpression of MdARF13 repressed anthocyanin accumulation in the apple callus
To examine whether MdARF13 affects anthocyanin accumulation, MdARF13 was overexpressed in red-fleshed apple calli. The color of the resulting transgenic calli changed from dark red to yellow, while the transcript levels of anthocyanin biosynthesis-related genes and the accumulation of anthocyanin decreased (Fig. 5). These observations implied that MdARF13 represses anthocyanin biosynthesis. To assess whether MdIAA121 inhibits MdARF13 activities, we generated calli cotransfected with MdARF13+MdIAA121 by A. tumefaciens-mediated transformation. The expression levels of anthocyanin structural genes and anthocyanin content were obviously higher in cotransfected calli than in calli overexpressing MdARF13 (Fig. 5). These results demonstrated that MdARF13 together with MdIAA121 mediate auxin signaling to regulate anthocyanin biosynthesis.
Fig. 5. Analysis of transgenic red-fleshed apple calli.
WT wild-type red-fleshed callus, 35S::GFP transgenic callus carrying the empty vector, 35S::MdARF13-GFP red-fleshed apple callus overexpressing MdARF13-GFP, 35S::MdARF13-GFP+MdIAA121-GFP red-fleshed apple callus overexpressing MdARF13-GFP and MdIAA121-GFP. a Appearance of the wild-type and transgenic red-fleshed apple calli. b Anthocyanin levels in wild-type and transgenic calli. c, d Expression of anthocyanin biosynthesis-related structural and transcription factor genes in wild-type and transgenic calli
MdARF13 interacts with MdMYB10
The MYB TFs are crucial for anthocyanin biosynthesis, and the expression of the corresponding genes is regulated by the external environment. In this study, we hypothesized that MdARF13 is responsive to auxin and affects MYB expression. To verify this hypothesis, we completed Y2H assays to investigate whether MdARF13 interacts with MdMYB9 and MdMYB10. We observed that MdARF13 interacts only with MdMYB10 (Fig. 6a). The physical interaction between MdARF13 and MdMYB10 was further confirmed in pull-down and BiFC assays (Fig. 6b, c). Thus, we speculated that interactions between MdARF13 and MdMYB10 affect anthocyanin biosynthesis.
Fig. 6. Interaction between MdARF13 and MdMYB10.
The interaction between MdARF13 and MdMYB10 was verified in a yeast two-hybrid, b pull-down, and c bimolecular fluorescence complementation assays
MdARF13 binds to the MdDFR promoter
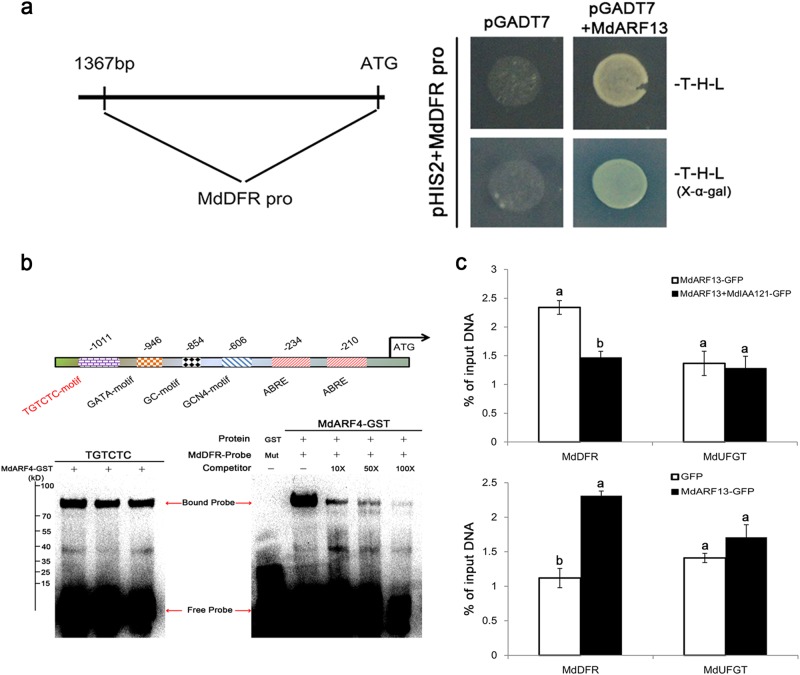
ARF TFs regulate their downstream target genes by binding to the TGTCTC AuxRE in the promoter region and subsequently promoting or repressing expression29. We analyzed the promoter elements of the anthocyanin biosynthesis structural genes (MdCHS, MdCHI, MdDFR, MdLDOX, MdFLS, and MdUFGT) using the PlantCARE cis-acting regulatory element database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) and determined that only MdDFR contains a putative AuxRE in its promoter region (Fig. 7b). We conducted an EMSA to clarify whether MdARF13 is recruited to the MdDFR promoter region. The assay results indicated that MdARF13 could interact with DNA probes containing the TGTCTC AuxRE (Fig. 7b). Moreover, Y1H assays confirmed that MdARF13 could interact with the MdDFR promoter (Fig. 7a), suggesting that MdARF13 suppresses the expression of MdDFR to decrease anthocyanin accumulation. Additionally, the ChIP–qPCR assay results suggested that the MdARF13-GFP fusion protein could bind to the DFR promoter, while the overexpression of MdIAA121 in MdARF13-overexpressing calli clearly inhibited the recruitment of MdARF13 to the MdDFR promoter (Fig. 7c).
Fig. 7. MdARF13 binds directly to the MdDFR promoter.
a The 1427-bp MdDFR promoter sequence was analyzed. MdARF13 interacted with MdDFR promoter fragments in a Y1H assay. b Schematic diagram of the MdDFR promoter with an ARF-binding site (AuxRE). The EMSA results indicated that the MdARF13-GST fusion protein could bind directly to the AuxRE in the MdDFR promoter. Biotin-labeled probes were incubated with the MdARF13-GST fusion protein, and the free and bound DNA fragments were separated in an acrylamide gel. Unlabeled probes were used as competitors. c ChIP–qPCR data revealed that MdARF13 interacts with the MdDFR promoter in vivo, while MdIAA121 inhibits this interaction. The ChIP signal was quantified by qPCR as the percentage of the total input DNA. An actin gene was used as an internal control
Discussion
Auxin has long been recognized for its essential role in plant growth and development. Previous studies concluded that auxin signaling regulates anthocyanin biosynthesis in plant calli, which is associated with changes in the external auxin concentration20,26. In separate studies, Ji et al.26 and Liu et al.47 observed that increasing auxin concentrations within a certain range can inhibit anthocyanin biosynthesis in apple and A. thaliana. In this study, we observed that increasing NAA concentrations increased the inhibition of anthocyanin accumulation. Deikman and Hammer48 reported that the application of exogenous auxins suppressed the expression of anthocyanin regulatory and structural genes to varying degrees. Similarly, we observed that the expression levels of most structural and regulatory genes decreased in calli treated with NAA. Furthermore, the expression of genes encoding two types of TFs (Aux/IAA and ARF) associated with auxin signaling also increased to varying degrees, especially MdARF13. Thus, we speculate that these auxin signaling factors help regulate the expression of genes related to the anthocyanin biosynthesis pathway.
The Aux/IAA and ARF TFs are an integral part of the complex and elaborate auxin signaling pathway. Genome-wide analyses of ARF families have been completed in many plant species, including rice, tomato, and maize49–51. In A. thaliana, in addition to ARF3, 13, 17, and 23, the ARF family mainly comprises proteins with the following three parts: an N-terminal DBD, a middle region that functions as an AD or RD, and a CTD29. In this study, we determined that MdARF13 is homologous to AtARF4 and has a similar structure. ARFs exist in the nucleus for a relatively short period29. Consistent with this previous report, we observed that MdARF13 is a nuclear protein. Most Aux/IAAs function as repressors and are believed to dimerize with ARF activators via their CTDs. However, the Aux/IAA repressors are rapidly degraded as auxin concentrations increase, thereby impairing their inhibitory effect on early auxin-responsive genes35. Our Y2H, BiFC, and pull-down assays revealed that MdARF13 can interact with MdIAA121. Additionally, exogenous auxin treatment induces the degradation of MdIAA121 by the 26S proteasome, suggesting that MdIAA121 might serve as a labile repressor of MdARF13. However, future studies will need to identify the specific ubiquitination mechanisms associated with MdIAA121 degradation.
To further investigate the functions of MdARF13 during anthocyanin biosynthesis, MdARF13 was overexpressed in red-fleshed apple calli. A previous study involving transfected protoplasts indicated that AtARF4 functions as a transcriptional repressor35. In the current study, qPCR data revealed that the transcript levels of auxin-regulated anthocyanin biosynthesis-related genes were down-regulated more in the calli with the 35S::MdARF13-GFP construct than in the wild-type control. Thus, MdARF13 is likely an inhibitor of anthocyanin biosynthesis in apple. Additionally, Aux/IAA can function as a repressor that dimerizes with the target ARF52. We observed that overexpressing MdIAA121 in calli overexpressing MdARF13 weakened the inhibitory effect of MdARF13 on anthocyanin biosynthesis, suggesting that MdARF13 may be targeted by MdIAA121. The interaction between these two proteins may attenuate the inhibitory effect of MdARF13 on anthocyanin biosynthesis.
The MYB TFs are crucial for regulating anthocyanin biosynthesis in apples. External environmental factors affect anthocyanin accumulation because the associated signaling elements regulate the expression of MYB genes. An earlier study indicated that the regulation of anthocyanin biosynthesis by MdMYB1 is responsive to light, with COP1 functioning as a negative regulator that degrades MdMYB1 in darkness53. The HY5 TF targets the MYBD promoter in cytokinin-induced signaling pathways, leading to the accumulation of anthocyanin54. In this study, Y2H, BiFC, and pull-down assays verified the in vitro and in vivo interaction between MdARF13 and MdMYB10, suggesting that MdARF13 might repress anthocyanin accumulation by down-regulating MdMYB10 expression. Moreover, ARF TFs contain DNA-binding motifs and play key roles in the regulation of downstream genes through TGTCTC sequence-specific interactions with promoters30. We detected an MdARF13-binding site upstream of the MdDFR gene (Fig. 7a). EMSA, ChIP, and Y1H assays were conducted to confirm the specific binding of MdARF13 to the MdDFR promoter. We also proved that MdIAA121 functions as a repressor of the auxin signal and interacts with MdARF13 to inhibit the recruitment of MdARF13 to the MdDFR promoter. Therefore, in apple, exogenous auxins repress anthocyanin biosynthesis most likely through the MdIAA121–MdARF13 signal transduction pathway.
ARF TFs are important for auxin signaling, and are involved in many processes related to plant growth and development. In this study, we observed that the inhibition of anthocyanin biosynthesis by high auxin concentrations involves MdARF13 (Fig. 8). Clarifying the role of auxin in the regulation of anthocyanin biosynthesis by ARFs may provide new insights into the regulation of anthocyanin metabolism by other hormones. During the cultivation of fruit trees, the rational application of hormones based on different environmental conditions can improve the appearance and nutritional value of fruit crops. Herein, we discussed the involvement of ARFs in the anthocyanin biosynthesis pathway, which may have implications for the development of new cultivation techniques aimed at improving fruit coloration under diverse environmental conditions.
Fig. 8.
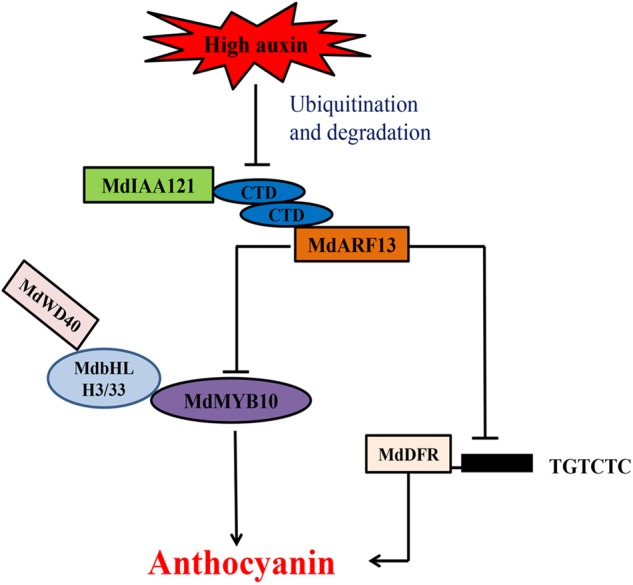
Model of the effects of auxins on anthocyanin accumulation via the Aux/IAA–ARF signaling pathway
Electronic supplementary material
Acknowledgements
This work was supported by grants from the National Key Research Project (2016YFC0501505) and the National Natural Science Foundation of China (31572091 and 31730080).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41438-018-0068-4).
References
- 1.King MC, Cliff MA. Development of a model for prediction of consumer liking from visual attributes of new and established apple cultivars. J. Am. Pom. Soc. 2002;56:223–229. [Google Scholar]
- 2.Horbowicz M, Kosson R, Grzesiuk A, Debski H. Anthocyanins of fruits and vegetables-their occurrence, analysis and role in human nutrition. Veg. Crops Res. Bull. 2008;68:5–22. [Google Scholar]
- 3.Mol J, Grotewold E, Koes R. How genes paint flowers and seeds. Trends Plant Sci. 1998;3:212–217. doi: 10.1016/S1360-1385(98)01242-4. [DOI] [Google Scholar]
- 4.Knekt P, et al. Dietary flavonoids and the risk of lung cancer and other malignant neoplasms. Am. J. Epidemiol. 1997;146:223–230. doi: 10.1093/oxfordjournals.aje.a009257. [DOI] [PubMed] [Google Scholar]
- 5.Middleton JE, Kandaswami C, Theoharides TC. The effects of plant flavonoids on mammalian cells: implications for inflammation, heart disease, and cancer. Pharmacol. Rev. 2000;52:673–751. [PubMed] [Google Scholar]
- 6.Hollman P, Katan M. Absorption, metabolism and health effects of dietary flavonoids in man. Biomed. Pharmacother. 1997;51:305–310. doi: 10.1016/S0753-3322(97)88045-6. [DOI] [PubMed] [Google Scholar]
- 7.Zafra-Stone S, et al. Berry anthocyanins as novel antioxidants in human health and disease prevention. Mol. Nutr. Food Res. 2007;51:675–683. doi: 10.1002/mnfr.200700002. [DOI] [PubMed] [Google Scholar]
- 8.Baudry A, et al. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–380. doi: 10.1111/j.1365-313X.2004.02138.x. [DOI] [PubMed] [Google Scholar]
- 9.Quattrocchio F, et al. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix–loop–helix transcription factors of the anthocyanin pathway. Plant Cell. 2006;18:1274–1291. doi: 10.1105/tpc.105.034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2394. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cone KC, Cocciolone SM, Burr FA, Burr B. Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell. 1993;5:1795–1805. doi: 10.1105/tpc.5.12.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aharoni A, et al. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 2001;28:319–332. doi: 10.1046/j.1365-313X.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- 13.Deluc L, et al. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008;147:2041–2053. doi: 10.1104/pp.108.118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mcghie TK, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in rosaceae. BMC Plant Biol. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takos AM, et al. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ban Y, et al. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007;48:958–970. doi: 10.1093/pcp/pcm066. [DOI] [PubMed] [Google Scholar]
- 17.Espley RV, et al. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–427. doi: 10.1111/j.1365-313X.2006.02964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cominelli E, et al. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J. Plant Physiol. 2008;165:886–894. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Rowan DD, et al. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009;182:102–115. doi: 10.1111/j.1469-8137.2008.02737.x. [DOI] [PubMed] [Google Scholar]
- 20.Ozeki Y, Komamine A. Effects of growth regulators on the induction of anthocyanin synthesis in carrot suspension cultures. Plant Cell Physiol. 1986;27:1361–1368. doi: 10.1093/oxfordjournals.pcp.a077234. [DOI] [Google Scholar]
- 21.Qi T, et al. The Jasmonate-ZIM-Domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23:1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer HJ, Staden J. The in vitro production of an anthocyanin from callus cultures of Oxalis linearis. Plant Cell Tissue Organ Cult. 1995;40:55–58. doi: 10.1007/BF00041119. [DOI] [Google Scholar]
- 23.Ozeki Y, Komamine A. Effects of inoculum density, zeatin and sucrose on anthocyanin accumulation in a carrot suspension culture. Plant Cell Tissue Organ Cult. 1985;5:45–53. doi: 10.1007/BF00033568. [DOI] [Google Scholar]
- 24.Zhou LL, Zeng HN, Shi MZ, Xie DY. Development of tobacco callus cultures over expressing Arabidopsis PAP1/MYB75 transcription factor and characterization of anthocyanin biosynthesis. Planta. 2008;229:37–51. doi: 10.1007/s00425-008-0809-y. [DOI] [PubMed] [Google Scholar]
- 25.Murthy HN, Lee EJ, Paek KY. Production of secondary metabolites from cell and organ cultures: strategies and approaches for biomass improvement and metabolite accumulation. Plant Cell Tissue Organ Cult. 2004;118:1–16. doi: 10.1007/s11240-014-0467-7. [DOI] [Google Scholar]
- 26.Ji XH, et al. Effect of auxin, cytokinin and nitrogen on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii f. niedzwetzkyana) Plant Cell Tissue Organ Cult. 2015;120:325–337. doi: 10.1007/s11240-014-0609-y. [DOI] [Google Scholar]
- 27.Ji XH, Zhang R, Wang N, Yang L, Chen XS. Transcriptome profiling reveals auxin suppressed anthocyanin biosynthesis in red-fleshed apple callus (Malus sieversii f. niedzwetzkyana) Plant Cell Tissue Organ Cult. 2015;123:389–404. doi: 10.1007/s11240-015-0843-y. [DOI] [Google Scholar]
- 28.Guilfoyle TJ, Ulmasov T, Hagen G. The ARF family of transcription factors and their role in plant hormone-responsive transcription. Cell. Mol. Life Sci. 1998;54:619–627. doi: 10.1007/s000180050190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulmasov T, Hagen G, Guilfoyle TJ. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.13.12.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guilfoyle TJ, Hagen G. Auxin response factors. J. Plant Growth Regul. 2001;10:281–291. doi: 10.1007/s003440010026. [DOI] [Google Scholar]
- 32.Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002;49:387–400. doi: 10.1023/A:1015255030047. [DOI] [PubMed] [Google Scholar]
- 33.Benfey PN. Auxin action: slogging out of the swamp. Curr. Biol. 2002;12:R389–R390. doi: 10.1016/S0960-9822(02)00881-3. [DOI] [PubMed] [Google Scholar]
- 34.Hellmann H, Estelle M. Plant development: regulation by protein degradation. Science. 2002;297:793–797. doi: 10.1126/science.1072831. [DOI] [PubMed] [Google Scholar]
- 35.Tiwari SB, Hagen G, Guilfoyle T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell. 2003;15:533–543. doi: 10.1105/tpc.008417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sessions A, et al. ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development. 1997;124:4481–4491. doi: 10.1242/dev.124.22.4481. [DOI] [PubMed] [Google Scholar]
- 37.Pekker I, Alvarez JP, Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell. 2005;17:2899–2910. doi: 10.1105/tpc.105.034876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hardtke CS, et al. Overlapping and nonredundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- 39.Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tian CE, et al. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004;40:333–343. doi: 10.1111/j.1365-313X.2004.02220.x. [DOI] [PubMed] [Google Scholar]
- 41.Vriezen WH, Feron R, Maretto F, Keijman J, Mariani C. Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 2008;177:60–76. doi: 10.1111/j.1469-8137.2007.02254.x. [DOI] [PubMed] [Google Scholar]
- 42.De JM, Woltersarts M, Feron R, Mariani C, Vriezen WH. The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 2009;57:160–170. doi: 10.1111/j.1365-313X.2008.03671.x. [DOI] [PubMed] [Google Scholar]
- 43.Sun J, et al. Effects of methyl jasmonate and abscisic acid on anthocyanin biosynthesis in callus cultures of red-fleshed apple (Malus sieversii, f. niedzwetzkyana) Plant Cell Tissue Organ Cult. 2017;130:227–237. doi: 10.1007/s11240-017-1217-4. [DOI] [Google Scholar]
- 44.An XH, et al. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015;56:650–662. doi: 10.1093/pcp/pcu205. [DOI] [PubMed] [Google Scholar]
- 45.Hu DG, et al. MdMYB1 regulates anthocyanin and malate accumulation by directly facilitating their transport into vacuoles in apples. Plant Physiol. 2016;170:1315–1330. doi: 10.1104/pp.15.01327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 47.Liu Z, Shi MZ, Xie DY. Regulation of anthocyanin biosynthesis in arabidopsis thaliana red pap1-d cells metabolically programmed by auxins. Planta. 2014;239:765–781. doi: 10.1007/s00425-013-2011-0. [DOI] [PubMed] [Google Scholar]
- 48.Deikman J, Hammer PE. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiol. 1995;108:47–57. doi: 10.1104/pp.108.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang D, et al. Genomewide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa) Gene. 2007;394:13–24. doi: 10.1016/j.gene.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 50.Kumar R, Tyagi AK, Sharma AK. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genomics. 2011;285:245–260. doi: 10.1007/s00438-011-0602-7. [DOI] [PubMed] [Google Scholar]
- 51.Xing H, et al. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics. 2011;12:178. doi: 10.1186/1471-2164-12-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiwari SB, Hagen G, Guilfoyle TJ. Aux/iaa proteins contain a potent transcriptional repression domain. Plant Cell. 2004;16:533–543. doi: 10.1105/tpc.017384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li YY, et al. Mdcop1 ubiquitin e3 ligases interact with mdmyb1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012;160:1011–1022. doi: 10.1104/pp.112.199703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen NH, et al. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 2015;84:1192–1205. doi: 10.1111/tpj.13077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.