“Memories warm you up from the inside. But they also tear you apart.” ― Haruki Murakami, Kafka on the Shore.
One of the characteristics of the human immune system, in addition to the recognition of nonself pathogens and the tolerance of self-antigens, is its memory response to previously encountered pathogens and the mounting of a stronger, faster secondary response against these same antigens. A long-held paradigm in immunologic study in vertebrates is that immune memory is restricted to the adaptive immune response via memory T and B cells, which possess two distinct features: specificity (recognizing pathogen antigen fragments through surface receptors) and longevity (proliferation and long-term survival after the first encounter).1–3 Unlike adaptive immunity, the innate immune system exhibits a rapid response against pathogens and transforming cells without a previous encounter, and this response depends on a series of default genes and proteins expressed on cell surfaces and in the cytoplasm that recognize the pathogen-associated molecular patterns (PAMPs) of pathogens. Therefore, it was surprising to find several models of pathogen infections in invertebrates, including insects, worms, and jawless fish, that lack fully developed adaptive immunity but possess the features of antigen specificity and longevity likely involving phagocytic cells that can “recall” prior infection with limited specificity. In fact, immune memory has recently been described in vertebrate myeloid lineages,4 in which epigenetic changes are proposed to occur in macrophages previously stimulated through pattern recognition receptors.5
Although derived from a common lymphoid precursor cell shared with B and T cells, NK cells lack somatically rearranged antigen receptors and instead rely on signals from germ line-encoded surface receptors for activation,6 and NK cells are considered to be part of the innate immune system. Since their discovery, abundant evidence has highlighted the importance of NK cells in the host defense against infections and tumors7–10 and in modulating adaptive immune responses through both direct interactions with T cells and through indirect mechanisms, such as induction of dendritic cell (DC) maturation.11–14 Recently, however, NK cells have been shown to possess traits of adaptive immunity and can acquire immunological memory in a similar manner to T and B cells.15
Many representative studies in mice have demonstrated that certain receptors expressed by NK cells can bind to the surfaces of infected cells with remarkably high affinity. During mouse cytomegalovirus (MCMV) infection in C57BL/6 mice, naïve NK cells that express the activating receptor Ly49H recognize the virally encoded glycoprotein m157 on the surface of the MCMV-infected host cells, resulting in robust activation and proliferation of antigen-specific NK cells. This process critically depends on pro-inflammatory IL-12 signaling through STAT4 and Zbtb32 and on costimulatory signaling through the activating receptor DNAM-1. During clonal proliferation, antigen-specific NK cells maintain viability by increasing the expression of miR155 to antagonize the pro-death factors Noxa and SOCS1; however, at the peak of virus-driven expansion, effector NK cells undergo BIM-mediated cell death to form a stable pool of long-lived-memory NK cells via a process that depends on endogenous IL-15. MCMV-elicited memory NK cells respond robustly to a secondary challenge with MCMV but are less responsive to heterologous infection. The detailed mechanisms of NK cell memory formation are reviewed in depth by Sun et al.16 and Peng and Tian.17
Although distinct transcriptional profiles of NK cells in the naïve, early effector, late effector, and memory (day 27 postinfection) stages following MCMV infection have been reported, how transcription is controlled at the epigenetic level in NK cells during these different stages is currently unknown. Furthermore, how chromatin profiling can be used to distinguish the distinct responses of NK cells and canonical adaptive immunity-associated CD8+ T cells to the same viral infection requires exploration.
In a recent study published in Nature Immunology, Lau et al.18 investigated the transcriptional profiles controlled at the epigenetic level in NK cells as they transition between the naïve, effector, and memory states. They performed parallel chromatin accessibility analysis via a transposase-accessible chromatin assay using high-throughput sequencing (ATAC-seq)19 and transcriptional profiling by RNA-seq on Ly49H+ NK cells during MCMV infection to investigate the role of chromatin modification at the epigenetic level in the determination of their transcriptional fate. In the first week of virus infection, regions near or within the Socs3, Cish, Pdcd1, Dnmt3a, and Il10 gene loci were among the top 10% of the most modulated regions. Most of the variable regions found near Tbx21, Klrg1, Ifng, and Zbtb32 showed transient changes in chromatin accessibility early during infection but returned to baseline or near baseline levels in the memory cells. A majority of these pathways involve genes related to the interleukin 2 (IL-2)-mediated and IL-12-mediated signaling pathways, the Jak–STAT signaling pathway, and the T cell antigen receptor signaling pathway.
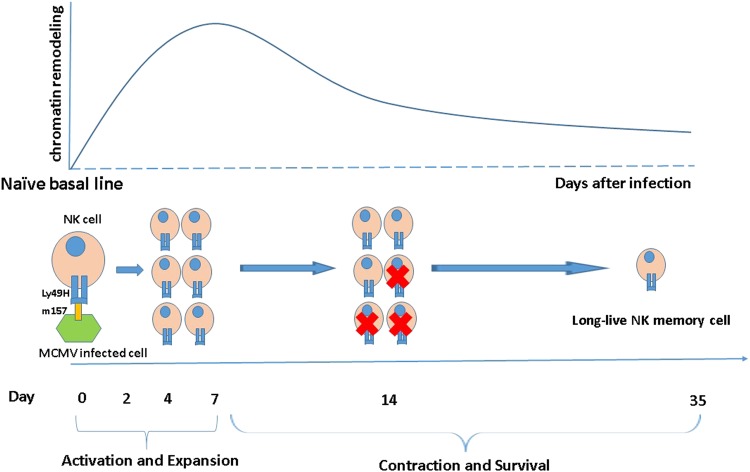
Interestingly, comparison of the chromatin accessibility and mRNA expression profiles showed that both profiles displayed similar patterns 1 week after infection, but that these transitions diverged during later infection stages (day 7 to day 35). As shown in Fig. 1, the majority of the chromatin remodeling appeared to be completed by days 14 and day 35, suggesting that an epigenetic-level effect on memory cell fate may occur far earlier than transcriptional alterations during lymphocyte differentiation. As in the generation of CD8+ T cell memory, NK cells transitioning from day 14 to day 35 continued to undergo gene expression changes, and the most strongly correlated pathways at the earliest time points after infection were type I and type II interferon (IFN) signaling.
Fig. 1.
Chromatin remodeling (alterations in the differentially accessible regions) in the process of NK cell transitioning from naïve to long-lived stages. A majority of these epigenetic changes occurred by day14 after infection and minimal changes continued to occur as the differentiating NK cells transitioned between days 14 and 35. While many of these changing regions returned to the naïve levels, a sizable portion of these regions (at least~30%) remained at a substantially altered state of accessibility. The greatest global changes occurred during the first week of viral infection and relatively little epigenetic modulation occurred between days 14 and 35
Lau and coworkers hypothesized that IL-12 and type-I IFN signaling through STAT4 and STAT1, respectively, may affect the epigenetic landscape in NK cells soon after infection.18 This analysis yielded 162 differentially expressed genes with a total of 207 peak regions near the loci that were simultaneously occupied by STAT4 and DA (differentially accessible) upon STAT4 deletion. In contrast, a similar STAT1 analysis revealed that only 28 genes with regions that were occupied by both STAT1 and DA were commonly regulated during infection. STAT4 and STAT1 promote chromatin remodeling through very different mechanisms, and proinflammatory cytokine-induced STAT occupancy may play a direct role in affecting both local and distal changes in chromatin accessibility to promote transcription.
The authors performed pairwise analysis of memory and naïve NK cells and used transcriptomic data to filter these regions to those that were assigned to any genes that showed significant changes in expression at any sequential transition time point during infection to enrich for the most relevant functional processes and active regions.18 Among the more accessible regions, the most enriched pathway was NK cell-mediated cytotoxicity, which increased chromatin accessibility in memory cells and was correlated with increased transcription. Among the less accessible regions, downregulation of accessible regions within gene bodies in memory cells was correlated with decreased transcript abundance. Furthermore, TCF–LEF-associated and NF-κ B-associated regions showed coordinated decreases in accessibility, and Tcf7 and Lef1 actually showed the highest expression levels among the common TCF–LEF proteins in naïve cells. Both Tcf7 and Nfkb1 showed decreased expression levels during the memory stage, consistent with the decreased accessibility of their genomic regions. These data suggested that memory NK cells are epigenetically poised for specific signaling pathways and transcription factor binding that may modify their effector function.
Because of the many parallel characteristics of NK cells and CD8+ T cells,6 Lau et al.18 investigated whether these two cytolytic lymphocytes possess any similar epigenetic attributes during their memory formation. They performed RNA-seq and ATAC-seq experiments with naïve, effector, and memory MCMV-specific CD8+ T cells during MCMV infection. The transitions from naïve to effector to memory stages followed similar trajectories in both cell types. PCA and hierarchical clustering of these normalized values revealed that naïve NK cells clustered closer to memory CD8+ T cells than to either naïve or effector CD8+ T cells. Visualization of DA-associated regions defined by CD8+ T cells showed that naïve NK cells generally exhibited higher degrees of accessibility compared with naïve T cells in regions that eventually opened up in the effector and memory stages of CD8+ T cells. The effector NK cells still clustered closest to effector CD8+ T cells, and memory NK cells clustered closest to memory CD8+ T cells, which indicated that there may be a common epigenetic trajectory during the generation of lymphocyte memory. In a more detailed analysis, evidence suggested that NK cells and CD8+ T cells possess shared underlying epigenetic signatures that may be commonly regulated by the AP-1 and TCF–LEF transcription factor families during the generation of immune memory.
As has been said, memory is sometimes sweet and sometimes bitter. However, without it, life has no meaning at all. Lau and coworkers expanded our current understanding of the molecular mechanisms underlying the immunological memory of NK cells in terms of the importance of functional and evolutional conservation, and they may have unveiled novel models for the design of targeted vaccines against infectious diseases and/or tumors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Lawrence Shih-Hsin Wu, Email: shwu@mail.cmu.edu.tw.
Jiu-Yao Wang, Email: a122@mail.ncku.edu.tw.
References
- 1.Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+T cells during an acute virus infection. Immunity. 1998;8:167–175. doi: 10.1016/S1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murali-Krishna K, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998;8:177–187. doi: 10.1016/S1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 3.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu. Rev. Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 4.Netea MG, et al. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saeed S, et al. Epigenetic programming of monocyteto- macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun JC, Lanier LL. NK cell development, homeostasis and function: parallels with CD8+ T cells. Nat. Rev. Immunol. 2011;11:645–657. doi: 10.1038/nri3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat. Immunol. 2016;17:1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 8.Brandstadter JD, Yang Y. Natural killer cell responses to viral infection. J. Innate Immun. 2011;3:274–279. doi: 10.1159/000324176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, et al. Suppressed expression of miR-378 targeting gzmb in NK cells is required to control dengue virus infection. Cell Mol. Immunol. 2016;13:700–708. doi: 10.1038/cmi.2015.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun C, Sun H, Zhang C, Tian Z. NK cell receptor imbalance and NK cell dysfunction in HBV infection and hepatocellular carcinoma. Cell Mol. Immunol. 2015;12:292–302. doi: 10.1038/cmi.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zingoni A, et al. Crosstalk between activated human NK cells and CD4+T cells via OX40-OX40 ligand interactions. J. Immunol. 2004;173:3716–3724. doi: 10.4049/jimmunol.173.6.3716. [DOI] [PubMed] [Google Scholar]
- 12.Zingoni A, Ardolino M, Santoni A, Cerboni C. NKG2D and DNAM-1 activating receptors and their ligands in NK-T cell interactions: role in the NK cell-mediated negative regulation of T cell responses. Front. Immunol. 2012;3:408. doi: 10.3389/fimmu.2012.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moretta A. The dialogue between human natural killer cells and dendritic cells. Curr. Opin. Immunol. 2005;17:306–311. doi: 10.1016/j.coi.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 14.Crouse J, Xu HC, Lang PA, Oxenius A. NK cells regulating T cell responses: mechanisms and outcome. Trends Immunol. 2015;36:49–58. doi: 10.1016/j.it.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Cerwenka A, Lanier LL. Natural killer cell memory in infection, inflammation and cancer. Nat. Rev. Immunol. 2016;16:112–123. doi: 10.1038/nri.2015.9. [DOI] [PubMed] [Google Scholar]
- 16.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng H, Tian Z. Natural killer cell memory: progress and implications. Front. Immunol. 2017;8:1143. doi: 10.3389/fimmu.2017.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau CM, et al. Epigenetic control of innate and adaptive immune memory. Nat. Immunol. 2018;19:963–972. doi: 10.1038/s41590-018-0176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]