Abstract
In this study, we investigated the expression of seven MYB transcription factors (a total of 17 genes that included Dof1.1, IQD1-1, MYB28, MYB29, MYB34, MYB51, and MYB122 and their isoforms) involved in aliphatic and indolic glucosinolate (GSL) biosynthesis and analyzed the aliphatic and indolic GSL content in different organs of Chinese cabbage (Brassica rapassp. Pekinensis). MYB28 and MYB29 expression in the stem was dramatically different when compared with the levels in the other organs. MYB34, MYB122, MYB51, Dof1.1, and IQD1-1 showed very low transcript levels among different organs. HPLC analysis showed that the glucosinolates (GSLs) consisted of five aliphatic GSLs (progoitrin, sinigrin, glucoalyssin, gluconapin, and glucobrassicanapin) and four indolic GSLs (4-hydroxyglucobrassicin, glucobrassicin, 4-methoxygluco-brassicin, and neoglucobrassicin). Aliphatic GSLs exhibited 63.3% of the total GSLs content, followed by aromatic GSL (19.0%), indolic GSLs (10%), and unknown GSLs (7.7%) in different organs of Chinese cabbage. The total GSL content of different parts (ranked in descending order) was as follows: seed > flower > young leaves > stem > root > old leaves. The relationship between GSLs accumulation and expression of GSLs biosynthesis MYB TFs genes in different organs may be helpful to understand the mechanism of MYB TFs regulating GSL biosynthesis in Chinese cabbage.
Keywords: Chinese cabbage, gene expression, glucosinolate, transcription factor
1. Introduction
Glucosinolates (GSLs), which are β-thioglucoside-N-hydroxysulfates (cis-N-hydroximinosulfate esters), are sulfur-rich anionic secondary metabolites that are derived from a glucose molecule and an amino acid. They occur as secondary metabolites of almost all plants of the order Brassicales (including the families Brassicaceae, Capparidaceae, and Caricaceae), and also in the genus Drypetes (family Euphorbiaceae) [1]. Approximately 200 different GSLs are known to occur naturally in plants [1,2]. GSLs play important roles in plant defense against herbivores and microbes [3] and have received considerable attention because of their anti-carcinogenic, anti-oxidative, and anti-microbial activities [4,5,6]. The amino acids methionine, tryptophan (Trp), and phenylalanine are the most prominent GSL biosynthetic precursors and are involved in the synthesis of aliphatic, indolic, and aromatic GSLs, respectively [3,7]. Indolic GSLs constitute an important group of Trp-derived secondary metabolites in brassicas where they function as defense compounds. Indolic GSLs and other GSLs are regulated by myrosinases, which convert them into biologically active nitrile, isothiocyanate, or thiocyanate forms [8]. During biotic and abiotic stress responses, GSL biosynthesis is regulated by a complex network of transcription factors (TFs) belonging to the R2R3-MYB family [9,10,11,12,13,14,15,16,17]. MYB28, MYB76, and MYB29 control the biosynthesis of high aliphatic GSLs [14,15,16,17], while MYB51, MYB122, and MYB34, alternatively called high indolic GSLs, can be manipulated to coordinately control the suite of enzymes that synthesize indolic GSLs (Figure 1) [9,12,13,18]. Skirycz et al. [12] reported that the DOF transcription factor AtDof1.1 (OBP2), which is inducible by herbivory and methyl jasmonate, is part of a regulatory network controlling GSL biosynthesis in Arabidopsis.
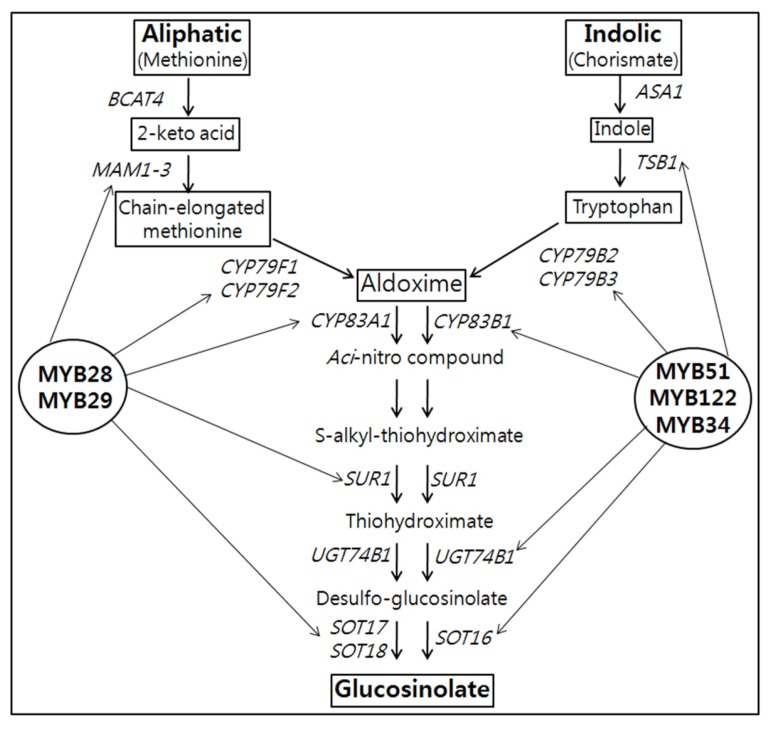
Figure 1.
Schematic representation of the aliphatic and indolic glucosinolate biosynthesis in B. rapa [20]. The key MYB transcription factors involved in aliphatic and indolic glucosinolates are shown in bold. BCAT4, MAM1-3-Side-chain elongation; CYP79F1, CYP79F2, CYP79B2, CYP79B3, CYP83A1, CYP83B1, SUR1, UGT7B1-Core structure formation; ASA1, anthranilate synthase alpha 1; TSB1, tryptophan synthase beta 1.
Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates GSL accumulation and plant defense [10]. The production of GSLs is intimately connected to primary sulfur metabolism. The final step of GSL core synthesis is the sulfation of the desulfo-GSL precursors [19]. Yatusevich et al. [20] showed that at least some of the ATPS (ATP sulfurylase), APK (APS kinase), and APR (APS reductase) genes are directly regulated by the R2R3-MYB TFs involved in the regulation of core GSL biosynthesis. Chinese cabbage (Brassica rapa ssp. pekinensis) belongs to the member of Brassicaceae family and is the most popular type of leafy vegetables widely used in East Asian cuisine. In Korean cuisine, it is the main ingredient of baechu Kimchi, the most common type of Kimchi. Although MYB TFs involved in GSL biosynthesis have been examined [9,10,11,12,13,14,15,16,17], gene expression studies on MYB TFs in different organs of Chinese cabbage have not been reported. In this study, we examined GSL accumulation and expression of R2R3-MYB TFs involved in the regulation of GSLs biosynthesis in different organs of Chinese cabbage.
2. Results and Discussion
2.1. Identification of Transcription Factors Involved in GSL Biosynthesis
Eighteen TFs known to regulate GSL biosynthesis were identified in a Brassica database (http://brassicadb.org/brad/glucoGene.php) [21]. The open reading frame of MYB29-1 had 99% homology with MYB29-2 and was therefore excluded in this study. Wang et al. [21] identified 52 homologous A. thaliana GLS (AtGS) biosynthetic genes using BLASTN and BLASTP, on the basis of the draft B. rapa genome v1.0 and 41,174 annotated genes. Brassica TF genes were found to share almost 72%–92% nucleotide sequence identity with their Arabidopsis orthologs [21] (Table 1).
Table 1.
Orthologs list of transcription factors involved in GSL biosynthesis in Chinese cabbage.
| Group name | AGI | BrID | References |
|---|---|---|---|
| Dof1.1 | At1g07640 | Bra031588 (Dof1.1-1) | Skirycz et al. [12] |
| Bra030696 (Dof1.1-2) | |||
| IQD1-1 | At3g09710 | Bra034081 (IQD1-1-1) | Levy et al. [10] |
| Bra001299 (IQD1-1-2) | |||
| MYB28 | At5g61420 | Bra012961 (MYB28-1) | Gigolashvili et al. [14]; Hirai et al. [16] |
| Bra035929 (MYB28-2) | |||
| Bra029311 (MYB28-3) | |||
| MYB29 | At5g07690 | Bra005949 (MYB29) | Gigolashvili et al. [15]; Hirai et al. [16] |
| MYB34 | At5g60890 | Bra013000 (MYB34-1) | Celenza et al. [9] |
| Bra035954 (MYB28-2) | |||
| Bra029350 (MYB28-3) | |||
| Bra029349 (MYB28-4) | |||
| MYB51 | At1g18570 | Bra025666 (MYB51-1) | Gigolashvili et al. [13] |
| Bra031035 (MYB51-2) | |||
| Bra016553 (MYB51-3) | |||
| MYB122 | At1g74080 | Bra015939 (MYB122-1) | Gigolashvili et al. [13] |
| Bra008131 (MYB122-2) |
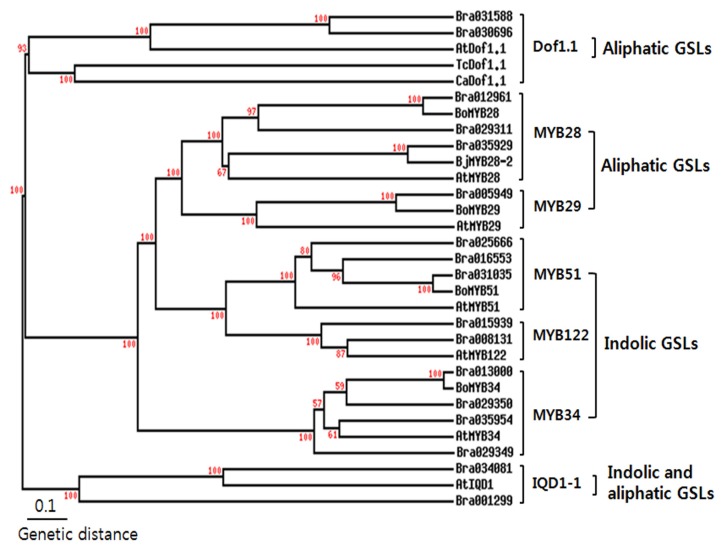
Arabidopsis MYB51/HIG1 (high indolic glucosinolate 1) did not contain a typical nuclear localization signal (NLS) as revealed by PredictNLS [22]; however, an amino acid residue stretch KKRLIKK was detected that might act as a SV40-type NLS [14]. Software prediction of protein subcellular localization showed that all the transcription factors were present in the nucleus, except for Dof1.1-2, which according to the TargetP and ChloroP software programs, was placed in the chloroplasts (data not shown). Precursor amino acid synthesis and side chain elongation are known to take place in the chloroplasts [23,24]. The oxidation reactions of the GLS core pathway, catalyzed by CYP79 and CYP83 enzymes, take place at the ER-cytosol interface, as showed by the targeting of CYP7979F1- and CYPF2-reporter fusion proteins to the ER [25]. A phylogenetic tree was then constructed using the deduced transcription factor amino acid sequences of A. thaliana and B. rapa (Figure 2). Wang et al. [21] reported that each transcription factor of B. rapa showed more than 70% sequence identity when compared to corresponding TFs in A. thaliana. As expected, MYB28 and MYB29-2, which are involved in aliphatic GSL production, were clustered together, whereas MYB51 and MYB122, which are involved in indolic GSL production, were clustered together. IQD1-1 and Dof1.1 were completely separate from the other TFs.
Figure 2.
Phylogenetic tree of MYB transcription factors of B. rapa with other plants. The phylogenetic tree was generated by applying the cluster algorithm using TreeTop Phylogenetic tree. Bootstrap values were resample 100 times. GenBank Accession No. TcDof1.1 (Theobroma cacao, EOY34069), CaDof1.1 (Cicer arietinum, XP_004505871), BjMYB28-2 (Brassica juncea, AFX96282), BoMYB28 (Brassica oleracea, CBI71385), BoMYB29 (Brassica oleracea, BAM78212), BoMYB51 (Brassica oleracea, ACB59198).
2.2. Gene Expression of MYB TFs in Different Organs
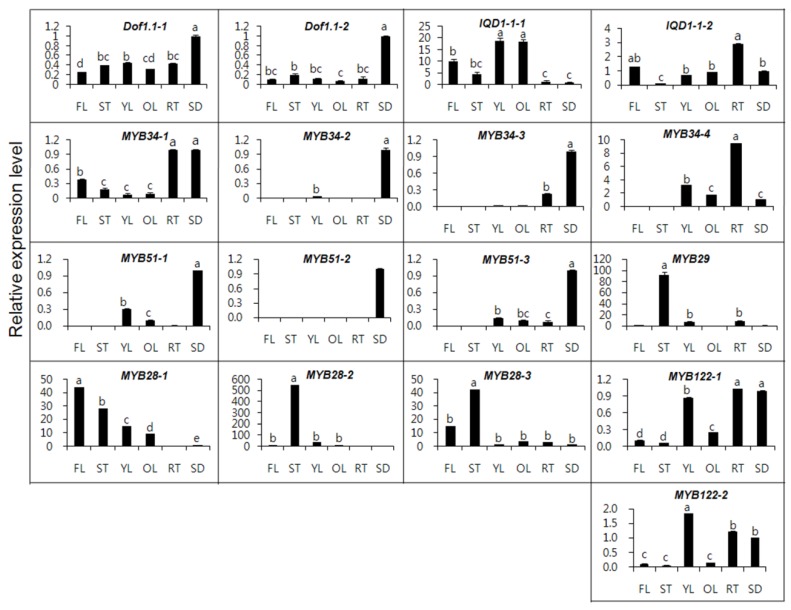
Data regarding the transcription of MYB TFs in different organs are presented in Figure 3. MYB28 and MYB29, involved in aliphatic GSLs showed extremely different patterns of expression among different organs, compared with other TFs. Gigolashvili et al. [14] reported that the R2R3-MYB TF MYB28/HAG1 (high aliphatic glucosinolate 1) represents a key component in the regulation of aliphatic methionine-derived GSL biosynthesis in A. thaliana. In addition, Hirai et al. [16] described that MYB29 probably plays an important role for methyl jasmonate-mediated induction of a set of aliphatic GSL biosynthetic genes in A. thaliana. The transcription of MYB29 in the stems was 46-, 11-, and 92-fold higher, respectively, than that of flowers, young leaves, and old leaves.
Figure 3.
Expression of MYB transcription factors in different Chinese cabbage organs. FL-flower, ST-stem, YL-young leaf, OL-old leaf, RT-root, SD-seed. The longitudinal axis indicates the expression levels of genes relative to that of actin. Each value is the mean of three replicates, and error bars indicate SDs. Mean values indicated by the same letter are not significantly different at least significant differences (LSD) (p = 0.05).
Additionally, the transcript level of MYB28-2 in the stems was also 552-, 184-, and 16-fold higher, respectively, than that of seeds, roots, and young leaves, whereas the transcript level of MYB28-1 in the flowers was 44-, 4.8- and 1.5-fold higher than that of seeds, old leaves, and stems. The highest MYB28 expression level was detected in inflorescences of flowering plants and in old leaves, the sites of accumulation of aliphatic GSLs in Arabidopsis [14]. Gigolashvili et al. [13] identified that HIG1 (high indolic glucosinolate)/MYB51 regulates indolic GSL biosynthesis in A. thaliana and plays a role in biotic stress responses. Arabidopsis HIG1/MYB51 was expressed in roots but not in mature flowers or siliques and MYB51 promoter activity was highest in the vegetative parts of the plants, mainly in mature rosette leaves [13]. In contrast to Arabidopsis, among B. rapa MYB51 isoforms, MYB51-2 was expressed only in seeds, while MYB51-1 and MYB51-3 were expressed in other organs, except for the flowers and stems. In the case of MYB122, MYB122-2 transcription was highest in young leaves, whereas among the MYB34 isoforms, MYB34-4 transcript level in the roots was 9, 4.5 and 3 times higher compared with seeds, old leaves, and young leaves. Transcription of MYB34-2 and MYB34-3 was only observed in seeds, young leaves, and roots, whereas MYB34-1 was expressed in all organs.
In Arabidopsis, MYB34 overexpressing plants exhibited defectiveness in the development of generative organs, the root system, as well as the vegetative biomass [13]. Jin et al. [26] reported that MYB51 and MYB122 could act as activators or repressors, depending on the context, in a similar way like AtMYB4. The expression of GSL biosynthetic genes, that is, CYP79B2, UGT74B1, CYP79F1, CYP79F2, IQD1, and Dof1.1 in A. thaliana often exhibited to be restricted to vascular tissues [10,12,25,27,28]. In spite of the common precursors between IAA and IG biosynthetic pathways, they could be specifically regulated by the different activities of MYB122, MYB34, and MYB51 [13].
Dof1.1-1 and Dof1.1-2 expression showed similar patterns in all organs, except for seeds. IQD1-1, which is involved in both indolic and aliphatic GSLs biosynthesis, showed the highest expression level in young and old leaves. The expression of IQD1-1 in the young leaves was 19-, 13.6-, and 4.2-fold higher, respectively, than in seeds, roots, and stems, whereas IQD1-1-2 transcript level in roots was expressed 29- and 2.9-fold higher, respectively, than in the stems and seeds. Zang et al. [29] described that the expression of B. rapa desulfoglucosinolatesulfotransferases (BrST) isoforms involved in core GSLs biosynthesis in mature leaf and root was higher than other tissues, displaying functional redundancy for differential expression. In this study, MYB28, MYB29, and Dof1.1 isoforms, involved in aliphatic GSLs biosynthesis, exhibited high expression level in the flowers, stems, and seeds, whereas indolic GSLs biosynthesis genes showed high level in a variety of organs. Therefore, we suggest that expression of MYB TFs isoforms involved in GSLs could differ tremendously depending on tissue type in Chinese cabbage.
2.3. GSLs Analysis of Different Organs
Individual GSLs were separated by HPLC and identified based on retention time of HPLC and our GSL database. Moreover, the GSLs were confirmed by LC-ESI-MS in positive mode (Table 2). Organs including flowers, stems, young leaves, old leaves, roots, and seeds of Chinese cabbage were quantified with an external standard, sinigrin, and response factor (Table 3) [2, ISO 1992]. Five aliphatic GSLs (progoitrin, sinigrin, glucoalyssin, gluconapin, and glucobrassicanapin), 2 indolic GSLs (glucobrassicin and 4-methoxyglucobrassicin), and 1 aromatic GSL (gluconasturtiin) were detected in all organs. Gluconapin and sinigrin, from the aliphatic GSLs, showed the highest levels in the seeds (60.13 and 3.58 μmol/g DW, respectively), whereas old leaves had the lowest levels (0.34 and 0.11 μmol/g DW, respectively). This result was consistent with gene expression level of Dof1.1 involving aliphatic GSL biosynthesis. Very recently, Cartea et al. [30] demonstrated that total GSL content ranged from 19 to 37.3 μmol/g DW in early and extra-late groups, respectively, and from 19.5 to 36.3 μmol/g DW for turnips and turnip greens groups, respectively in leaves of B. rapa.
Table 2.
Glucosinolates identified from Chinese cabbage by LC-ESI-MS.
| Trivial name * | Systematic names | Compound groups | [M+H]+ m/z | Response factor [2, ISO 1992] |
|---|---|---|---|---|
| Progoitrin | 4-Methylsulfinylbutyl GSL | Aliphatic | 310 | 1.09 |
| Sinigrin | 2-Propenyl GSL | Aliphatic | 280 | 1.00 |
| Glucoalyssin | 5-Methylsufinylpentyl GSL | Aliphatic | 372 | 1.07 |
| Gluconapin | 3-Butenyl GSL | Aliphatic | 294 | 1.11 |
| Glucobrassicanapin | Pent-4-enyl GSL | Aliphatic | 308 | 1.15 |
| Unknown 1 | Unknown | Unknown | 327 | 1.00 |
| Unknown 2 | Unknown | Unknown | 292 | 1.00 |
| Glucobrassicin | 3-Indolymethyl GSL | Indolic | 369 | 0.29 |
| 4-Methoxyglucobrassicin | 4-Methoxy-3-indolylmethyl GSL | Indolic | 399 | 0.25 |
| Neoglucobrasscin | N-Methoxy-3-indolylmethyl GSL | Indolic | 399 | 0.20 |
| Gluconasturtiin | 2-Phenethyl GSL | Aromatic | 344 | 0.95 |
* The elution order of HPLC chromatogram (data not shown) was as follow: progoitrin-sinigrin-glucoalyssin-unknown 1-gluconapin-unknown2-glucobrassicanapin-glucobrassicin-4-methoxyglucobrassicin-gluconasturtiin-neoglucobrasscin.
Table 3.
Glucosinolate content (µmol/g DW) in different organs of Chinese cabbage.
| Trivial name | Flower | Stem | Young leaf | Old leaf | Root | Seed |
|---|---|---|---|---|---|---|
| Progoitrin | 5.53 ± 0.04 | 3.78 ± 0.02 | 4.03 ± 0.10 | 1.11 ± 0.05 | 1.11 ± 0.03 | 3.26 ± 0.21 |
| Sinigrin | 2.23 ± 0.22 | 1.17 ± 0.02 | 0.34 ± 0.03 | 0.11 ± 0.03 | 0.12 ± 0.00 | 3.58 ± 0.79 |
| Glucoalyssin | 0.68 ± 0.22 | 0.18 ± 0.12 | 0.22 ± 0.10 | 0.19 ± 0.01 | 0.11 ± 0.00 | 0.47 ± 0.21 |
| Gluconapin | 4.51 ± 0.25 | 1.35 ± 0.10 | 1.57 ± 0.19 | 0.34 ± 0.03 | 0.42 ± 0.02 | 60.13 ± 1.76 |
| Glucobrassicanapin | 10.85 ± 0.20 | 4.61 ± 0.18 | 4.32 ± 0.16 | 0.89 ± 0.04 | 1.73 ± 0.04 | 4.92 ± 0.90 |
| Unknown 1 | 1.08 ± 0.11 | 0.99 ± 0.08 | 0.99 ± 0.10 | 0.29 ± 0.04 | 0.62 ± 0.07 | 0.41 ± 0.08 |
| Unknown 2 | 0.52 ± 0.23 | 0.47 ± 0.24 | 0.50 ± 0.22 | 0.16 ± 0.00 | ND | 7.34 ± 1.34 |
| Glucobrassicin | 1.96 ± 0.44 | 0.92 ± 0.14 | 0.43 ± 0.05 | 0.37 ± 0.20 | 0.72 ± 0.05 | 0.90 ± 0.09 |
| 4-Methoxyglucobrassicin | 0.20 ± 0.03 | 0.41 ± 0.05 | 1.14 ± 0.06 | 0.15 ± 0.04 | 0.19 ± 0.02 | 1.27 ± 0.03 |
| Neoglucobrassicin | 0.14 ± 0.02 | 0.08 ± 0.04 | 0.14 ± 0.04 | 0.09 ± 0.04 | 0.30 ± 0.00 | 0.12 ± 0.02 |
| Gluconasturtiin | 2.10 ± 0.13 | 0.80 ± 0.09 | 1.89 ± 0.16 | 0.65 ± 0.23 | 8.42 ± 0.16 | 0.91 ± 0.12 |
| Total | 29.81 ± 0.69 | 14.77 ± 0.53 | 15.59 ± 0.42 | 4.30 ± 0.38 | 13.66 ± 0.23 | 83.38 ± 2.98 |
Gluconapin was the major glucosinolate in 113 varieties of turnip greens (B. rapa) from northwestern Spain grown at two sites [30]. Progoitrin and glucobrassicanapin were detected mainly in the stems, young leaves, old leaves, and flowers. Padilla et al. [31] demonstrated that glucoraphanin presenting in 27 of 113 varieties of turnip greens (B. rapa) should be studied more exhaustively since this aliphatic glucosinolate is the precursor of sulforaphane, a potent anti-cancer isothiocyanate. In this study, progoitrin exhibited 18.2% of the total GSLs content. Kim et al. [32] reported that glucobrassicanapin has the greatest proportion (21.1%) of total GSLs in the leaves. However, the glucobrassicanapin content was found to be highest in the flowers in this study.
From the aromatic GSLs, the level of gluconasturtiin in the roots (8.42 μmol/g DW) was 4–15 fold higher than that in the other organs. The total GSL content according to the site (ranked in descending order) is as follows: seeds > flowers > young leaves > stems > roots > old leaves. Aliphatic GSLs showed 63.3% of the total GSLs content, followed by aromatic GSL (19.0%), indolic GSLs (10%), and unknown GSLs (7.7%) in different organs of Chinese cabbage. In addition, total aliphatic, indolic, aromatic, and unknown GSLs content showed 65.7, 11.7, 12.7, and 9.7%, respectively in leaves. Cartea et al. [30] reported that aliphatic GSLs were predominantly exhibited 95.5% of the total GSLs content, followed by indolyl GSLs (2.4%) and aromatic GSL (2.1%) in leaves of vegetable turnip rape (B. rapa L. var. rapa).
In our study, progoitrin and glucobrassicanapin, which belongs to the aliphatic GSLs group, were highly detected in flowers (Table 3). The transcript level of MYB28-1 in flowers was higher compared to other organs (Figure 3). It seems that there is a relationship between aliphatic GSLs content and gene expression level of MYB28-1. The rank of total aliphatic GSLs content was as follows: seeds (72.36 μmol/g DW) > flowers (23.80 μmol/g DW) > stems (11.09 μmol/g DW) > young leaves (10.48 μmol/g DW) > roots (3.49 μmol/g DW)> old leaves (2.64 μmol/g DW). On the other hand, the rank of total indolic GSLs content was flowers (2.30 μmol/g DW) > seeds (2.29 μmol/g DW) > young leaves (1.71 μmol/g DW) > stems (1.41 μmol/g DW) > roots (1.21 μmol/g DW) > old leaves (0.61 μmol/g DW). In this study, the transcript levels of MYB34, MYB51, and MYB122 were higher in the seeds and roots compared to other organs. Furthermore, the two indolic GSLs, glucobrassicin and 4-methoxyglucobrassicin, were also highly accumulated in the flowers and seeds. These results reveal that there is somewhat a correlation between the expression levels of indolic GLSs responsive genes and their related compounds. The reason of having relationship between gene expression and GSL content is somewhat, might be due to presence of many genes relating in amino acid chain elongation, core structure formation, and secondary modification taking part in GSL biosynthesis as well as MYB transcription factors.
3. Experimental
3.1. Plant Materials and Growth Conditions
Seeds of Chinese cabbage (B. rapa ssp. Pekinensis cv. Asia Alpine), were purchased from Asia Seed Co., Ltd (Seoul, Korea). Chinese cabbages were grown at the experimental farm of Chungnam National University (Daejeon, Korea). After 4 months, different organs (seeds, flowers, stems, young leaves, old leaves, and roots) were excised from several plants. All samples were immediately frozen in liquid nitrogen and then stored at −80 °C and/or freeze-dried for RNA isolation and/or high-performance liquid chromatography (HPLC) analysis.
3.2. Bioinformatic Analysis
For sequence data of B. rapa ssp. pekinensis, Brassica genome database (http://brassicadb.org/brad/glucoGene.php) [21] was utilized. In MYB29 isoform, Bra009245 (MYB29-1) is almost similar to Bra005949 (MYB29-2). Thus, Bra009245 gene was not used in this study. For protein subcellular localization prediction, TargetP (http://www.cbs.dtu.dk/services/TargetP/) [33], ChloroP (http://www.cbs.dtu.dk/services/ChloroP/) [33], and Wolf PSORT (http://wolfpsort.org/) [34] were used. The phylogenetic relationships of MYB TFs were analyzed with the TreeTop Phylogenetic tree prediction program (http://www.genebee.msu.su/services/phtree_reduced.html) [35] by applying the cluster algorithm. In the bootstrap, a multiple Sequence Alignment was resampled 100 times.
3.3. cDNA Synthesis and Quantitative Real-time PCR Analysis
Total RNA was isolated from different organs using Total RNA extraction Kit (Geneaid, New Taipei, Taiwan). For qRT-PCR, first-strand cDNA was synthesized from the total RNA with ReverTra Ace-α-(Toyobo, Osaka, Japan) Kit and oligo (dT)20 primer. The protocol for the reverse transcriptase polymerase chain reaction (RT-PCR) is as follows: Eleven micro liters of RNase-free water was mixed with 1 µg of total RNA (1 µL), 2 µL of 10° buffer, 1 µL of 10 mM each dNTP, 2 µL of 10 µM oligo dT primer, 1 µL of 40 µ/µL RNase inhibitor, and 1 µL of 4 u/µL reverse transcriptase to a final volume of 20 µl. The mixture was reamplified for 20 min at 42 °C and heated for 5 min at 99 °C. In order to design the primers for real time PCR, we performed DNA sequence alignment for each MYB transcription factor in Chinese cabbage and gene-specific primers were designed using an online program (http://web.bioneer.co.kr/tools/tmcalculator.jsp) [36] (Table S1). The SYBR Green qRT-PCR assay was carried out in a total volume of 20 µL, containing 10 µL of 2X SYBR Green Real time PCR master mix (Toyobo, Osaka, Japan), 0.5 µM (each) of specific primers, and template cDNA was diluted 10-fold. The amplification program consisted of one cycle of 95 °C for 3 min, followed by 40 cycles of 95 °C for 15 s, 72 °C for 20 s and annealing temperature of each gene was shown (Table S1). The reaction was performed in triplicate on a Mini Opticon Real-Time PCR System (Bio-Rad, Hercules, CA, USA) with the SYBR Green Real-time PCR Master Mix (Toyobo, Osaka, Japan). The actin gene (GenBank accession No. FJ969844) was selected as a reference gene because actin gene exhibited expression stability in different organs. The actin gene has also been used as a reference gene in several studies [37,38,39].
3.4. Chemicals
(−)-Sinigrin (2-propenyl glucosinolate) hydrate from horseradish for use as an external standard, and aryl sulfatase (Type H-1, EC 3.1.6.1) for desulfation of glucosinolates were purchased from Sigma–Aldrich (St. Louis, MO, USA). DEAE-Sephadex A-25 for loading into a mini-column (1,000 µL-Pasteur pipette) was provided by Amersham Biosciences (Uppsala, Sweden). HPLC-grade acetonitrile (CH3CN) and methanol (MeOH) were supplied by J. T. Baker (Phillipsburg, NJ, USA). Ultrapure water having a resistivity of 18.2 MΩ/cm was produced by a PureLab Option from ELGA Labwater (Model LA 621, Marlow, UK).
3.5. Extraction of Desulfo-glucosinolates (DS-GSLs) and HPLC Analysis
DS-GSLs were extracted using a slight modification of the procedures reported in a previous study [40,41]. Briefly, crude GSLs from 100 mg of freeze-dried powder were extracted with 1.5 of boiling 70% (v/v) MeOH at 70 °C for 5 min at a water-bath. After centrifugation at 12,000 rpm at 4 °C for 10 min, the supernatant was collected into a 5 mL test tube, and the residue was re-extracted twice as described above. The combined supernatants were taken as the crude GSL extracts. The extracts were loaded into a mini-column previously packed with DEAE-Sephadex A-25 and desulfated by the addition of 75 μL of an aryl sulfatase solution. DS-GSLs samples were eluted into a 2 mL microcentrifuge tube with 0.5 mL (×3) of ultrapure water.
The separation of DS-GSLs was carried out on a reversed-phase Inertsil ODS-3 column (150 × 3.0 mm i.d., particle size 3 μm; GL Sciences, Tokyo, Japan) with an E type cartridge guard column (10 × 2.0 mm i.d., 5 μm) using an Agilent Technologies 1,200 series HPLC system (Palo Alto, CA, USA). The detection wavelength, column oven temperature and flow rate were set at 227 nm, 40 °C and 0.2 mL/min, respectively. The mobile phase consisted of ultrapure water (solvent A) and CH3CN (solvent B). The gradient programs were as follows: A linear step from 7% to 24% of solvent B for 18 min, 24% of solvent B for the next 14 min, then kept constant at solvent B 24% for 3 min followed by a rapid drop to 7% solvent B at 32.1 min, and kept constant at solvent B 7% for 8 min (total 40 min). The individual GSLs were identified based on their HPLC retention times and our data base and quantified with the external standard, sinigrin (0.5 mg/5 mL for its desulfation, but it was diluted 4-times before HPLC injection) passed through the same extraction process together with sample preparation, with their HPLC area and response factor (ISO 9167-1, 1992).
3.6. LC/ESI-MS Analysis for Quantitation of Desulfoglucosinolates (DS-GSLs)
The MS data were acquired by electrospray ionization (ESI)- mass spectrometry with an API 4000 Q TRAP system (Applied Biosystems, Foster City, CA, USA) in positive ion mode ([M+H]+) that was equipped with an Agilent 1,200 series HPLC system. The MS operating conditions were as follows: scan range, m/z 100–800 (scan time, 1.0 s); curtain gas (20 psi), nebulizing gas (50 psi), heating gas (50 psi) by high purity nitrogen (N2); heating gas temperature, 550 °C; ion spray voltage, 5,500 V; declustering potential, 100 V; entrance potential, 10 V.
3.7. Statistical Analysis
The data were analyzed by using the computer software Statistical Analysis System (SAS version 9.2, SAS Institute Inc., Cary, NC, USA, 2009). All data are given as the mean and standard deviation of triplicate experiments. Treatment mean comparisons were performed with the Least Significant Difference (LSD).
4. Conclusion
In this study, we performed qRT-PCR of TFs factors involved GSLs biosynthesis and analyzed GSLs content in different organs of Chinese cabbage. Even though expression levels of MYB28 and MYB29 in seeds was not high, transcript levels of flowers and stems were higher compared with other organs. Our results showed distinctly that MYB28-1 regulate aliphatic GSLs biosynthesis in Chinese cabbage, whereas MYB34, MYB51, and MYB122 involving in indolic GSLs biosynthesis could differ depending on different organ in Chinese cabbage. Especially, there is a correlation between aliphatic GSLs content and gene expression level of MYB28-1, Dof1.1-1, and Dof1.1-2. Therefore, our results may be helpful in understanding the mechanism of MYB TFs regulating GSL biosynthesis in Chinese cabbage.
Acknowledgments
This work was financially supported by grants from Cabbage Genomics assisted breeding supporting Center (CGC) research programs and Technology Development Program for Agriculture and Forestry (No. 2011-1607) funded by Ministry for Food, Agriculture, Forestry and Fisheries of the Korean Government.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/18/7/8682/s1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: In general, samples of the compounds analyzed herein are unavailable from the authors due to their isolation on a small scale. They are readily analyzed using the procedures described.
References
- 1.Rodman J.E., Karol K.G., Price R.A., Sytsma K.J. Molecules, morphology, and Dahlgren’s expanded order Capparales. Syst. Bot. 1996;21:289–307. doi: 10.2307/2419660. [DOI] [Google Scholar]
- 2.Clarke D.B. Glucosinolates, structures and analysis in food. Anal. Methods. 2010;2:310–325. doi: 10.1039/b9ay00280d. [DOI] [Google Scholar]
- 3.Halkier B.A., Gershenzon J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006;57:303–333. doi: 10.1146/annurev.arplant.57.032905.105228. [DOI] [PubMed] [Google Scholar]
- 4.Fahey J.W., Zhang Y., Talalay P. Broccoli sprouts: An exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc. Natl. Acad. Sci. USA. 1997;94:10367–10372. doi: 10.1073/pnas.94.19.10367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahey J.W., Haristoy X., Dolan P.M., Kensler T.W., Scholtus I., Stephenson K., Talalay P., Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc. Natl. Acad. Sci. USA. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talalay P., Fahey J.W. Phytochemicals from cruciferous plants protect against cancer by modulating carcinogen metabolism. J. Nutr. 2001;131:3027S–3033S. doi: 10.1093/jn/131.11.3027S. [DOI] [PubMed] [Google Scholar]
- 7.Grubb C.D., Abel S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006;11:89–100. doi: 10.1016/j.tplants.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Wittstock U., Halkier B.A. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002;7:263–270. doi: 10.1016/S1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- 9.Celenza J.L., Quiel J.A., Smolen G.A., Merrikh H., Silvestro A.R., Normanly J., Bender J. The Arabidopsis ATR1Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy M., Wang Q., Kaspi R., Parrella M.P., Abel S. Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 2005;43:79–96. doi: 10.1111/j.1365-313X.2005.02435.x. [DOI] [PubMed] [Google Scholar]
- 11.Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., Zanor M.I., Gershenzon J., Strnad M., Szopa J., Mueller-Roeber B., Witt I. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- 13.Gigolashvili T., Berger B., Mock H.P., Müller C., Weisshaar B., Flügge U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- 14.Gigolashvili T., Yatusevich R., Berger B., Müller C., Flügge U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;51:247–261. doi: 10.1111/j.1365-313X.2007.03133.x. [DOI] [PubMed] [Google Scholar]
- 15.Gigolashvili T., Engqvist M., Yatusevich R., Müller C., Flügge U.I. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008;177:627–642. doi: 10.1111/j.1469-8137.2007.02295.x. [DOI] [PubMed] [Google Scholar]
- 16.Hirai M.Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K., et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sønderby I.E., Hansen B.G., Bjarnholt N., Ticconi C., Halkier B.A., Kliebenstein D.J. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS One. 2007;2:e1322. doi: 10.1371/journal.pone.0001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malitsky S., Blum E., Less H., Venger I., Elbaz M., Morin S., Eshed Y., Aharoni A. The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 2008;148:2021–2049. doi: 10.1104/pp.108.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Underhill E.W., Wetter L.R., Chisholm M.D. Biosynthesis of glucosinolates. Biochem. Soc. Symp. 1973;38:303–326. [PubMed] [Google Scholar]
- 20.Yatusevich R., Mugford S.G., Matthewman C., Gigolashvili T., Frerigmann H., Delaney S., Koprivova A., Flügge U.I., Kopriva S. Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 2010;62:1–11. doi: 10.1111/j.1365-313X.2009.04118.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang H., Wu J., Sun S., Liu B., Cheng F., Sun R., Wang X. Glucosinolate biosynthetic genes in Brassica rapa. Gene. 2011;487:135–142. doi: 10.1016/j.gene.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 22.Nair R., Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005;348:85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 23.Falk K.L., Vogel C., Textor S., Bartram S., Hick A., Pickett J.A., Gershenzon J. Glucosinolate biosynthesis: demonstration and characterization of the condensing enzyme of the chain elongation cycle in Eruca sativa. Photochemistry. 2004;65:1073–1084. doi: 10.1016/j.phytochem.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Textor S., Bartram S., Kroymann J., Falk K.L., Hick A., Pickett J.A., Gershenzon J. Biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana: Recombinant expression and characterization of methylthioalkylmalate synthase, the condensing enzyme of the chain-elongation cycle. Planta. 2004;218:1026–1035. doi: 10.1007/s00425-003-1184-3. [DOI] [PubMed] [Google Scholar]
- 25.Reintanz B., Lehnen M., Reichelt M., Gershenzon J., Kowalczyk M., Sandberg G., Godde M., Uhl R., Palme K. Bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell. 2001;13:351–367. doi: 10.1105/tpc.13.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin H., Cominelli E., Bailey P., Parr A., Mehrtens F., Jones J., Tonelli C., Weisshaar B., Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grubb C.D., Zipp B.J., Ludwig-Muller J., Masuno M.N., Molinski T.F., Abel S. Arabidopsisglucosyltranserase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 2004;40:893–908. doi: 10.1111/j.1365-313X.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- 28.Mikkelsen M.D., Hansen C.H., Wittstock U., Halkier B.A. Cytochrome P450cyp79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indoleglucosinolates and indole-3-acetic acid. J. Biol. Chem. 2000;275:33712–33717. doi: 10.1074/jbc.M001667200. [DOI] [PubMed] [Google Scholar]
- 29.Zang Y.X., Kim H.U., Kim J.A., Lim M.H., Jin M., Lee S.C., Kwon S.J., Lee S.I., Hong J.K., Park T.H., Mun J.H., Seol Y.J., Hong S.B., Park B.S. Genome-wide identification of glucosinolate synthesis genes in Brassica rapa. FEBS J. 2009;276:3559–3574. doi: 10.1111/j.1742-4658.2009.07076.x. [DOI] [PubMed] [Google Scholar]
- 30.Cartea M.E., de Haro A., Obregon S., Soengas P., Velasco P. Glucosinolatevariation in leaves of Brassica rapacrops. Plant Foods Hum. Nutr. 2012;67:283–288. doi: 10.1007/s11130-012-0300-6. [DOI] [PubMed] [Google Scholar]
- 31.Padilla G., Cartea M.E., Velasco P., Haro A., Ordás A. Variation of glucosinolates in vegetable crops of Brassica rapa. Photochemistry. 2007;68:536–545. doi: 10.1016/j.phytochem.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Kim J.K., Chu S.M., Kim S.J., Lee D.J., Lee S.Y., Lim S.H., Ha S.H., Kweon S.J., Cho H.S. Variation of glucosinolates in vegetable crops of Brassica rapa L. ssp. pekinensis. Food Chem. 2010;119:423–428. doi: 10.1016/j.foodchem.2009.08.051. [DOI] [Google Scholar]
- 33.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 34.Paul H., Keun-Joon P., Takeshi O., Naoya F., Hajime H., Adams-Collier C.J., Kenta N. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chumakov K.M., Yushmanov S.V. The maximum topological similarity principle in molecular systematic. Mol. Genet. Microbiol. Virusol. 1988;3:3–9. [PubMed] [Google Scholar]
- 36.Triinu K., Maido R. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 37.Wang K.L., Bolitho K., Garryn K., Kortstee A., Karunairetnam S., McGhie T.K., Espley R.V., Hellens R.P., Allan A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Y.Z., Hu F.C., Hu G.B., Li X.J., Huang X.M., Wang H.C. Differential expression of anthocyanin biosynthetic genes in relation to anthocyanin accumulation in pericarp of Litchi Chinensis Sonn. PLoS One. 2011;6:e19455. doi: 10.1371/journal.pone.0019455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan Y., Chiu L.W., Li L. Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta. 2009;230:1141–1153. doi: 10.1007/s00425-009-1013-4. [DOI] [PubMed] [Google Scholar]
- 40.Kim S.J., Kawaharada C., Jin S., Hashimoto M., Ishii G., Yamauchi H. Structural elucidation of 4-(cystein-S-yl) butyl glucosinolate from the leaves of Eruca sativa. Biosci. Biotechnol. Biochem. 2007;71:114–121. doi: 10.1271/bbb.60400. [DOI] [PubMed] [Google Scholar]
- 41.International Standards Organization (ISO) ISO 9167–1 (E). Rapeseed: Determination of Glucosinolates Content. ISO; Geneva, Switzerland: 1992. Part 1: Method using high performance liquid chromatography; pp. 1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.