Abstract
4-Deoxybostrycin is a natural anthraquinone compound isolated from the Mangrove endophytic fungus Nigrospora sp. collected from the South China Sea. Nigrosporin is the deoxy-derivative of 4-deoxybostrycin. They were tested against mycobacteria, especially Mycobacterium tuberculosis. In the Kirby-Bauer disk diffusion susceptibility test, they both had inhibition zone sizes of over 25 mm. The results of the absolute concentration susceptibility test suggested that they had inhibitory effects against mycobacteria. Moreover, 4-deoxybostrycin exhibited good inhibition which was even better than that of first line anti-tuberculosis (TB) drugs against some clinical multidrug-resistant (MDR) M. tuberculosis strains. The gene expression profile of M. tuberculosis H37Rv after treatment with 4-deoxybostrycin was compared with untreated bacteria. One hundred and nineteen out of 3,875 genes were significantly different in M. tuberculosis exposed to 4-deoxybostrycin from control. There were 46 functionally known genes which are involved in metabolism, information storage and processing and cellular processes. The differential expressions of six genes were further confirmed by quantitative real-time polymerase chain reaction (qRT-PCR). The present study provides a useful experiment basis for exploitation of correlative new drugs against TB and for finding out new targets of anti-mycobacterial therapy.
Keywords: anti-mycobacterial activity, 4-deoxybostrycin, differentially expressed genes
1. Introduction
Tuberculosis (TB) remains an international public health burden around the World. Current global estimates indicate that one third of the global population was infected with TB. In 2010 there were still 8.8 million new cases of TB, 1.1 million deaths from TB among HIV-negative people and an additional 0.35 million deaths from HIV-associated TB [1]. China has the largest population account for 20% of the world total. TB remains a big threat to this country. The report of the 4th National Epidemiological Sampling Survey of TB in China stated that the infection rate of TB in the country was 44.5% (about 550 million) [2]. Although currently available drugs kill most isolates of Mycobacterium tuberculosis, strains resistant to each of them have emerged, and multiply resistant strains are increasingly widespread, 500,000 of whom are multidrug-resistant (MDR) [3]. The growing problem of drug-resistance combined with the deadly link between TB and HIV infection underscore the urgent need for new anti-TB agents.
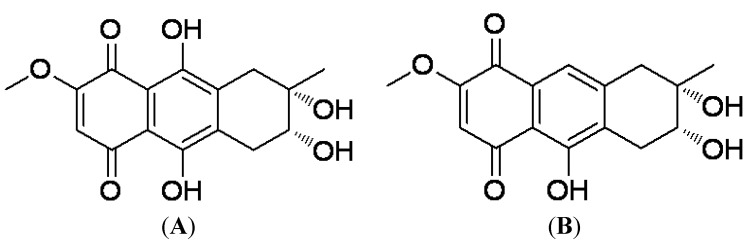
4-Deoxybostrycin (Figure 1A), a natural anthraquinone compound, was first isolated from the fungus Alternaria eichhorniae [4]. Its structure was identified by interpretation of spectral data [5,6]. Nigrosporin (Figure 1B) is the deoxy-derivative of 4-deoxybostrycin. It was also a natural product which was first isolated from the fungus Nigrospora oryzae. Its structure was identified by interpretation of spectral data [7]. Recently we obtained 4-deoxybostrycin and nigrosporin isolated from the fungus Nigrospora sp. [8,9]. It has been reported that 4-deoxybostrycin has various biological properties, including antibacterial [8], phytotoxic [4,10], antimalarial [11], and cytotoxic activities [8,12]. However, its anti-mycobacterial activity is not well investigated.
Figure 1.
The structure of 4-deoxybostrycin (A) and nigrosporin (B).
In this study, we tested it and its derivative’s anti-mycobacterial activity in vitro. We first screened and quantitated the anti-mycobacterial activity of the compound and its derivative with the Kirby-Bauer disk diffusion susceptibility test and the absolute concentration susceptibility test respectively. Then we detected the differential expression of M. tuberculosis genes after treatment with 4-deoxybostrycin by gene chips and confirmed the results with quantitative real-time polymerase chain reaction (qRT-PCR) experiments.
2. Results and Discussion
2.1. Preparation of 4-Deoxybostrycin and Nigrosporin
4-deoxybostrycin and nigrosporin were isolated from the mangrove endophytic fungus Nigrospora sp. collected from the South China Sea as previously reported [8,9].
2.2. The In Vitro Anti-Mycobacterial Activity of the Compounds
The in vitro anti-mycobacterial activity of the compounds was first screened with the Kirby-Bauer disk diffusion susceptibility test. 4-Deoxybostrycin and nigrosporin showed an inhibition zone size of 30 mm and 27 mm, respectively, against M. bovis BCG in 4 weeks-cultivated Middlebrook 7H11 agar plates. Their minimum inhibitory concentration (MIC) values against a series of mycobacterial strains were also determined in Middlebrook 7H11 agar slants with the absolute concentration susceptibility test, and these were compared with commercial control drugs including streptomycin (SM), isoniazid (INH), rifampicin (RFP), and ethambutol (EMB). As shown in Table 1, the two compounds both showed inhibitory effects against mycobacteria. Moreover, against two MDR M. tuberculosis clinical isolates K2903531 and 0907961, 4-deoxybostrycin exhibited a good inhibition, which even better than that of the first line anti-TB agents.
Table 1.
MIC of 4-deoxybostrycin and nigrosporin to BCG, M. tuberculosis and other mycobacterial strains.
| Bacterial strains | MIC (μg/mL) a | |||||
|---|---|---|---|---|---|---|
| 4-deoxybostrycin | Nigrosporin | Control drugs | ||||
| SM | INH | RFP | EMB | |||
| M. bovis BCG (strain Pasteur, ATCC 35734) | 39 | 15 | 0.1 | 0.1 | 0.05 | 1.6 |
| M. tuberculosis H37Rv reference strain (ATCC 27294) | 15 | 20 | 0.4 | 0.05 | <0.1 | 6.4 |
| Clinical MDR M. tuberculosis strain (K2903531, resistant to SM, INH, RFP and EMB) | <5 | 30 | >20 | >3.2 | >20 | 7.5 |
| Clinical MDR M. tuberculosis strain (0907961, resistant to SM and EMB) | 10 | 20 | 20 | 0.1 | 0.1 | >6.4 |
| Clinical drug-resistant M. tuberculosis strain (K0903557, resistant to INH) | 30 | 30 | 0.2 | 2.5 | 0.25 | <1.6 |
| Clinical drug-sensitive M. tuberculosis strain (0907762) | 10 | ND b | <0.1 | 0.025 | 10 | <1.6 |
| M. avium reference strain (ATCC 25291) | >60 | >60 | 20 | 5 | 30 | 3 |
| M. intracellulare reference strain (ATCC 13950) | >60 | >60 | >20 | 8 | >50 | 2.5 |
| Clinical extensively drug-resistant (XDR) M. avium-intracellulare strain (K0803182, resistant to SM, INH, RFP, levofloxacin [LVFX], protionamide and isoniazid aminosalicylate) | >60 | >60 | 0.1 | 5 | 6.4 | 7.5 |
a MIC (minimum inhibitory concentration) is defined as the lowest concentration inhibiting 100% of the inoculum relative to controls. b ND: not done.
2.3. 4-Deoxybostrycin-Induced Alterations in Gene Expression in M. tuberculosis H37Rv
According to the study above, 4-deoxybostrycin exhibited a better anti-mycobacterial activity. To find out the differentially expressed genes after treatment with 4-deoxybostrycin and investigate the possible anti-mycobacterial mechanism of the compound at the gene level, we tested the differences of gene expression in M. tuberculosis H37Rv just exposed to 4-deoxybostrycin by using the M. tuberculosis cDNA microarrays which consists of 3,875 predicted open reading frames of the M. tuberculosis H37Rv strain (13). Of all genes tested, 119 genes were significantly different in M. tuberculosis H37Rv exposed to 4-deoxybostrycin compared to untreated control. We found that 52 were significantly increased, and 67 were significantly decreased in the 4-deoxybostrycin treatment group. There were 46 functionally known genes, including 24 up-regulated genes and 22 down-regulated genes involved in nucleotide, lipid, energy, coenzyme, carbohydrate metabolism, information storage and processing, and other cellular processes (Table 2).
Table 2.
Detection of number and functions of differentially expressed M. tuberculosis H37Rv genes induced by 4-deoxybostrycin with M. tuberculosis cDNA microarray and their functions.
| Gene name | Synonym | Mean of ratio | SD | Functional Category |
|---|---|---|---|---|
| Rv0907 | - | 0.46 | 0.03 | Cellular processes: Cell envelope biogenesis, outer membrane |
| Rv1518 | - | 0.51 | 0.05 | Cellular processes: Cell envelope biogenesis, outer membrane |
| cmaA1 | Rv3392c | 1.88 | 0.08 | Cellular processes: Cell envelope biogenesis, outer membrane |
| Rv1212c | - | 1.98 | 0.21 | Cellular processes: Cell envelope biogenesis, outer membrane |
| fecB | Rv3044 | 1.66 | 0.04 | Cellular processes: Inorganic ion transport and metabolism |
| ctpG | Rv1992c | 1.60 | 0.05 | Cellular processes: Inorganic ion transport and metabolism |
| cysA | Rv2397c | 1.78 | 0.08 | Cellular processes: Inorganic ion transport and metabolism |
| Rv0282 | - | 0.47 | 0.04 | Cellular processes: Posttranslational modification, protein turnover, chaperones |
| Rv3673c | - | 1.78 | 0.09 | Cellular processes: Posttranslational modification, protein turnover, chaperones |
| Rv2264c | - | 1.79 | 0.02 | Cellular processes: Posttranslational modification, protein turnover, chaperones |
| htpX | Rv0563 | 1.90 | 0.15 | Cellular processes: Posttranslational modification, protein turnover, chaperones |
| narL | Rv0844c | 0.45 | 0.10 | Cellular processes: Signal transduction mechanisms |
| Rv3132c | - | 1.58 | 0.04 | Cellular processes: Signal transduction mechanisms |
| Rv1354c | - | 2.59 | 0.25 | Cellular processes: Signal transduction mechanisms |
| ogt | Rv1316c | 0.42 | 0.04 | Information storage and processing: DNA replication, recombination and repair |
| ligB | Rv3062 | 1.78 | 0.06 | Information storage and processing: DNA replication, recombination and repair |
| Rv0922 | - | 1.99 | 0.21 | Information storage and processing: DNA replication, recombination and repair |
| rpsD | Rv3458c | 0.42 | 0.03 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| rpsS | Rv0705 | 0.44 | 0.07 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| prfB | Rv3105c | 0.45 | 0.02 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| rplQ | Rv3456c | 0.45 | 0.03 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| truA | Rv3455c | 0.45 | 0.03 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| infB | Rv2839c | 0.46 | 0.02 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| rplT | Rv1643 | 0.47 | 0.04 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| Rv0881 | - | 1.68 | 0.07 | Information storage and processing: Translation, ribosomal structure and biogenesis |
| gnd2 | Rv1122 | 0.44 | 0.03 | Metabolism: Carbohydrate transport and metabolism |
| Rv2039c | - | 0.46 | 0.02 | Metabolism: Carbohydrate transport and metabolism |
| Rv1200 | - | 1.63 | 0.04 | Metabolism: Carbohydrate transport and metabolism |
| pgk | Rv1437 | 1.65 | 0.07 | Metabolism: Carbohydrate transport and metabolism |
| Rv2471 | - | 1.83 | 0.12 | Metabolism: Carbohydrate transport and metabolism |
| Rv2040c | - | 2.11 | 0.19 | Metabolism: Carbohydrate transport and metabolism |
| panB | Rv2225 | 0.52 | 0.03 | Metabolism: Coenzyme metabolism |
| Rv1335 | - | 1.66 | 0.04 | Metabolism: Coenzyme metabolism |
| nadA | Rv1594 | 1.73 | 0.09 | Metabolism: Coenzyme metabolism |
| cobU | Rv0254c | 1.78 | 0.03 | Metabolism: Coenzyme metabolism |
| appC | Rv1623c | 0.42 | 0.05 | Metabolism: Energy production and conversion |
| Rv0247c | - | 0.54 | 0.05 | Metabolism: Energy production and conversion |
| ctaC | Rv2200c | 0.37 | 0.10 | Metabolism: Energy production and conversion |
| Rv1257c | - | 1.74 | 0.04 | Metabolism: Energy production and conversion |
| pks18 | Rv1372 | 0.44 | 0.04 | Metabolism: Lipid metabolism |
| fadD32 | Rv3801c | 0.47 | 0.01 | Metabolism: Lipid metabolism |
| Rv3815c | - | 1.77 | 0.08 | Metabolism: Lipid metabolism |
| deoC | Rv0478 | 0.53 | 0.05 | Metabolism: Nucleotide transport and metabolism |
| dgt | Rv2344c | 0.59 | 0.04 | Metabolism: Nucleotide transport and metabolism |
a Genes were classified as differentially expressed if the Cy5/Cy3 intensity ratios were outside the range of 0.67–1.5.
The tuberculosis cDNA gene chips employed in our study were based on the M. tuberculosis H37Rv gene sequence. The functions of these differentially expressed genes included nucleotide, lipid, coenzyme, and carbohydrate metabolism; energy production; information storage and processing; and cellular processes. Therefore, we conclude that 4-deoxybostrycin exerts its anti-TB function through multiple targets. We screened several differentially expressed genes from the compound. 4-deoxybostrycin successfully inhibited drug-resistant M. tuberculosis isolates, which allows for new study into the genes affected by the compound to promote the discovery of new anti-M. tuberculosis mechanisms and targets. Meanwhile, we can justify and predict the compound’s toxicity and determine if there will be cross-resistance with other anti-TB drugs by examining the affected genes.
2.4. 4-Deoxybostrycin-Induced Expression Changes of Some Genes Were Confirmed with qRT-PCR
Total RNA from M. tuberculosis H37Rv from either 4-deoxybostrycin-treated or untreated control cultures used for microarray experiments was converted to cDNA, and the expressions of 36 functionally known genes were confirmed with qRT-PCR. Gene expression was normalised with M. tuberculosis housekeeping genes. There were only six genes (Rv1518, Rv0282, gnd2, Rv3044 [fecB], Rv3673c, and Rv1372 [pks18]) that were significantly differentially expressed compared to untreated control cultures (Table 3).
Table 3.
qRT-PCR results demonstrating 4-deoxybostrycin-induced differential expression of M. tuberculosis H37Rv genes.
| Gene names | Rv1518 | gnd2 | Rv1212c | appC | rpsS | nadA | dgt | prfB | cmaA1 | Rv0247c | Rv3815c | Rv0922 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ratio a | 0.642 | 0.539 | 1.187 | 0.83 | 0.793 | 1.137 | 1.243 | 1.01 | 0.964 | 0.739 | 0.933 | 0.961 |
| Rv0282 | Rv3673c | Rv3132c | cobU | narL | rplT | cysA | rplQ | fadD32 | Rv2040c | Rv1354c | Rv1200 | |
| 1.989 | 9.219 | 0.742 | 0.919 | 0.937 | 0.728 | 0.805 | 0.807 | 1 | 0.817 | 0.801 | 0.924 | |
| Rv3044 | Rv1372 | Rv1257c | htpX | pgk | ctaC | infB | rpsD | ogt | Rv2264c | Rv0907 | Rv1335 | |
| 15.46 | 0.59 | 1.023 | 0.846 | 0.708 | 1.069 | 0.787 | 0.733 | 0.826 | 1.275 | 0.878 | 1.26 |
a The ratio was calculated by dividing the amplified gene copies of 4-deoxybostrycin-treated bacteria by the amplified gene copies of untreated bacteria; genes were classified as differentially expressed if the ratios were outside the range of 0.67–1.5.
We performed qRT-PCR to verify the gene chips. The results showed that their consistency was just 12.3%, indicating that screening through gene chips involves high experimental error that necessitates further confirmation. This array consists of 3,875 predicted open reading frames of the M. tuberculosis H37Rv strain [13]. We identified six genes that were significantly differentially expressed compared to untreated control cultures. Gene product of Rv1518 is conserved hypothetical protein, possible glycosyl transferase involved in exopolysaccharide synthesis, was identified in the cell membrane fraction of M. tuberculosis H37Rv [14]. Gene product of Rv0282 is identified in the membrane fraction of M. tuberculosis H37Rv using 1D-SDS-PAGE and uLC-MS/MS [15], and its function is not clearly understood. Rv3044, probable fecB, FeIII dicitrate-binding periplasmic lipoprotein (see citation below), identified in immunodominant fractions of M. tuberculosis H37Rv culture filtrate using 2D-LPE, 2D-PAGE, and LC-MS or LC-MS/MS [13]. Rv3673c, possible membrane protein, thioredoxin-like protein (thiol-disulfide interchange protein) [16]. Rv1372, conserved hypothetical protein, function of which is not clearly understood [17]. The present study provides a useful experiment basis for exploitation of correlative new drugs against TB and for finding out new targets of anti-mycobacterial therapy.
3. Experimental
3.1. Preparation and Structure of 4-Deoxybostrycin and Nigrosporin
4-deoxybostrycin and nigrosporin were extracted from the fungal fermentation broth by solvent extraction method then purified through chromatography including silica gel column chromatography, ODS column chromatography and HPLC. All reagents and solvents used in experiments were commercially available. Melting points were measured on an X-4 micromelting point apparatus and were uncorrected. IR spectra were measured on a Bruker Vector 22 spectrophotometer (Bruker Corporation, Fremont, CA, USA) using KBr pellets. NMR spectra were determined on a Varian Inova-500 NB spectrometer or Bruker AV-400 NB spectrometer (Bruker Corporation) in CDCl3 or DMSO-d6 using TMS as internal standard, and coupling constants (J) are in Hz. EI mass spectra were recorded on a DSQ mass spectrometer and ESI mass spectra were obtained on a LCQ DECA XP LC-MS mass spectrometer.
4-deoxybostrycin Red solid (MeOH); mp: 224–225 °C; [α +90.9 (c 1.1 × 10−4, MeOH); 1H-NMR (300 MHz, DMSO-d6): δ 13.18 (s, 1H, 9-OH), 12.62 (s, 1H, 10-OH), 6.42 (s, 1H, 7-H), 4.82 (d, J = 5.1 Hz, 1H, 2-OH), 4.48 (s, 1H, 3-OH), 3.88 (s, 3H, 6-OMe), 3.61 (dt, J = 7.3, 5.1 Hz, 1H, 2-H), 2.82 (dd, J = 18.3, 5.1 Hz, 1H, 1-Hb), 2.77 (d, J = 18.1 Hz, 1H, 4-Hb), 2.64 (dd, J = 18.3, 7.3 Hz, 1H, 1-Ha), 2.56 (d, J = 18.1 Hz, 1H, 4-Ha), 1.18 (s, 3H, 3-CH3); 13C-NMR (125 MHz, DMSO-d6): δ 183.39 (C-8), 176.35 (C-5), 161.31 (C-10), 160.38 (C-6), 159.67 (C-9), 138.88 (C-9a), 136.33 (C-4a), 109.47(C-7), 108.97 (C-10a), 106.85 (C-8a), 70.12 (C-2), 68.81 (C-3), 56.95 (6-OMe), 35.72 (C-4), 29.91 (C-1), 25.39 (3-CH3); EIMS m/z 320 [M]+ (100), 302 (41), 287 (30), 259 (61), 247 (96), 234 (20), 219 (45), 205 (10).
Nigrosporin Yellow solid (MeOH); mp: >300 °C; [α +131.4 (c 1.75 × 10−4, MeOH); 1H-NMR (400 MHz, DMSO-d6): δ 12.67 (s, 1H, 9-OH), 7.26 (s, 1H, 10-H), 6.29 (s, 1H, 7-H), 4.76 (d, J = 3.6 Hz, 1H, 2-OH), 4.39 (s, 1H, 3-OH), 3.88 (s, 3H, 6-OMe), 3.66 (m, 1H, 2-H), 2.93(d, J = 17.5 Hz, 1H, 4-Hb), 2.85 (dd, J = 18.4, 4.9 Hz, 1H, 1-Hb), 2.77 (d, J = 17.5 Hz, 1H, 4-Ha), 2.69 (dd, J = 18.4, 7.1 Hz, 1H, 4-Ha), 1.17 (s, 3H, 3-CH3); 13C-NMR (100 MHz, DMSO-d6): δ 190.76 (C-8), 178.79 (C-5), 161.11 (C-6), 158.31 (C-9), 144.01 (C-4a), 131.70 (C-9a), 127.95 (C-10a), 119.37 (C-10), 110.65 (C-8a), 109.33 (C-7), 70.51 (C-2), 69.41 (C-3), 56.78 (6-OMe), 41.67 (C-4), 29.75 (C-1), 25.07 (3-CH3); EIMS m/z 304 [M]+ (34), 286 (85), 271 (100), 257 (57), 243 (94), 229 (45), 215(54), 201 (23).
3.2. Bacterial Strains and Culture Conditions
The following mycobacterial strains were used in this study, M. bovis BCG (strain Pasteur, ATCC 35734) and a virulent reference strain of M. tuberculosis H37Rv (ATCC 27294) which were obtained from the China Center of Medical Culture Collection; two MDR M. tuberculosis clinical isolates (K2903531, resistant to SM, INH, RFP and EMB; 0907961, resistant to SM and EMB), a drug-resistant M. tuberculosis clinical isolate (K0903557, resistant to INH), a drug-sensitive M. tuberculosis clinical isolate (0907762), and an extensively drug-resistant (XDR) clinical M. avium-intracellulare isolate (K0803182, resistant to SM, INH, RFP, LVFX, protionamide and isoniazid aminosalicylate), and M. avium reference strain (ATCC 25291) and M. intracellulare reference strain (ATCC 13950), which were kindly provided by the Guangzhou Chest Hospital, China.
There were two culture media used in our experiments including Middlebrook 7H9 (Bio-Rad, Richmond, CA, USA) and Middlebrook 7H11 (Bio-Rad). Middlebrook 7H9 broth was used for the cultivation of M. tuberculosis H37Rv to test the differential expression of genes in vitro. Middlebrook 7H11 was used for the cultivation of all mycobacterial strains in Kirby-Bauer disk diffusion susceptibility test (agar plate) or the absolute concentration susceptibility test (agar slant) to screen or quantify in vitro the anti-mycobacterial activity (including MIC) of the compounds.
3.3. The Kirby-Bauer Disk Diffusion Susceptibility Test
Bacterial suspension was prepared from 10-days old M. bovis BCG or other mycobacterial strain cultures grown on Middlebrook 7H11 agar slant, supplemented with 10% oleic acid, bovine serum albumin (fraction V), dextrose, and catalase (OADC; Remel, Lenexa, KS, USA) and 0.05% Tween 80. The turbidity of the suspension was adjusted to a McFarland no. 3 (9 × 108 CFU/mL) in sterile normal saline. The bacteria was spread on Middlebrook 7H11 agar plates, then discs with 100 μg different kinds of compounds were placed on the plates. All plates were incubated at 37 °C for 4–8 weeks before measuring the diameter of the zone of inhibition of each chemical.
3.4. The Absolute Concentration Susceptibility Test
Middlebrook 7H11 agar medium was prepared and sterilized by autoclaving at 121 °C for 15 min. Sterilized medium was supplemented with 10% of sterile OADC at 55–60 °C. The final desired concentrations (100, 60, 50, 40, 30, 20, 15, 10 and 5 μg/mL, respectively, or finer range between 0.1–60 μg/mL) of chemicals were added to respective flasks and poured into 15 mL sterile screwcap plastic test tubes respectively. Upon solidification of medium and preparation of agar slant, the tubes, with or without chemical, were inoculated with 30 μL of 10-fold dilution (3 × 107 CFU/mL) of each mycobacterial strain’s suspension equivalent to a McFarland 1.0 standard (3 × 108 CFU/mL). All the tubes were then incubated at 37 °C for 4–8 weeks. The slants were read when all the cultures had grown on the control slant. If there was no colony grown in the presence of the compound, the lowest concentration of the compound was considered as the MIC.
3.5. Determination of the Differential Expressed Genes by Gene Chips and qRT-PCR
3.5.1. RNA Isolation of M. tuberculosis H37Rv
Bacterial liquid from the Middlebrook 7H9 medium at log growth period was taken. The experimental group was treated with 4-deoxybostrycin at the final concentration of MIC concentration. The control group was treated with equal volume of solvent which was used to dissolve and dilute the compound. Both groups were incubated for 2 h at 37 °C and then harvested by centrifugation (6000 × g, 5 min). Pellets were resuspend in 95 °C preheated 200 μL MaxBacterial Enhancement Reagent (Invitrogen, Carlsbad, CA, USA) and incubated at 95 °C for 4 min, then transferred to 2 mL screwcap tubes containing 1 ml Trizol reagent (Invitrogen) and 0.5 mL of 0.1-mm-diameter glass beads. Cells were disrupted by strenuous vibration [18]. The RNA was extracted according to the Trizol manufacturer’s instructions.
To remove residual DNA, samples were treated with Turbo DNAse (Ambion, Austin, TX, USA). The integrity of all RNA samples was checked by nondenaturing agarose gel electrophoresis, with RNA concentration quantified by spectrophotometry.
3.5.2. Preparation of Labelled cDNA of Bacteria for Gene Chips and Hybridization
cDNA was synthesized and fluorescently labelled by a direct procedure. The RNA was labelled by Cy5-dUTP in untreated control group and Cy3-dUTP in treated group respectively. After heating at 70 °C for 3 min and chilling on ice, the RNA was reverse transcribed. The labelled cDNA probes were then purified and concentrated using the MiniElute Cleanup kit (Qiagen, Valencia, CA, USA). The total purified cDNA probes were added to the arrays in a hybridization solution containing a final concentration of 5 × SSPE, 2% SDS, 5 × Denhardt’s regent. The arrays were pre-hybridized with denatured salmon sperms DNA at 65 °C for 1 h and then were hybridized overnight at 65 °C, using roller bottles at 6 r.p.m. After removing the hybridized mixture, the chips were washed with preheated Panorama® Microbial Array wash solution (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer’s instructions.
3.5.3. Microarray Data Analysis
Microarray slides were scanned using a Typhoon™ FLA 9000 Biomolecular Imager (GE Healthcare, Uppsala, Sweden). Images were processed and the fluorescent intensity of each spot was quantified using the ImageQuant TL v2005 software. Four independent biological replicates were analyzed for each compound group, one swap-dye experiment was included. Median intensity values were corrected by background subtraction. Further analysis was performed using SPSS software (version 16.0 for Windows; SPSS Inc., Chicago, IL, USA). Data was represented by the spot signal as a percentage of the total signal from all spots on the array. Cy5/Cy3 intensity ratios were determined using normalized values. For each gene, the geometric mean was calculated from the intensity ratios of the 2 replicates and the result value was used to determine differences in mRNA abundance between treated group and untreated group. Genes were classified as differentially expressed if the Cy5/Cy3 intensity ratios were out of the range of 0.67–1.5. Statistical significance of the chosen genes was verified by following real-time PCR.
3.6. Detection of Differential Expressed Genes by qRT-PCR
Total RNA was extracted from the treated and untreated M. tuberculosis H37Rv as mentioned above. cDNA was synthesized from total RNA using random hexamers. The design of primers (Table 4) used for real-time PCR was based on the published genome sequence of M. tuberculosis H37Rv [13]. Real-time PCR was performed using SYBR Green (Roche, Mannheim, Germany) in a LightCycler 4800 (Roche, Germany). The end point used in the real-time RT-PCR quantification, Ct, was defined as the PCR cycle number at which each assay target reached the threshold. The data represented the fold change in mRNA expression relative to M. tuberculosis incubated with medium alone. To calibrate each amplification result, three M. tuberculosis housekeeping genes including 10Sa RNA (http://www.ncbi.nlm.nih.gov/pubmed/1371186), MPT70 (http://www.ncbi.nlm.nih.gov/protein/BAA07184.1) and diaminopimelate decarboxylase (http://www.ncbi.nlm.nih.gov/protein/YP_006514669.1) served as correction factors. The ratio of the same gene expression between the treated and untreated M. tuberculosis H37Rv was calculated by the amplified gene copies of 4-deoxybostrycin-treated bacteria divided by the amplified gene copies of untreated bacteria. The criteria used to justify differential expression was the same as above gene chips, i.e., genes were classified as differentially expressed if the ratios were out of the range of 0.67–1.5.
Table 4.
The primers of qRT-PCR.
| Gene names | PCR product length (bp) | Forward primer (5′→3′) | Reverse primer (5′→3′) |
|---|---|---|---|
| Rv1518 | 145 | gcctcaaccgaaaccacaa | gcgaaagccattccgaca |
| Rv0282 | 117 | ccaacgcacgcaccactt | cggatgttctcccgcttca |
| gnd2 | 111 | gccaaaggtggacacgactg | ctgagacaactcacgcaacgag |
| Rv3044 | 102 | gggtttgacgccgcagtt | ccgacaccacgcaggttatt |
| Rv3673c | 99 | cacgatctcgtcggcactg | cgggtcgcaaatgtgatgc |
| Rv1372 | 69 | ggtggtagtccgcagtagtttc | cgaaataagcgttgagttggtc |
| Rv0247c | 91 | ctgttgtagacggaggatgacg | cggagacgaaagctgtggc |
| cobU | 94 | cagatgtacctcatcgcagacg | ggtggtgccatcccattctt |
| htpX | 70 | gactggcatcctgcgtatcct | gacgtgagacagctcgtggc |
| rpsS | 147 | acttcatcggccatacctttgc | gcttgctctttcggtcgtctttt |
| narL | 68 | cacgttcaccgagccactca | gcgacgaccacccgttattt |
| Rv0907 | 114 | acaacgtcgtgacctgggata | cggagcgatgcgagtagag |
| Rv1200 | 112 | gcttcggcttcgtctacctg | cacagcagtccacccagca |
| Rv1212c | 126 | tcgtcggcgtcgtaatgc | gctgggtatcgtaaacctggaa |
| Rv1335 | 86 | tatccattccgaccatcctgc | gctgatgacggcacccaag |
| pgk | 84 | aaggcggacagcattgtgatt | cagcagcgatgtgccaacc |
| nadA | 133 | catgttgcaccagcttcgc | ttcgtcggcaccctctacc |
| Rv2040c | 117 | caccagcaagatggcgaaca | ggtcccgagacggctacctat |
| ctaC | 137 | agccaaacttccaattccactg | caccgtcataccgttcctcatc |
| Rv2264c | 104 | gccaccgaagtcgcatacc | agccactgtcactgctccct |
| dgt | 93 | tgcgttccaaccgaccct | ccgctgcgttcaataccg |
| cysA | 85 | gcgcttacggatcttcaacc | cggattcgtcttccagcacta |
| infB | 132 | ccgagtagtagcggatctcca | gcggcattaccgaaaccaa |
| prfB | 119 | cgcttgcgttccaacaact | gtgcggctcacccacatt |
| Rv3132c | 94 | acaaggctggtcgctgaatg | tcgatggtctggtggaggc |
| cmaA1 | 68 | tcaggaaacgagcgaaggtg | cgggttgcatccgaaagag |
| rplQ | 92 | gaggtcaccgtcttctcccg | ggctacacccgtatcatcaaaa |
| rpsD | 117 | acgccgttgacgttgaaatg | actgctgaagatcctcgaaagc |
| fadD32 | 107 | tcagtccgaagtggcgaaga | gaacctccagcggcaagat |
| Rv3815c | 60 | gcctgggatggtccttggt | cgattgtgcccgtcgttgt |
| ogt | 242 | cgaaataagcgttgagttggtc | ggcatggctcggtgttga |
| Rv1354c | 441 | ctggcggtatgtaggtcttgc | ggcgggtttgttgacttcg |
| Rv0922 | 387 | ccgcaatgggtttggtcg | ggtggatttggctggaggc |
| Rv1257c | 354 | ggtccatgatttcgccgtac | caatacccacccgttgctg |
| 10Sa RNA (control) | 85 | ttcgctatgcctctgctcg | ggactcctcgggacaacca |
| MPT70 (control) | 135 | tgaccagcatcctgacctacc | cggcgttaccgaccttga |
| diaminopimelate decarboxylase(control) | 142 | ccttactgctattcgctgtcga | ggcacgggtcacctcactt |
4. Conclusions
4-Deoxybostrycin is a natural anthraquinone compound isolated from the mangrove endophytic fungus Nigrospora sp. collected from the South China Sea. It showed a good anti-TB activity in vitro. Moreover, 4-deoxybostrycin could affect the expression of M. tuberculosis H37Rv genes which are involved in nucleotide, lipid, energy, coenzyme, carbohydrate metabolism, information storage and so on. Perhaps 4-deoxybostrycin could be a candidate for the development of new agents to treat TB.
Acknowledgments
This work was supported by the 863 Funds of China (2007AA09Z414, to X. Lai), the National Natural Science Foundation of China (20972197 and 41276146, to Z. She), the Key Science and Technique Research Project of Guangdong Province of China (2010B030600004 and 2011A080403006, to Z. She) and Fundamental Research Funds for the Central Universities of China (11lgjc01, to Z. She).
Conflicts of Interest
We declare that we have no associations with commercial or other associations that might pose a conflict of interest.
Footnotes
Sample Availability: Samples of the title compound are available from the authors.
References
- 1.WHO Global tuberculosis control 2011. [(accessed on 4 October 2011)]; Available online: http://www.who.int/tb/publications/global_report/2011/gtbr11_full.pdf.
- 2.Duanmu H. Report on the 4th national epidemiological sampling survey of tuberculosis. Zhonghua Jie He Hu Xi Za Zhi. 2002;25:3–7. [PubMed] [Google Scholar]
- 3.Matteelli A., Carvalho A.C., Dooley K.E., Kritski A. TMC207: the first compound of a new class of potent anti-tuberculosis drugs. Future Microbiol. 2010;5:849–858. doi: 10.2217/fmb.10.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charudattan R., Rao K.V. Bostrycin and 4-deoxybostrycin: Two nonspecific phytotoxins produced by Alternaria eichhorniae. Appl. Environ. Microbiol. 1982;43:846–849. doi: 10.1128/aem.43.4.846-849.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noda T., Take T., Watanabe T., Abe J. The structure of bostrycin. Tetrahedron. 1970;26:1339–1346. doi: 10.1016/S0040-4020(01)93004-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen H., Zhong L., Long Y., Li J., Wu J., Liu L., Chen S., Lin Y., Li M., Zhu X., et al. Studies on the synthesis of derivatives of marine-derived bostrycin and their structure-activity relationship against tumor cells. Mar. Drugs. 2012;10:932–952. doi: 10.3390/md10040932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka M., Fukushima T., Tsujino Y., Fujimori T. Nigrosporins A and B, New phytotoxic and antibacterial metabolites produced by a fungus Nigrospora oryzae. Biosci. Biotech. Bioch. 1997;11:1848–1852. doi: 10.1271/bbb.61.1848. [DOI] [PubMed] [Google Scholar]
- 8.Xia X., Li Q., Li J., Shao C., Zhang J., Zhang Y., Liu X., Lin Y., Liu C., She Z. Two new derivatives of griseofulvin from the mangrove endophytic fungus Nigrospora sp. (Strain No. 1403) from Kandelia candel (L.) Druce. Planta Med. 2011;77:1735–1738. doi: 10.1055/s-0030-1271040. [DOI] [PubMed] [Google Scholar]
- 9.Yang K.L., Wei M.Y., Shao C.L., Fu X.M., Guo Z.Y., Xu R.F., Zheng C.J., She Z.G., Lin Y.C., Wang C.Y. Antibacterial anthraquinone derivatives from a sea anemone-derived fungus Nigrospora sp. J. Nat. Prod. 2012;75:935–941. doi: 10.1021/np300103w. [DOI] [PubMed] [Google Scholar]
- 10.Rao K.V., Donia M.S., Peng J., Garcia-Palomero E., Alonso D., Martinez A., Medina M., Franzblau S.G., Tekwani B.L., Khan S.I., et al. Manzamine B and E and ircinal A related alkaloids from an Indonesian Acanthostrongylophora sponge and their activity against infectious, tropical parasitic, and Alzheimer’s diseases. J. Nat. Prod. 2006;69:1034–1040. doi: 10.1021/np0601399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommart U., Rukachaisirikul V., Sukpondma Y., Phongpaichit S., Sakayaroj J., Kirtikara K. Hydronaphthalenones and a dihydroramulosin from the endophytic fungus PSU-N24. Chem. Pharm. Bull. (Tokyo) 2008;56:1687–1690. doi: 10.1248/cpb.56.1687. [DOI] [PubMed] [Google Scholar]
- 12.Ge H.M., Song Y.C., Shan C.Y., Ye Y.H., Tan R.X. New and cytotoxic anthraquinones from Pleospora sp. IFB-E006, an endophytic fungus in Imperata cylindrical. Planta Med. 2005;71:1063–1065. doi: 10.1055/s-2005-864190. [DOI] [PubMed] [Google Scholar]
- 13.Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., 3rd, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 14.Mawuenyega K.G., Forst C.V., Dobos K.M., Belisle J.T., Chen J., Bradbury E.M., Bradbury A.R., Chen X. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol. Biol. Cell. 2005;16:396–404. doi: 10.1091/mbc.e04-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu S., Chen J., Dobos K.M., Bradbury E.M., Belisle J.T., Chen X. Comprehensive proteomic profiling of the membrane constituents of a Mycobacterium tuberculosis strain. Mol. Cell. Proteomics. 2003;2:1284–1296. doi: 10.1074/mcp.M300060-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Camus J.C., Pryor M.J., Médigue C., Cole S.T. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 17.Sankaranarayanan R., Saxena P., Marathe U.B., Gokhale R.S., Shanmugam V.M., Rukmini R. A novel tunnel in mycobacterial type III polyketide synthase reveals the structural basis for generating diverse metabolites. Nat. Struct. Mol. Biol. 2004;11:894–900. doi: 10.1038/nsmb809. [DOI] [PubMed] [Google Scholar]
- 18.Manganelli R., Voskuil M.I., Schoolnik G.K., Smith I. The Mycobacterium tuberculosis ECF sigma factor sigmaE: Role in global gene expression and survival in macrophages. Mol. Microbiol. 2001;41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]