Abstract
Vetch (Vicia sativa L.) is one of the most important annual forage legumes in the World due to its multiple uses (i.e., hay, grain, silage and green manure) and high nutritional value. However, detrimental cyanoalanine toxins in its plant parts including seeds and its vulnerability to hard winter conditions are currently reducing the agronomic values of vetch varieties. Moreover, the existence in the public domain of very few genomic resources, especially molecular markers, has further hampered breeding efforts. Polymorphic simple sequence repeat markers from transcript sequences (cDNA; simple sequence repeat [SSR]) were developed for Vicia sativa subsp. sativa. We found 3,811 SSR loci from 31,504 individual sequence reads, and 300 primer pairs were designed and synthesized. In total, 65 primer pairs were found to be consistently scorable when 32 accessions were tested. The numbers of alleles ranged from 2 to 19, frequency of major alleles per locus were 0.27–0.87, the genotype number was 2–19, the overall polymorphism information content (PIC) values were 0.20–0.86, and the observed and expected heterozygosity values were 0.00–0.41 and 0.264–0.852, respectively. These markers provide a useful tool for assessing genetic diversity, population structure, and positional cloning, facilitating vetch breeding programs.
Keywords: cDNA-SSR, genetic diversity, 454 sequencing, Viciastiva subsp. sativa
1. Introduction
Vicia sativa subsp. sativa, known as the common vetch, is one of the most commonly grown winter cover crops, or green manure, and is also used as pasture, silage, and hay [1,2]. It is cultivated with mixtures of cereal grains, providing cool-weather weed suppression and preventing fall N scavenging. It has been successfully applied in vineyards and orchards [1,2]. Due to its economic and ecological advantages, common vetch is now widespread through many parts of World, including the Mediterranean basin, west and central Asia, China, eastern Asia, India, and the USA [1,2,3].
The common vetch produces seeds that are quite similar to those of lentils in physical appearance and are highly nutritious [3,4,5,6,7]. However, due to the presence of cyanoalanine toxin in the seeds, which is detrimental to mono-gastric animals, including humans, the common vetch is currently tightly restricted as a feed or food source [3,4,5,6]. Moreover, its vulnerability to severe winter conditions (<−10 °C) further reduces its true agricultural potential [8,9]. Thus, to address these drawbacks, it is imperative to genetically improve this legume species either through conventional breeding or biotechnology approaches. However, a severe lack of genomic resources in the public domain has hampered such efforts.
Next-generation transcriptome sequencing is an excellent solution for enriching relevant genomic resources for non-model crop species such as the common vetch, providing functional annotations as well as genetic marker information [10,11,12]. In particular, cDNA-SSR markers generated from this approach can facilitate marker-assisted selection for vetch improvement programs, because these may be associated with functionally annotated transcribed genes, are cost-effective, and are easily transferable to related species [10,11,12,13]. Recently, we sequenced transcriptomes of common vetch using 454 pyrosequencing technology, and found 3,811 SSR loci from 31,504 individuals. In the present study, we developed and characterized polymorphic cDNA-SSR markers based on these transcriptome sequences to further contribute to breeding and molecular genetic studies of this species.
2. Results and Discussion
V. sativa subsp. sativa transcriptome sequencing yielded about 28 Mb and GS De Novo yielded 86,532 raw sequencing reads, based on the GS-FLX sequencer. SSRs are one of the most popular marker systems, consisting of various numbers of tandem-repeat di-, tri-, or tetra-nucleotide DNA motifs [14].
To identify SSR markers, we used the ARGOS program with default settings for V. sativa subsp. sativa singleton collections. In total, 3,811 potential SSR motifs were identified, with the majority being trinucleotide (76.3%) and dinucleotide (14.6%) repeats. There was a low rate (9%) of all other types of SSRs (e.g., tetra-, penta-, and hexa-nucleotide motifs) and the majority of trinucleotide SSRs had the GGT/GTG//TGG motif, followed by those with the ACC/CCA/CAC motif. In addition, CT/TC, AT/TA, and GA/AG motifs were abundant among the dinucleotide cDNA-SSRs. The relative proportion of SSR motif types was comparable to that of other plant species [15,16,17,18,19,20]. Kaur et al. [18] reported in theory, the frequencies of di-, tri-, tetra-, penta-, and hexanucleotide repeats should progressively decrease, based on the relative probability of replication slippage events. However, trinucleotide repeat units were predominant, followed by tetra-, di-, hexa-, and pentanucleotide repeat units.
Among the identified SSR loci, we selected 100 primer pairs on the basis of same annealing temperature, only 65 primer pairs produced single dominant polymerase chain reaction (PCR) products that were scorable for 32 accessions (Table 1 and Figure S1), The selected 65 polymorphic primer pairs sequences that were deposited in GenBank to provide a foundation for community genomic resources for vetch breeding and biotechnology research. The number of alleles (NA) per locus varied widely among the markers (Table 2) and ranged from 2 to 19, with an average of 6.6 alleles. The frequency of major alleles (MAF) per locus was 0.27–0.87 with an average of 0.508. In addition, the HO values were 0.00–0.86 with an average of 0.106, and the HE values were 0.264–0.852 with an average of 0.670. Lastly, polymorphic index content (PIC) values were 0.20–0.91, with an average of 0.59 Table 2. Considering the relatively high polymorphism levels, the cDNA-SSR markers developed in the present study will be useful for marker-assisted selection and population genetic studies to improve vetch varieties.
Table 1.
Chaμracteristics of the 65 cDNA-SSR markers for common vetch (Vicia sativa subsp. sativa).
| Marker | Primer sequence (5'-3') | Motif | GenBank Acc. No. | Ta (°C) | BLAST top hit Acc. No. | Description | E-value |
|---|---|---|---|---|---|---|---|
| GBSSR-VSspS-020 | F: CATTTGGCTGATCCTGTCA R: GGCTTCATCATGAGACAAGAA |
(TAT)5 | KF008486 | 55 | L19651.1 | Pisum sativum chloroplast photosystem I 24 kDa light harvesting protein (lhca3) mRNA, complete cds | 7e-92 |
| GBSSR-VSspS-023 | F: CCTGCATTCACAACCATTT R: CGCCATCGATGTTTTGTT |
(CAT)5 | KF008487 | 55 | None | None | None |
| GBSSR-VSspS-024 | F: ACGGTGTTAACGGTCACG R: TCTCCAAACCGACACCAG |
(AGA)5 | KF008488 | 55 | AF262939.1 | Pisum sativum chloroplast protein import component Toc159mRNA. complete cds | 1e-160 |
| GBSSR-VSspS-028 | F: TGAGCCGTTGACACAACA R: GGCGATCCTCCTACTTGAA |
(TGC)5 | KF008489 | 55 | None | None | None |
| GBSSR-VSspS-037 | F: GAAACAAGCTGAAGGCCC R: TCAGGAAATGACCAAACCA |
(GAA)5 | KF008490 | 55 | X76774.1 | P.sativum mRNA for HMG1 protein | 2e-123 |
| GBSSR-VSspS-038 | F: CTCCCCAACTTGTTCCCT R: GGGAACTTGTCGATGTGG |
(TTC)5 | KF008491 | 55 | BT143793 | Medicago truncatula clone JCVI-FLMt-3P13 unknown mRNA | 3e-41 |
| GBSSR-VSspS-042 | F: GGTTCGAGAGCTTTGCTG R: CTGTGCCACTTGACCTCC |
(CGT)5 | KF008492 | 55 | JF965421.1 | Medicago sativa ethylene response factor 11 (ERF11) mRNA, complete cds | 2e-37 |
| GBSSR-VSspS-057 | F: GAGGTTTCCGGTGAGGAG R: GTTCCAGCAGGTGAAGCA |
(GGT)7 | KF008493 | 55 | XM_003548374.1 | PREDICTED: Glycine max DEAD-box ATP-dependent RNA helicase 31-like (LOC100799999), mRNA | 7e-28 |
| GBSSR-VSspS-066 | F: AGGAGAGGCAAGGACCAG R: CACGGCTATTTTCTTCTTTTTC |
(GA)10 | KF008494 | 55 | AB078603.1 | Pisum sativum SN4TDR mRNA for 110kDa 4SNc-Tudor domin protein, complet cds | 1e-93 |
| GBSSR-VSspS-067 | F: CAAACTTGTCACCACATATACAA R: GGTGGTCACTAGTGGAGGTG |
(CCA)5 | KF008495 | 55 | KC218790.1 | Vicia faba clone NAC-VF- 218 microsatellite sequence | 3e-39 |
| GBSSR-VSspS-071 | F: GTATTCCTCTGGTGGTGGG R: CACCACCAAGACCTCCAA |
(TGG)5 | KF008496 | 55 | XM 003617084.1 | Medicago truncatula Heterogeneous nuclear ribonucleoprotein DO (MTR 5g088220) mRNA, complete cds | 9e-108 |
| GBSSR-VSspS-073 | F: CCTCCCAATCCTCCATTC R: CCCTAGTCCTCCAATTTCG |
(GTT)5 | KF008497 | 55 | XM_004506417.1 | PREDICTED: Cicer arietinum scarecrow-like protein 6-like (LOC101497219), mRNA | 2e-58 |
| GBSSR-VSspS-075 | F: TTCAGCAAGCCCATCATT R: CGTCCGTCCAATCAACAA |
(TTA)6 | KF008498 | 55 | JF768700.1 | Lens culinaris microsatellite LcSSR535 sequence | 1e-71 |
| GBSSR-VSspS-076 | F: CCTGGTCCCAGAAATGGT R: AAGCCAGAGGGCATTGAT |
(CTG)6 | KF008499 | 55 | XM_003602555.1 | Medicago truncatula NAC domin protein (MTR_3g096140) mRNA complete cds | 1e-136 |
| GBSSR-VSspS-079 | F: AAAGCAAATTGTTAAAGAAAGGG R: GAGGATGCTGCACATATGTAGTT |
(AAT)5 | KF008500 | 55 | None | None | None |
| GBSSR-VSspS-080 | F: AATGCATGGATCGAGGTG R: GAATCCATCGGCAACGTA |
(TGG)5 | KF008501 | 55 | XM_004510745.1 | PREDICTED: Cicer arietinum uncharacterized LOC 101512367 (LOC 101512367), transcript variant X3, mRNA | 6e-110 |
| GBSSR-VSspS-088 | F: CGAAGAGGTAAATGACGCC R: AGTGACCTATATTTAGCATCGTT |
(TGG)5 | KF008502 | 55 | None | None | None |
| GBSSR-VSspS-090 | F: AGACGCACCACAACAGAAA R: GGGCTAGACATGGCACAA |
(AGC)6 | KF008503 | 55 | BT149661.1 | Medicago truncatula clone JCVI-FLMt-1507 unknown mRNA | 7e-62 |
| GBSSR-VSspS-091 | F: CCAAACCAGCAAGAGCAG R: GAGCAGCGTTGTCTCGTC |
(CTT)5 | KF008504 | 55 | XM_003594377.1 | Medicago trancatula Endoglucanase (MTR_2g028480) mRNA complete cds | e-122 |
| GBSSR-VSspS-099 | F: ATCCATGCCTCTTTTGCC R: AGCCTCATTTCAGCAGCA |
(TCT)5 | KF008505 | 55 | BT137674.1 | Medicago truncatula clone JCVIMt-1708 unknown mRNA | 3e-77 |
| GBSSR-VSspS-102 | F: TTCAACGGAGATGGATCG R: CGTCTTCTTTCAGAGGCG |
(GTT)5 | KF008506 | 55 | X59773.1 | Pisum sativum mRNA for P protein. a part of glycine cleavage complex | 0.0 |
| GBSSR-VSspS-107 | F: TGGTTTCTTTCTAAAGGGGTG R: CGGCTCGATGGACAGTAG |
(GTT)5 | KF008507 | 55 | BT146949.1 | Medicago truncatula clone JCVI-FLMt-19I1 unknown | e-127 |
| GBSSR-VSspS-115 | F: CATAAACAAGGGCAAGAAAA R: GAGGAAAACATTGGTGGGA |
(TGC)6 | KF008508 | 55 | XM_004501895.1 | PREDICTED: Cicer arietinum nucleobase- ascorbate transporter 6-like (LOC101504609), transcript X2, mRNA | 6e-59 |
| GBSSR-VSspS-117 | F: CGGTGCACTAAGTGGGAA R: TTAATGATGGTGGCGAGG |
(AGG)5 | KF008509 | 55 | BT135350.1 | Medicago truncatula clone JCVI-FLMt-9H4 unknown mRNA | 2e-114 |
| GBSSR-VSspS-118 | F: GCATTTCCCTTGGTCTCC R: CAGAAAGAGCAACCGTGC |
(TGG)5 | KF008510 | 55 | XM_004514512.1 | PREDICTED: Cicer arietinum N-alpha-acetyltransferase 10-like (LOC101504041), mRNA | 3e-68 |
| GBSSR-VSspS-119 | F: CACCACCAAGACCTCCAA R: CCATCATCATCACCAGCC |
(ACC)5 | KF008511 | 55 | XM_003617084.1 | Medicago truncatula Heterogeneous nuclear ribonucleoprotein D0 (MTR_5g088220), complete cds | 9e-98 |
| GBSSR-VSspS-125 | F: GGCCGGTATTCGTCAACT R: CCCCGTATTTTCTCGGTC |
(TGG)5 | KF008512 | 55 | XM_004510745.1 | PREDICTED: Cicer arietinum uncharacterized LOC 101512367 (LOC101512367), transcript variant X3, mRNA | 2e-134 |
| GBSSR-VSspS-126 | F: TGGCGCTTATCGCTATGT R: TCCACTCATTCCACTCGT |
(TG)7 | KF008513 | 55 | XM_003623363.1 | Medicago truncatula Patellin-6 (TR_7G070480) mRNA, complete cds | 5e-81 |
| GBSSR-VSspS-129 | F: AGGAGAGGCAAGGACCAG R: CTTTTTCTCTAACTCATTCATGTC |
(GA)10 | KF008514 | 55 | AB0786031 | Pisum sativum SN4TDR mRNA for 110kDa 4SNc-Tudor domain protein, complete cds | 8e-117 |
| GBSSR-VSspS-135 | F: TGGTGGAGATTTGTTGGG R: CTTCATCTTCCCACACCG |
(TGG)5 | KF008515 | 55 | BT136030.1 | Medicago truncatula clone JCVI-FLMt-16L14unknown mRNA | 4e-40 |
| GBSSR-VSspS-138 | F: CGGAGTTCACATAAAACATACTAC R: TGGGAGTGTTGAGATGGG |
(TTA)7 | KF008516 | 55 | AB176563.1 | Vicia faba MET mRNA for type 2 metallothionein, complete cds | 6e-98 |
| GBSSR-VSspS-140 | F: TTGCTTTGATGTTTGGAGC R: CCCTAAATTCCCAACCCA |
(GGT)7 | KF008517 | 55 | XM_003613856.1 | Medicago truncatula Cysteine-rich receptor-like protein kinase (MTR_5g042440) mRNA, complete cds | 3e-86 |
| GBSSR-VSspS-156 | F: GGCCAATTTAGCGAGCTT R: CACTATCATCAACCTCTAACGGA |
(GTG)5 | KF008519 | 55 | BT134176.1 | Medicago truncatula clone JCVI-FLMt-15D24 unknown mRNA | 2e-118 |
| GBSSR-VSspS-158 | F: TGAGCTTATTGCCAGTGGA R: CCATGTCATCATCGGATTC |
(TGG)5 | KF008520 | 55 | KC218603.1 | Vicia faba clone NAC-VF-31 microsatellite sequence | 7e-124 |
| GBSSR-VSspS-162 | F: GAGACAGTGGAAGTATCGGC R: CACAGCAAATGCATCGGT |
(AAG)6 | KF008521 | 55 | BT146412.1 | Medicago truncatula clone JCVI-FLMt-21014 unknown mRNA | 1e-75 |
| GBSSR-VSspS-166 | F: GTGGCCATGATCCATTTG R: TTCCTCGAGAGGGAAAGC |
(TGG)5 | KF008522 | 55 | XM_003605932.1 | Medicago truncatula DnaJ (MTR_4g50420) mRNA complete cds | 0.0 |
| GBSSR-VSspS-172 | F: GCTTTGGAAGAGCCCAAT R: TCCAGGATTGTAACCCCC |
(TGG)5 | KF008523 | 55 | XM_003617084.1 | Medicago truncatula Heterogeneous nuclear ribonucleoprotein D0 (MTR_5g088220) mRNA, complete cds | 5e-90 |
| GBSSR-VSspS-173 | F: GGGCACGGTGGTCACTA R: TGACTACCACCACCTCCG |
(TGG)5 | KF008524 | 55 | AJ831469.1 | Pisum sativum mRNA for putative glycine rich protein precursor (grp1 gene) | 2e-13 |
| GBSSR-VSspS-179 | F: AGCTATGCGAGAGGCTCC R: CTGTGGGAAGGCACATCT |
(TGA)6 | KF008525 | 55 | None | None | None |
| GBSSR-VSspS-181 | F: CACTGTGACTCAGTTTCGTTG R: CGATTTTGAACCCTAACCG |
(TTC)5 | KF008526 | 55 | None | None | None |
| GBSSR-VSspS-182 | F: GCGTTGTGGCGTATTTCT R: TGGAGGAAAGGAAACTACTCA |
(GCA)6 | KF008527 | 55 | AB676029.1 | Lathyrus japonicas DNA, 61 locus, haplotype: D | 4e-62 |
| GBSSR-VSspS-185 | F: CTCCTCAATTTTCCCCCA R: TTTGGTGCGATTGTTTCC |
(CAT)5 | KF008528 | 55 | Ap009676.1 | Lotus japonicas genomic DNA, clone: LjT30I08, TM0492 | 4e-26 |
| GBSSR-VSspS-187 | F: CCAGGTTGCTTTCCTTACTTT R: TTAGCCCTCAAAGCCTCC |
(ATC)5 | KF008529 | 55 | X54359.1 | P.sativum mRNA of cDNA clone 26g | 2e-154 |
| GBSSR-VSspS-192 | F: AGGGTCTTCCTTCCCACA R: TATGGTGACACGTTCGCA |
(ATC)5 | KF008530 | 55 | XM_004505980.1 | PREDICTED: Cicer arietinum uncharacterized LOC101500025 (LOC101500025), transcript variant X2, mRNA | 1e-100 |
| GBSSR-VSspS-203 | F: TCCATCTGGTTGGTGGTG R: GAAAGCCAATTTTTCAGCAA |
(GTT)7 | KF008531 | 55 | BT147294.1 | Medicago truncatula clone JCVI-FLMt-15A20 unknown mRNA | 2e-50 |
| GBSSR-VSspS-217 | F: CCATCGCCACCACCA R: TCCCGGAACAAAAATCAA |
(AAC)7 | KF008532 | 55 | EF447278.1 | Pisum sativum cultivar Finale SYM8 (SYM8) gene, partial cds | 5e-58 |
| GBSSR-VSspS-245 | F: CAATAGGGGGACCCTTCA R: GCTGCAAGCTGCTACCAT |
(GGA)5 | KF008533 | 55 | HQ439603.1 | Phalaenopsis hybrid cultivar candidate developmental transcription factor TCP1 mRNA, partial cds | 5e-04 |
| GBSSR-VSspS-247 | F: GGTTCAATACGATCCATAGAATA R: TGATCGCCAATTCTGGAC |
(CAC)6 | KF008534 | 55 | KC294548.1 | Aeschynomene ciliate voucher IRRI 13078 cyclophilin 1 (CYP1) gene, complete cds | 3e-46 |
| GBSSR-VSspS-249 | F: AAAACATGGTTGAGTGTTTTTG R: TAACCCTCTCGGTTTCGG |
(ATA)5 | KF008535 | 55 | KC218749.1 | Vicia faba clone NAC-VF-177 microsatellite sequence | 2e-55 |
| GBSSR-VSspS-251 | F: TGGTGGACGTCACTATGGA R: CATGGTGCTTCCGACAAT |
(TGG)5 | KF008536 * | 55 | KC218790.1 | Vicia faba clone NAC-VF-218 microsatellite sequence | 5e-30 |
| GBSSR-VSspS-252 | F: CATGGTGCTTCCGACAAT R: TCGAAATCAGGACTTACCACA |
(CCA)5 | KF008536 * | 55 | KC218790.1 | Vicia faba clone NAC-VF-218 microsatellite sequence | 5e-30 |
| GBSSR-VSspS-262 | F: ATTGGGCCCTCTTTTTGA R: GGGGGTAGAAAAGTTGCG |
(AT)7 | KF008537 | 55 | XM_004500781.1 | PREDICTED: Cicer arietinum dolichyl- diphoshooligosaccharide-protein glycosyltransferase subunit 1A-like (LOC101502563), mRNA | 1e-37 |
| GBSSR-VSspS-268 | F: AAATTTGTCTGACGAAAAACG R: TGCTTGAGAGTGCCATCA |
(TAC)5 | KF008538 | 55 | BT144509.1 | Medicago truncatula clone JCVI-FLMt-13M6 unknown mRNA | 2e-47 |
| GBSSR-VSspS-269 | F: TTCCATTTATCCTCCTATCCTCT R: CTTGAATGCGAAACGAGG |
(CGC)5 | KF008539 | 55 | XM_004507695.1 | PREDICTED: Cicer arietinum eukaryotic translation initiation factorisoform 4G-1-like (LOC101492356), mRNA | 1e-56 |
| GBSSR-VSspS-284 | F: TGGAAGGAAATGGCAGTG R: ATCCGTTTCGGATTGGTT |
(GCA)5 | KF008540 | 55 | JX539287.1 | Vigna radiata cultivar MCV-1 clone GGSSR_911 microsatellit sequence | 2e-120 |
| GBSSR-VSspS-291 | F: CCCAACCGAACCACTTATT R: TAATAGCTCCGGCCCAGT |
(CTA)5 | KF008541 | 55 | None | None | None |
| GBSSR-VSspS-301 | F: AACCAAACAACAATGGGTT R: TCAACCGGTGAAAGATGG |
(CAA)5 | KF008542 | 55 | JN849865.1 | Medicago falcata voucher PI494662A caffeic acid-O- methyltransferase (COMT) gene.exon 1 and partial cds | 2e-128 |
| GBSSR-VSspS-304 | F: CCGTTCTACGCAATTCTCC R: CGACCAAGAACACCAGGA |
(TTC)5 | KF008543 | 55 | XM_003593951.1 | Medicago truncatula hypothetical protein (MTR_2g020190) mRNA,complete cds | 1e-55 |
| GBSSR-VSspS-305 | F: CATGAAAGAGTTTTGCACCTT R: CCGACGACGAGATTGAGA |
(GCA)5 | KF008544 | 55 | None | None | None |
| GBSSR-VSspS-308 | F: TGAGAGCATAGACAGCAAACA R: TGGATTTGGTCGCATAGC |
(AAC)5 | KF008545 | 55 | None | None | None |
| GBSSR-VSspS-309 | F: TCTTCAAAAGAGTACAAAAGGGA R: GAATTGGACACCTTGGCA |
(AAT)5 | KF008546 | 55 | BT137399.1 | Medicago truncatula clone JCVI-FlMt-19L22 unknown mRNA | 5e-124 |
| GBSSR-VSspS-310 | F: GGGTGCCCTAGCATTTGT R: ATCTCCGGCGTCAGTTTC |
(CTC)6 | KF008547 | 55 | M69105.1 | Pisum sativum outer membrane protein (OM14) mRNA, complete cds | 9e-96 |
| GBSSR-VSspS-311 | F: TTGAGGCGGTGTTGGTAG R: ATGTCATGGCCAACTGCT |
(GGA)6 | KF008548 | 55 | None | None | None |
| GBSSR-VSspS-313 | F: GAACAATGCAGCCTGGAA R: GCTGCAATCGCATTCTCT |
(TTG)5 | KF008549 | 55 | XM_003613196.1 | Medicago truncatula U-box domin containing protein (MTR_5g034440) mRNA, complete cds | 2e-112 |
TA, annealing temperature. * same sequence two primers identified.
Table 2.
Diversity statistics from initial primer screening in 32 accessions of common vetch (Vicia sativa subsp. sativa).
| Marker | NA | MAF | HO | HE | PIC |
|---|---|---|---|---|---|
| GBSSR-VSspS-020 | 4 | 0.63 | 0.00 | 0.51 | 0.43 |
| GBSSR-VSspS-023 | 7 | 0.34 | 0.00 | 0.75 | 0.71 |
| GBSSR-VSspS-024 | 7 | 0.47 | 0.00 | 0.71 | 0.67 |
| GBSSR-VSspS-028 | 4 | 0.38 | 0.00 | 0.73 | 0.68 |
| GBSSR-VSspS-037 | 4 | 0.38 | 0.00 | 0.71 | 0.65 |
| GBSSR-VSspS-038 | 7 | 0.28 | 0.00 | 0.79 | 0.76 |
| GBSSR-VSspS-042 | 2 | 0.84 | 0.00 | 0.26 | 0.23 |
| GBSSR-VSspS-057 | 8 | 0.36 | 0.03 | 0.77 | 0.74 |
| GBSSR-VSspS-066 | 7 | 0.25 | 0.00 | 0.83 | 0.81 |
| GBSSR-VSspS-067 | 6 | 0.50 | 0.00 | 0.67 | 0.62 |
| GBSSR-VSspS-071 | 3 | 0.50 | 0.00 | 0.62 | 0.54 |
| GBSSR-VSspS-073 | 8 | 0.31 | 0.00 | 0.81 | 0.79 |
| GBSSR-VSspS-075 | 6 | 0.38 | 0.03 | 0.76 | 0.72 |
| GBSSR-VSspS-076 | 3 | 0.56 | 0.00 | 0.57 | 0.50 |
| GBSSR-VSspS-079 | 6 | 0.55 | 0.00 | 0.64 | 0.60 |
| GBSSR-VSspS-080 | 3 | 0.50 | 0.00 | 0.59 | 0.51 |
| GBSSR-VSspS-088 | 5 | 0.38 | 0.00 | 0.74 | 0.70 |
| GBSSR-VSspS-090 | 7 | 0.41 | 0.00 | 0.75 | 0.71 |
| GBSSR-VSspS-091 | 4 | 0.47 | 0.00 | 0.68 | 0.64 |
| GBSSR-VSspS-099 | 4 | 0.56 | 0.00 | 0.60 | 0.54 |
| GBSSR-VSspS-102 | 8 | 0.44 | 0.03 | 0.75 | 0.73 |
| GBSSR-VSspS-107 | 3 | 0.66 | 0.00 | 0.51 | 0.45 |
| GBSSR-VSspS-115 | 4 | 0.48 | 0.03 | 0.54 | 0.44 |
| GBSSR-VSspS-117 | 6 | 0.41 | 0.00 | 0.71 | 0.67 |
| GBSSR-VSspS-118 | 3 | 0.53 | 0.00 | 0.61 | 0.54 |
| GBSSR-VSspS-119 | 2 | 0.65 | 0.00 | 0.46 | 0.35 |
| GBSSR-VSspS-125 | 3 | 0.41 | 0.00 | 0.63 | 0.56 |
| GBSSR-VSspS-126 | 7 | 0.25 | 0.00 | 0.82 | 0.79 |
| GBSSR-VSspS-129 | 7 | 0.25 | 0.00 | 0.81 | 0.79 |
| GBSSR-VSspS-135 | 5 | 0.50 | 0.00 | 0.62 | 0.55 |
| GBSSR-VSspS-138 | 11 | 0.32 | 0.06 | 0.83 | 0.81 |
| GBSSR-VSspS-140 | 8 | 0.22 | 0.03 | 0.81 | 0.79 |
| GBSSR-VSspS-151 | 5 | 0.67 | 0.03 | 0.52 | 0.48 |
| GBSSR-VSspS-156 | 5 | 0.66 | 0.00 | 0.53 | 0.50 |
| GBSSR-VSspS-158 | 5 | 0.59 | 0.00 | 0.57 | 0.52 |
| GBSSR-VSspS-162 | 6 | 0.53 | 0.00 | 0.66 | 0.63 |
| GBSSR-VSspS-166 | 6 | 0.38 | 0.00 | 0.74 | 0.70 |
| GBSSR-VSspS-172 | 3 | 0.41 | 0.00 | 0.65 | 0.57 |
| GBSSR-VSspS-173 | 4 | 0.58 | 0.00 | 0.58 | 0.53 |
| GBSSR-VSspS-179 | 8 | 0.56 | 0.03 | 0.64 | 0.61 |
| GBSSR-VSspS-181 | 11 | 0.28 | 0.00 | 0.85 | 0.84 |
| GBSSR-VSspS-182 | 7 | 0.38 | 0.00 | 0.76 | 0.73 |
| GBSSR-VSspS-185 | 10 | 0.31 | 0.03 | 0.82 | 0.80 |
| GBSSR-VSspS-187 | 5 | 0.44 | 0.00 | 0.69 | 0.64 |
| GBSSR-VSspS-192 | 3 | 0.63 | 0.00 | 0.51 | 0.43 |
| GBSSR-VSspS-203 | 6 | 0.34 | 0.00 | 0.76 | 0.72 |
| GBSSR-VSspS-217 | 11 | 0.28 | 0.00 | 0.83 | 0.81 |
| GBSSR-VSspS-245 | 7 | 0.28 | 0.00 | 0.81 | 0.78 |
| GBSSR-VSspS-247 | 6 | 0.69 | 0.03 | 0.49 | 0.46 |
| GBSSR-VSspS-249 | 6 | 0.56 | 0.00 | 0.64 | 0.61 |
| GBSSR-VSspS-251 | 5 | 0.45 | 0.00 | 0.70 | 0.66 |
| GBSSR-VSspS-252 | 3 | 0.50 | 0.00 | 0.61 | 0.53 |
| GBSSR-VSspS-262 | 9 | 0.47 | 0.00 | 0.72 | 0.70 |
| GBSSR-VSspS-268 | 3 | 0.47 | 0.00 | 0.64 | 0.57 |
| GBSSR-VSspS-269 | 4 | 0.50 | 0.00 | 0.58 | 0.49 |
| GBSSR-VSspS-284 | 6 | 0.44 | 0.00 | 0.66 | 0.60 |
| GBSSR-VSspS-291 | 5 | 0.53 | 0.00 | 0.64 | 0.59 |
| GBSSR-VSspS-301 | 3 | 0.53 | 0.00 | 0.58 | 0.50 |
| GBSSR-VSspS-304 | 7 | 0.46 | 0.00 | 0.68 | 0.63 |
| GBSSR-VSspS-305 | 3 | 0.69 | 0.00 | 0.48 | 0.43 |
| GBSSR-VSspS-308 | 6 | 0.54 | 0.00 | 0.66 | 0.63 |
| GBSSR-VSspS-309 | 8 | 0.34 | 0.00 | 0.78 | 0.75 |
| GBSSR-VSspS-310 | 8 | 0.38 | 0.03 | 0.75 | 0.71 |
| GBSSR-VSspS-311 | 7 | 0.44 | 0.00 | 0.73 | 0.70 |
| GBSSR-VSspS-313 | 5 | 0.44 | 0.00 | 0.71 | 0.67 |
| Mean | 5.7 | 0.460 | 0.006 | 0.670 | 0.624 |
NA, number of alleles; MAF, major allele frequency; HO, observed heterozygosity; HE, expected heterozygosity; PIC, polymorphic information content.
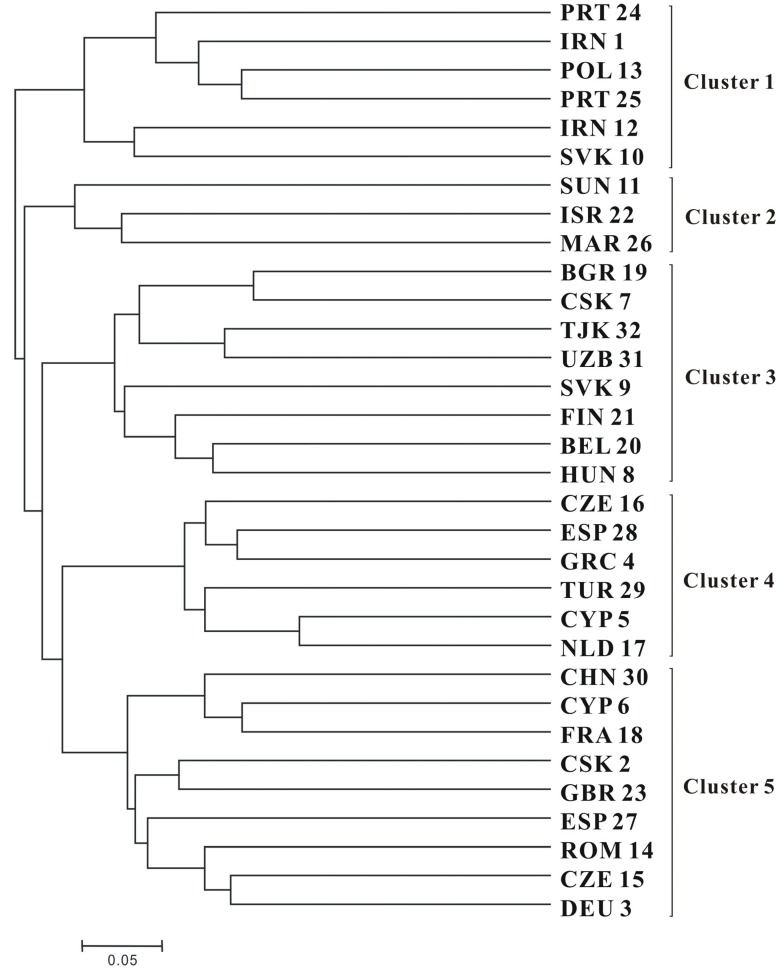
The dendrogram showed that the 32 common vetch accessions fell into five distinct clusters (Figure 1). Cluster 1 comprised accessions from South west Europe, West Asia and Central Europe each regions had 2 accessions. Cluster 2contained 3 accessions one from Eurasian, one from middle East Asia and one from North Africa. Cluster 3 contained eight accessions, five accessions from Central Europe and remaining accessions from South East Europe, north Europe and West Europe each region had one accessions. Cluster 4 contained six accessions, two accessions from South East Europe and one accession from each geographical region (Central Europe, South West Europe, Europe and North West Europe). Cluster 5 included nine accessions, two accessions from central Europe and two accessions from Europe regions, with rest of the accessions from East Asia, West Europe, South West Europe, Central and South East Europe and West General Europe, North Europe and West Europe each having one accession.
Figure 1.
Dendrogram generated using UPGMA cluster analysis based on genetic diversity of 32 common vetch (Vicia sativa subsp. sativa) accessions.
Dongi et al [21] reported cluster analysis of Trigonella foenumgraecum there was no clear clustering pattern of geographically closer accessions indicating that the association between genetic similarity and geographical distance was less significant. However, it is necessary to use more number of accessions from each geographical location to confirm the available pattern.
3. Experimental
3.1. Plant Material
Vicia sativa sativa seeds were selected from the National Agrobiodiversity Center, Rural Development Administration, Suwon, Korea (Table 3). Seedlings were germinated and grown in a glasshouse. The leaves of young seedlings were used to extract the mRNA required to synthesize the cDNA library and for 454 sequencing.
Table 3.
List of common vetch (Vicia sativa subsp. sativa) accessions.
| No. | Temp. ID | USDA-ARS No. | Country of origin | Geographical region of origin |
|---|---|---|---|---|
| 1 | K193581 | PI 226487 | Iran | West Asia |
| 2 | K193582 | PI 284058 | Czech Republic | Central Europe |
| 3 | K193583 | PI 284068 | Germany | Western Central Europe |
| 4 | K193584 | PI 284078 | Greece | South East Europe |
| 5 | K193585 | PI 284402 | Cyprus | Europe |
| 6 | K193586 | PI 284409 | Cyprus | Europe |
| 7 | K193587 | PI 284470 | Czech Republic | Central Europe |
| 8 | K193588 | PI 284471 | Hungary | Central Europe |
| 9 | K193589 | PI 308111 | Slovakia | Central Europe |
| 10 | K193590 | PI 308118 | Slovakia | Central Europe |
| 11 | K193591 | PI 325513 | Soviet Union | Eurasian |
| 12 | K193592 | PI 381065 | Iran | West Asia |
| 13 | K193593 | PI 393870 | Poland | Central Europe |
| 14 | K193594 | PI 393871 | Romania | Central and South East Europe |
| 15 | K193595 | PI 393872 | Czech Republic | Central Europe |
| 16 | K193596 | PI 393873 | Czech Republic | Central Europe |
| 17 | K193597 | PI 393877 | Netherlands | North West Europe |
| 18 | K193598 | PI 393878 | France | West Europe |
| 19 | K193599 | PI 393891 | Bulgaria | South East Europe |
| 20 | K193600 | PI 393904 | Belgium | West Europe |
| 21 | K193601 | PI 393907 | Finland | North Europe |
| 22 | K193602 | PI 393909 | Israel | Middle East Asia |
| 23 | K193603 | PI 393910 | United Kingdom | Europe |
| 24 | K193604 | PI 393912 | Portugal | South West Europe |
| 25 | K193605 | PI 493307 | Portugal | South West Europe |
| 26 | K193606 | PI 517191 | Morocco | North Africa |
| 27 | K193607 | PI 533741 | Spain | South West Europe |
| 28 | K193608 | PI 533743 | Spain | South West Europe |
| 29 | K193609 | PI 557498 | Turkey | South East Europe |
| 30 | K193610 | PI 577751 | China | East Asia |
| 31 | K193611 | PI 628288 | Uzbekistan | Central Europe |
| 32 | K193612 | PI 664293 | Tajikistan | Central Europe |
(Temp ID), Korean GeneBank ID; (ARS No.), USDA-ARS Number.
3.2. cDNA Preparation
Total RNA was extracted from Vicia sativa subsp. sativa leaves that were frozen in liquid nitrogen, ground into a powder, and then extracted using an RNeasy Plant Mini kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. The integrity of total RNA was determined using a BIOSPEC-NANO spectrophotometer (Shimadzu, Kyoto, Japan) and agarose gel electrophoresis. mRNA was purified using the PolyATract mRNA Isolation System IV (Promega, Madison, WI, USA), and the purified products were used to synthesize full-length cDNAs using a ZAP-cDNA Synthesis kit (Stratagene, Santa Clara, CA, USA). Finally, cDNA was fragmented by nebulization for library construction.
3.3. Library Preparation
Approximately 1 µg cDNA was used to generate a DNA library to use with the Genome Sequencer GS-FLX Titanium System (Roche, 454 Life Science, Branford, CT, USA). The cDNA fragment ends were polished (blunted), and two short adapters were ligated to both ends according to standard procedures described previously. The adapters, along with the sequencing key, a short sequence of four nucleotides used by the system’s software for base calling, provided priming of the sequences for both the amplification and sequencing of the sample library fragments. Following the repair of any nicks in the double-stranded library, the unbound strand of each fragment was released (with 5-Adaptor A). Finally, the quality of this single-stranded template DNA library was assessed using a 2100 BioAnalyzer (Agilent, Waldbronn, Germany). The library was quantified to determine the optimal amount needed as input for emulsion-based clonal amplification.
3.4. 454 Pyrosequencing
Single effective copies of template species from the DNA library to be sequenced were hybridized to DNA capture beads. Then the immobilized library was resuspended in an amplification solution, and the mixture was emulsified, followed by PCR amplification. The DNA-carrying beads were recovered from the emulsion and enriched after amplification. The second strands of the amplified products were melted, leaving the amplified single-stranded DNA library bound to the beads. Then the sequencing primer was annealed to the immobilized amplified DNA templates. After amplification, a single DNA-carrying bead was placed into each well of a PicoTiterPlate (PTP) device. Simultaneous sequencing with multiple samples on a single PTP (four-region gasket) was used. Then the PTP was inserted into the FLX Genome Titanium sequencer for pyrosequencing [22,23], and sequencing reagent was flowed sequentially over the plate. Information from the PTP wells was captured simultaneously by a camera, and the images were processed in real-time by an onboard computer. Multiplex identifiers were used to specifically tag unique samples in a GS FLX Titanium sequencing run, which were recognized by the GS data analysis software after the sequencing run and provided high confidence for assigning individual sequencing reads to the correct sample. Sequence assembly was performed after sequencing using GS De Novo Assembler software (Roche) to produce contigs and singletons. All sequence data were conformed to references using GS Reference Mapper software (Roche).
3.5.Discovery of cDNA-SSR Markers
All contigs and singletons from both transcriptomes were used to mine SSR motifs, and SSR motifs were identified using the ARGOS pipeline program (version 1.46) at the default settings to survey the molecular markers present in the V. sativa subsp. sativa accessions. Parameters were designed for identifying perfect di-, tri-, tetra-, penta-, and hexa-nucleotide motifs with a minimum of six repeats. The primer design parameters were set as follows: length range, 18–23 nucleotides with 21 as optimum; PCR product size range, 100–400 bp; optimum annealing temperature, 55 °C; and GC content 40–60%, with 50% as optimum. Vicia sativa subsp. sativa genomic DNA was extracted from 32 diverse common vetch accessions for cDNA-SSR marker validation using a DNeasy® Plant Mini kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s instructions. Fresh leaf tissue from each accession was used for each extraction and ground well using liquid nitrogen. DNA was resuspended in 100 μL water, and dilutions were made to 10 ng/μL followed by storage at either −20 °C or −80 °C. Randomly selected cDNA-SSR primer pairs were validated experimentally, and forward primers were synthesized by adding the M13 sequence to enable the addition of a fluorescent tail through the PCR amplification process [24]. PCR conditions included a hot-start at 95 °C for 10 min, followed by 10 cycles at 94 °C for 30 s, 60–50 °C for 30 s and 72 °C for 30 s, followed by 25 cycles at 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 30 s, with a final elongation step of 72 °C for 10 min. PCR products were separated and visualized using the QIAxcel Gel Electrophoresis System (Qiagen).
3.6. Data Analysis
The amplified SSR loci were scored for 32 accessions. The total number of alleles (NA), major allele frequency (allele with the highest frequency) (MAF), observed heterozygosity (counting heterozygocity) (HO), expected heterozygosity (HE), number of genotypes (NG), and polymorphic information content (PIC) were calculated using PowerMarker and GenAlEx (version 6.5) [25].
The expected heterozygosity formula is as follows:
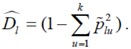 |
(1) |
A closely related diversity measure is the polymorphism information content (PIC) [26]:
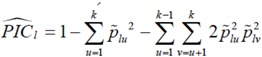 |
(2) |
The cluster analysis of 32 accessions was carried out based onunweighted pair group method with arithmetic mean (UPGMA ) phylogenetic and uprooted tree construction, based on the “CS chord 1967” distance method [27] in powermarker
4. Conclusions
We developed 65 cDNA-SSR markers, which were used successfully to investigate the genetic diversity among 32 accessions of Vicia stiva subsp. sativa. Considering the relatively high PIC values (0.59 in average), cDNA-SSR in Vicia sativa subsp. sativa is suggested to be an informative genetic marker system, which can also be applied to population genetic studies and marker-assisted selection to mine and accumulate useful alleles to increase the agronomic potential of vetch varieties.
Acknowledgments
This study was carried out with the support of the “Research Program for Agricultural Science & Technology Development (Project No. PJ008623)”, National Academy of Agricultural Science, Rural Development Administration, Korea.
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/18/7/8376/s1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Available from the authors Institute.
References
- 1.Hueze V., Tran G., Baumont R. Common vetch (Vicia sativa) Feedipedia. 2011;12:53–62. [Google Scholar]
- 2.Sullivan P. Overview of cover crops and green manures. ATTRA. 2003:1–16. [Google Scholar]
- 3.Tate M., Ennenking D. Common vetch (Vicia sativa ssp. sativa): Feed or future food. Grain Legumes. 2006:16–17. [Google Scholar]
- 4.Tate M.E., Enneking D. A mess of red pottage. Nature. 1992;359:357–358. doi: 10.1038/359357a0. [DOI] [PubMed] [Google Scholar]
- 5.Tate M.E., Rathjen J., Delaere I., Enneking D. Covert trade in toxic vetch continues. Nature. 1999;400:207–207. doi: 10.1038/22198. [DOI] [PubMed] [Google Scholar]
- 6.Thavarajah P., Thavarajah D., Premakumara G.A., Vandenberg A. Detection of Common Vetch (Vicia sativa L.) in Lentil (Lens culinaris L.) using unique chemical fingerprint markers. Food Chem. 2012;135:2203–2206. doi: 10.1016/j.foodchem.2012.06.124. [DOI] [PubMed] [Google Scholar]
- 7.Uzun A., Gucer S., Acikgoz E. Common vetch (Vicia sativa L.) germplasm: correlations of crude protein and mineral content to seed traits. Plant Foods Hum. Nutr. 2011;66:254–260. doi: 10.1007/s11130-011-0239-z. [DOI] [PubMed] [Google Scholar]
- 8.Firincioglu H.K., Unal S., Erberkas E., Dogruyol L. Relationships between seed yield and yield components in common vetch (Vicia sativa ssp. sativa) populations sown in spring and autumn in central Turkey. Field Crop. Res. 2010;116:30–37. doi: 10.1016/j.fcr.2009.11.005. [DOI] [Google Scholar]
- 9.Firincioglu H.K., Erbektas E., Dogruyol L., Mutlu Z., Unal S., Karakurt E. Phenotypic variation in autumn and spring-sown vetch (Vicia sativa ssp.) populations in central Turkey. Span. J. Agric. Res. 2009;7:596–606. [Google Scholar]
- 10.Mutz K.O., Heilkenbrinker A., Lonne M., Walter J.G., Stahl F. Transcriptome analysis using next-generation sequencing. Curr. Opin. Biotechnol. 2012;24:22–30. doi: 10.1016/j.copbio.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Ronaghi M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 2001;11:3–11. doi: 10.1101/gr.11.1.3. [DOI] [PubMed] [Google Scholar]
- 12.Varshney R.K., Close T.J., Singh N.K., Hoisington D.A., Cook D.R. Orphan legume crops enter the genomics era! Curr. Opin. Plant Biol. 2009;12:202–210. doi: 10.1016/j.pbi.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Luro F.L., Costantino G., Terol J., Argout X., Allario T., Wincker P., Talon M., Ollitrault P., Morillon R. Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics. 2008;9:287–310. doi: 10.1186/1471-2164-9-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim T.S., Booth J.G., Gauch H.G., Jr., Sun Q., Park J, Lee Y.H., Lee K.G. Simple sequence repeats in Neurospora crassa: Distribution, Polymorphism and evolutionary inference. BMC Genomics. 2008;9:31–48. doi: 10.1186/1471-2164-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Fang B., Chen J., Zhang X., Luo Z., Huang L., Chen X., Li Y. De novo assembly and characterization of root transcriptome using Illumina paired-end sequencing and development of cSSR markers in sweet potato (Ipomoea batatas) BMC Genomics. 2010;11:726–739. doi: 10.1186/1471-2164-11-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blanca J., Canizares J., Roig C., Ziarsolo P., Nuez F., Pico B. Transcriptome characterization and high throughput SSRs and SNPs discovery in Cucurbita pepo (Cucurbitaceae) BMC Genomics. 2011;12:104–119. doi: 10.1186/1471-2164-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moe K.T., Chung J.W., Cho Y.I, Moon J.K., Ku J.H., Jung J.K., Lee J., Park Y.J. Sequence information on simple sequence repeats and single nucleotide polymorphisms through transcriptome analysis of mungbean. J. Integr. Plant Biol. 2011;53:63–73. doi: 10.1111/j.1744-7909.2010.01012.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaur S., Pembleton L.W., Cogan N.O., Savin K.W., Leonforte T., Paull J., Materne M., Forster J.W. Transcriptome sequencing of field pea and faba bean for discovery and validation of SSR genetic markers. BMC Genomics. 2012;13:104–116. doi: 10.1186/1471-2164-13-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanase K., Nishitani C., Hirakawa H., Isobe S., Tabata S., Ohmiya A., Onozaki T. Transcriptome analysis of carnation (Dianthus caryophyllus L.) based on next-generation sequencing technology. BMC Genomics. 2012;13:292–303. doi: 10.1186/1471-2164-13-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang T., Bao S.Y., Ford R., Jia T.J., Guan J.P., He Y.H., Sun X.L., Jiang J., Hao J., Zhang X., Zong X. High-throughput novel microsatellite marker of faba bean via next generation sequencing. BMC Genomics. 2012;13:602–613. doi: 10.1186/1471-2164-13-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dangi R.S., Lagu M.D., Choudhary L.B., Ranjekar P.K., Gupta V.S. Assessment of genetic diversity in Trigonella foenum-graecum and Trigonella caerulea using ISSR and RAPD markers. BMC Plant Biol. 2004;4:13–24. doi: 10.1186/1471-2229-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elahi E., Ronaghi M. Pyrosequencing: a tool for DNA sequencing analysis. Methods Mol. Biol. 2004;255:211–219. doi: 10.1385/1-59259-752-1:211. [DOI] [PubMed] [Google Scholar]
- 23.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben LA., Berka J., Braverman MS., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley M. Functions of the gene products of Escherichia coli. Microbiol. Mol. Biol. Rev. 1993;57:862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peakall R., Smouse P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botstein D., White R.L., Skolnick M., Davis R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 27.Cavalli-Sforza L.L., Edwards A.W.F. Phylogenetic Analysis: Models and Estimation Procedures. Am. J. Hum. Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.