Abstract
Extracellular and intracellular oxidants or electrophiles are key contributors to the damages in cellular macromolecules, such as DNA, proteins and lipids. Nrf2 is a master transcription factor that modulates a cellular antioxidant response program and plays an important role in the protection against oxidants and electrophiles. Keap1 is a regulator of Nrf2 by serving as a substrate adaptor for Cullin3-dependent E3 ubiquitin ligase. While Nrf2 activation is a feasible strategy for treatment of age-related diseases, aberrant Nrf2 activation also confers a selective growth advantage of tumor cells during chemotherapy or radiotherapy. In the present review, we provide an overview of the Keap1-Nrf2-ARE system, the domain organization of Nrf2 and Keap1, and the regulatory mechanisms of Nrf2 proteolysis by Keap1. We also discuss how Nrf2 prevents tumor promotion, hampers the sensitivity of selected tumors against chemotherapy or radiotherapy, and reprograms the metabolism to facilitate the tumor proliferation. Finally, we illustrate the current status in the development of Nrf2 chemical activators and inhibitors for the use of potential chemopreventive agents and chemotherapeutic adjuvants, respectively.
Keywords: Keap1, Nrf2, ARE, oncogene, tumor suppressor gene, metabolism
1. Regulation of Cellular Homeostasis by Nrf2 and Keap1
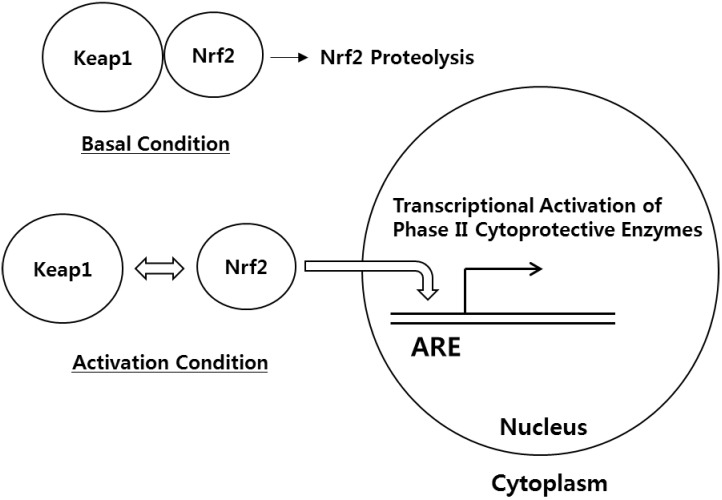
Living cells maintain homeostasis by adjusting the intracellular oxidation and reduction levels. However, this intricate balance is constantly challenged by various environmental oxidants and electrophiles, such as pollutants, food additives, ultraviolet, and ionizing radiations [1]. In addition, the generation of intracellular reactive oxygen species (ROS) as by-products during oxidative phosphorylation, including superoxide anion (O2−), hydroxyl radical (OH−), and hydrogen peroxide (H2O2) contributes to the perturbation of homeostasis [2]. It is well accepted that chronic exposure of human to electrophilic and oxidative stress contributes to a broad range of age-related pathogenesis, such as cancer, cardiovascular, and neurodegenerative diseases [3]. In order to combat against these insults, mammalian cells have developed the elaborate defense mechanisms to confer appropriate adaptive responses [4]. The ubiquitously expressed NF-E2-related factor-2 (Nrf2) is a transcription factor that modulates the basal and inducible expression of phase II cytoprotective enzymes, such as heme oxygenase-1 (HO-1), NAD[P]H:quinone oxidoreductase-1 (NQO1), superoxide dismutase (SOD), glutathione S-transferase (GST), and γ-glutamyl cysteine ligase (γ-GCL) [5]. Nrf2 regulates the transcriptional activation of these enzymes by binding to a nucleotide motif sequence existing in 5'-upstream promoter region, e.g., the antioxidant response element (ARE) [6]. ARE contains a consensus sequence 5'-TGACnnnGC-3' and was first identified in the regulatory regions of rat and mouse Gst and Nqo1 genes [7]. Nrf2 is sequestered by the Kelch-like ECH-associated protein 1 (Keap1) in the cytosol and constitutively targeted for poly-ubiquitination under a basal condition. Exposure of cells to electrophilic and oxidative stress, however, releases Nrf2 from Keap1, which enables Nrf2 to translocate to the nucleus and activate a battery of phase II cytoprotective genes by binding to the ARE (Figure 1). In addition to phase II cytoprotective genes, Nrf2 is known to control the transcription of various drug metabolizing enzymes (DMEs), transporters, and cellular reducing equivalents (GSH and NADPH) [8].
Figure 1.
Regulation of ARE-dependent Gene Expression by Keap1 and Nrf2.
2. Domain Architecture of Nrf2 and Keap1 and Their Functional Interplay
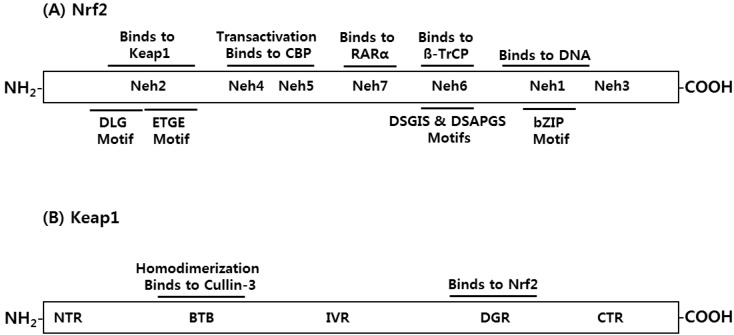
In addition to the transcriptional activation of phase II cytoprotective enzymes, Nrf2 is linked to a number of other cellular processes, such as differentiation, proliferation, cell growth, apoptosis and hematopoiesis by closely interacting with diverse intracellular regulatory proteins [9]. Nrf2 is a basic leucine zipper (bZIP) transcription factor that contains a Cap’n’Collar (CNC) structure and most transcription factors that harbor the CNC structure are transcriptional activators [10]. Domain analysis of Nrf2 has illustrated that Nrf2 is composed of seven conserved Nrf2-ECH homology (Neh) domains (Figure 2A). The Neh1 domain contains the bZIP motif, which enables Nrf2 to bind to the ARE sequence. In addition, this domain can interact with UbcM2, E2 ubiquitin conjugating enzyme, to regulate the Nrf2 protein stability [11]. The Neh2 domain, located in the most N-terminal region, acts as a negative regulatory domain by binding to Keap1 [12]. The Neh3 domain, located in the most C-terminal region, is known to assist Nrf2 transactivation through the interaction with chromatin remodeling protein CHD6 [13], while the tandem Neh4 and Neh5 domains are critical for Nrf2-mediated ARE transactivation by binding to a transcriptional coactivator, e.g., CBP [14]. The redox-insensitive Neh6 domain possesses DSGIS and DSAPGS motifs and provides a phosphorylation-dependent binding platform for β-TrCP [15]. Finally, Wang et al. have recently identified the novel domain in Nrf2, e.g., the Neh7 domain, that interacts with the retinoic acid receptor α (RARα) and represses Nrf2 target gene expression [16].
Figure 2.
Conserved Domains of Nrf2 and Keap1 Proteins. (A) Nrf2 contains seven Neh domains (Neh1-7), in which the Neh1 domain binds to DNA using the bZIP motif and the Neh2 domain interacts with Keap1 using the DLG and ETGE motifs. The Neh4 and Neh5 domains are required for gene transactivation. The Neh6 domain binds to β-TrCP using DSGIS and DSAPGS motifs. The Neh7 domain binds to RARα and suppresses the Nrf2 activity; (B) Keap1 contains five different domains (NTR, BTB, IVR, DGR and CTR), in which the BTB domain forms a homodimer for binding to Cullin-3 and the DGR domain forms a six-blade propeller with 6x Kelch motifs for the interaction with Nrf2.
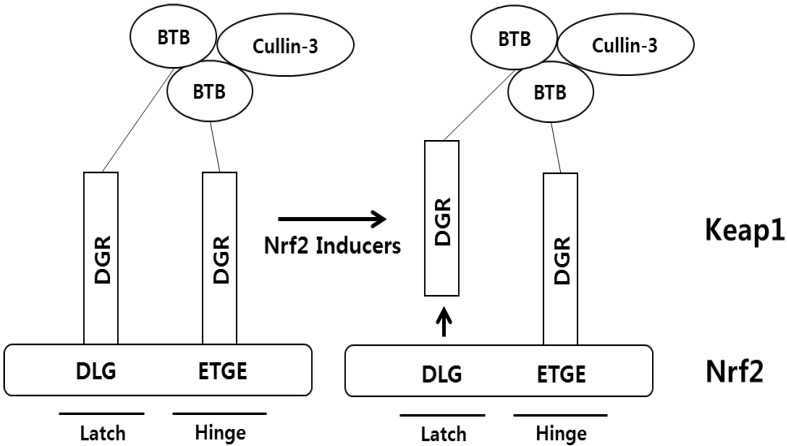
Keap1 is a cytosolic protein that inhibits the ARE-dependent gene expression by binding to the Neh2 domain of Nrf2. In fact, Keap1 was initially identified by yeast 2-hybrid assay, using the Neh2 domain of Nrf2 as bait [17]. Keap1 consists of 5 different domains: an amino-terminal region (NTR), a Broad complex, Tramtrack and Bric a brac domain (BTB), an intervening region (IVR), six Kelch/double glycine repeats (DGR), and a carboxyl terminal region (CTR) (Figure 2B). The cytoplasmic location of Keap1 can be explained, at least in part, by its binding ability to a cytoplasmic actin or myosin VIIa through the DGR domain [18]. Keap1 also employs the DGR regions to recognize two primary sequences, e.g., the ETGE and DLG motifs, existing in the Neh2 domain of Nrf2 protein by forming a six-bladed propeller [19]. In addition, two interesting features underlying the interaction between Nrf2 and Keap1 exists. First, Keap1 can dimerize with each other, using the BTB domain to interact with Cullin-3. Second, two Keap1 proteins bind to a single Nrf2 protein at a ratio of 2:1 [20], in which the overlapping ETGE and DLG motifs in Nrf2 bind to two Keap1 proteins with a differential affinity: a single Keap1 strongly binds to the ETGE motif of Nrf2 (Ka = 20 × 107 M−1) and, at the same time, another Keap1 interacts with the DLG motif with a weak affinity (Ka = 0.1 × 107 M−1) [21]. Based on these observations, so called the “hinge and latch” hypothesis was proposed to explain the regulatory mechanism of Nrf2 by Keap1 (Figure 3), in which the “hinge” mediates a high-affinity interaction between the ETGE motif of Nrf2 and Keap1 and this interaction is unaffected by stress inducers, whereas the “latch” mediates displacement of the DLG motif of Nrf2 from Keap1 in response to treatment of Nrf2 inducers [22].
Figure 3.
The “Hinge and Latch” Hypothesis. Under basal conditions, Keap1 forms a homodimer and associates with Cullin-3 protein. At the same time, the DGR domains of two Keap1 bind to the DLG (latch) and the ETGE (hinge) domains in a single Nrf2. In response to Nrf2 inducers, the DLG motif in Nrf2, but not ETGE motif in Nrf2, is released from the DGR domain in Keap1.
The cellular Nrf2 protein level is mediated, largely in part, by the ubiquitin-mediated proteolysis [23]. Ubiquitin is a 76 amino-acid protein whose main function is to mark proteins for degradation. The ubiquitin-mediated proteolysis requires a cascade of three enzymes: E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin-ligase) enzymes [24]. The E3 ubiquitin ligases contain either the homologous to E6-associated protein (E6-AP) COOH-terminus (HECT) domain or the really interesting new gene (RING) finger domain [25]. While the HECT-type E3 ubiquitin ligases display a catalytic activity by itself, the RING finger-type E3 ubiquitin ligase promotes the poly-ubiquitination of substrates by positioning substrates in a close proximity to the activated E2 enzymes (ROC1 or ROC2) through molecular assembly by Cullin proteins [26]. Cullins (Culs) consist of seven different isotypes in human (Cul1, 2, 3, 4A, 4B, 5, and 7) and serve as scaffold proteins to assemble the Cullin-RING E3 ubiquitin ligases [27]. Since Keap1 possesses the BTB domain, Keap1 behaves as an adaptor module for Cul3-type E3 ubiquitin ligase complex, contributing to a constant poly-ubiquitination of Nrf2 in a stretch of lysine (K) residues that exist in the ETGE-DLG intervening region of Nrf2 [28]. Additionally, recent studies have illustrated that the β-transducin repeat-containing protein (β-TrCP), an adaptor protein for the Cul1-Skp1-ROC1 E3 ubiquitin ligase complex, also targets Nrf2 for poly-ubiquitination and proteolysis, following GSK3β-mediated phosphorylation upon the DSGIS motif existing in the Neh6 domain of Nrf2 (Figure 2A) [29]. A follow-up study illustrated that the Neh6 domain possesses an additional recognition motif for β-TrCP, e.g., DSAPGS motif and that the binding of β-TrCP to this motif is not influenced by GSK activity (Figure 2A) [30]. At present, the detailed mechanisms how Keap1 and β-TrCP discriminate Nrf2 for poly-ubiquitination are largely unclear.
Dinkova-Kostova and colleagues have recently attempted to visualize the molecular interaction between EGFP-Nrf2 and Keap1-mCherry in cells by using a quantitative fluorescence resonance energy transfer (FRET) technique, coupled with fluorescence lifetime imaging microscopy (FLIM). FRET is a physical mechanism that describes the energy transfer between two light-sensitive molecules and frequently used to interrogate the proximity between two proteins in cells [31]. Based on the FLIM-FRET observations, they found that the Keap1/Nrf2 complex exists in two different conformations: (1) an open conformation in which only the high affinity “ETGE” motif, but not the low affinity “DLG” motif in Nrf2 is bound to Keap1 dimers and (2) a closed conformation in which both the “DLG” and “ETGE” motifs are bound to Keap1 dimers [32]. Contrary to the “hinge and latch hypothesis”, they observed that treatment of Nrf2 inducers [sulforaphane and sulfoxythiocarbamate (STCA)] increased the abundance of closed conformation in cells. Based on these data, authors proposed a new model, related to the Keap1/Nrf2 interaction and referred it to as “conformational cycling model” or “cyclic sequential attachment and regeneration model” According to this model, Nrf2 inducers function to promote Nrf2 stabilization by facilitating the formation of closed conformation, but not open conformation, of the Keap1-Nrf2 complex [33]. Although the Keap1-Nrf2 complex exists in the closed conformation, the direct binding of Nrf2 inducers to Keap1 seems to uncouple Nrf2 for targeted degradation possibly due to Keap1 conformational changes. As a result, Nrf2 is not released from Keap1 and, therefore, the free Keap1 dimer is not regenerated in cells. Because the newly synthesized Nrf2 is unable to bind to Keap1 due to the lack of free Keap1 dimers, Nrf2 is free to translocate into the nucleus and activates ARE-dependent gene expression.
3. Nrf2: Tumor Suppressor Gene or Oncogene?
Nrf2 knock-out mice are highly susceptible for drug-induced toxicity and oxidative stress-related diseases, such as cardiovascular diseases, acute lung injury, diabetes, and inflammation [34]. In addition, there are a number of studies illustrating that Nrf2 knock-out mice are more prone to develop chemically-induced tumors, compared with their wild-type littermates [35,36,37]. Similar to this observation, a conditional Keap1 disruption in mice reduced the level of cell damages, caused by oxidative stress and prevented the onset and exacerbation of stress-induced diseases [38]. Together, these results indicate that Nrf2 is a critical oxidative stress regulator. In addition, the attempts to elucidate novel Nrf2 or Keap1 binding proteins and their functions in cells have illustrated that Nrf2 is a tumor suppressor, important for the inhibition of tumor initiation. It is well established that, in response to mild genotoxic or oxidative stresses, p53 activates the transcriptional expression of p21, which in turn causes the cell cycle arrest or senescence at the G1/S boundary in normal cells by binding to G1/S cyclin-dependent kinases [39]. Interestingly, Zhang and colleagues have identified that p21 protein can bind to the DLG motif in Nrf2 and increase the Nrf2 stability by displacing Nrf2 from Keap1-dependent poly-ubiquitination [40]. This result implies that the anti-tumorigenic effects of Nrf2 can be attributed, at least in part, to its interaction with the p53-p21 axis and the resulting cell cycle arrest that favors the DNA damage repair [41]. The other Keap1 binding proteins, such as the Wilms tumor gene on the X chromosome (WTX) [42], PALB2/FANCN [43], and dipeptidyl peptidase 3 (DPP3) [44] also contain the ETGE motif in common and can promote the Nrf2 protein stability by competitively binding to Keap1. In particular, WTX and PALB2 are considered as bona fide tumor suppressors [45]. Therefore, it can be concluded that WTX and PALB2-mediated tumor suppression can be ascribed, at least in part, to maintaining an increased cellular Nrf2 level.
Although it seems counter-intuitive, Nrf2 activity is highly up-regulated in various types of tumors and the prognosis of patients with tumors expressing high levels of Nrf2 is poor [46]. In addition, somatic gain of function mutations in Nrf2 gene were identified in tumors and these mutations are clustered in the proximity of the DLG and ETGE motifs, suggesting that the hinge and latch hypothesis holds in the clinical setting [47,48,49]. Moreover, somatic heterozygous loss of function mutations in Keap1 were also identified in various tumor tissues [46]. Using a genetically-engineered mouse model, Suzuki et al. have demonstrated that Keap1 heterozygous mutation is sufficient to cause an increased Nrf2 activity [50]. These observations illustrate that cancer cells can hijack the Keap1/Nrf2 pathway to keep their intracellular ROS burden within a range, thereby permitting a proper growth and survival for tumor cells. Supporting this view, DeNicola et al. have reported that the activation of oncogenes, such as K-Ras, B-Raf, and c-Myc stimulates Nrf2 transcription via MAPK pathway and this event contributes to oncogene-stimulated cell proliferation in vivo by maintaining the intracellular ROS level below a threshold [51]. However, it appears that a simple accumulation of Nrf2 is not sufficient to incur a spontaneous tumor development [52]. Therefore, it would be the most appropriate to interpret that Nrf2 exhibits tumor-suppressive functions in normal cells, e.g., “a good side of Nrf2”, but cancer cells hijack the survival property of Keap1-Nrf2 pathway to confer a selective advantage to sustain their proliferation and stress resistance, e.g., “a bad side of Nrf2”. This conjecture is well supported by a recent observation that Nrf2 prevents initiation, but accelerates progression of tumors during urethane-induced lung carcinogenesis in mice [53].
A growing amount of evidence indicates that Nrf2 can rewire metabolic programs to promote cell growth and proliferation in cancer [54]. Mitsuishi et al. have demonstrated that Nrf2 can redirect glucose and glutamine into anabolic pathways [55]. In addition, Nrf2 regulates the expression of glucose-6-phosphoate dehydrogenase (G6PD), 6-phosphogluconate dehydrogenase (PGD), malic enzyme-1 (ME1), and isocitrate dehydrogenase-1 (IDH1), all of which can convert oxidized NADP+ into its reduced form, NADPH [56]. Because NADPH provides a reducing power in cells and is required for the biosynthesis of lipids and nucleotides, it is possible to assume that Nrf2 might contribute to an increased cell proliferation in tumors by providing a sufficient amount of NADPH. Another interesting target gene of Nrf2 is the pyruvate kinase (PK), a critical and final glycolytic enzyme that irreversibly converts phosphoenolpyruvate (PEP) into pyruvate. Interestingly, Nrf2 was shown not to increase, but rather decrease the PK expression [57]. Although the detailed mechanism is unclear yet, it is possible to surmise that a decreased PK expression would have promoted the accumulation of glycolytic intermediates, which were in turn diverted towards the pentose phosphate pathway. In addition to the regulatory of Nrf2 in metabolic genes, fumarate, a metabolite in the tricarboxylicacid (TCA) cycle, was shown to form a direct adduct with Keap1 and activates the Nrf2-dependent gene expression in the hereditary form of type 2 papillary renal carcinoma [58]. Likewise, an accumulation of fumarate modified selected cysteine residues in Keap1 and abrogated the function of Keap1 to promote Nrf2 proteolysis and it led to an increase in the renal cyst formation of fumarate hydratase (FH)-deficient mice [59]. These observations fit well with the postulated tumor suppressor functions of certain Krebs cycle enzymes, such as isocitrate dehydrogenase (IDH), succinate dehydrogenase (SD) and fumarate hydratase (FH) [60]. Further detailed mechanistic studies, addressing how accumulated metabolites in the intermediary metabolism cooperate with the Keap1/Nrf2 pathway to assist the tumor development will be an interesting theme of future investigations.
4. Development of Chemical Nrf2 Inducers and Inhibitors
Chemoprevention has emerged as an important research discipline in recent years [61]. Natural chemopreventive agents, such as curcumin, sulforaphane are strong chemical inducers of ARE-dependent phase II cytoprotective enzymes [62]. Accumulating evidence indicates that ARE-dependent gene activation by chemopreventive compounds is mediated by modulating the activities of intracellular signaling kinase pathways. For example, the mitogen-activated protein kinases (MAPKs) are implicated in the regulation of ARE-dependent gene expression, in which ERK and JNK are positive regulators while p38 MAPK is a negative regulator in response to treatment of chemopreventive agents [63]. On the other hand, there exist numerous intracellular kinases that are involved in ARE-dependent gene regulation as well, such as phosphatidylinositol 3'-kinase (PI3K), PKR-like endoplasmic reticulum kinase (PERK), protein kinase C (PKC), Fyn kinase, and glycogen synthase kinase-3β (GSK3β) [64]. However, the detailed molecular mechanisms underlying how Nrf2 phosphorylations contribute to ARE-dependent gene expression are largely unclear except for PKCδ and Fyn kinases. Niture et al. have reported that PKCδ activation results in phosphorylation of Nrf2 at Ser40 and it contributes to the escape or release of Nrf2 from Keap1 [65]. On the other hand, Jain et al. have reported that activation of GSK3β directly phosphorylates Fyn, leading to a nuclear localization of Fyn and Nrf2 phosphorylation at Tyr568 [66]. Interestingly, they have observed that Fyn-mediated Nrf2 phosphorylation at Tyr568 suppressed Nrf2 transactivation by facilitating a nuclear export of Nrf2.
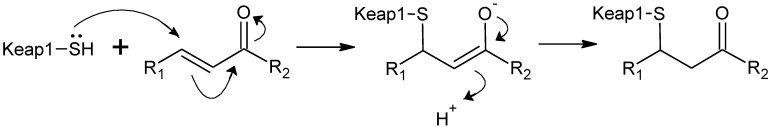
In an insightful review, Magesh et al. have classified around 90 natural or synthetic Nrf2 chemical activators as follows: (1) Michael acceptors; (2) oxidizable phenols and quinones; (3) isothiocyanates and sulfoxythiocarbamates; (4) dithiolethiones and diallyl sulfides; (5) trivalent arsenicals; (6) vicinal dimercaptans; (7) selenium-based compounds; (8) polyenes; (9) hydroxyperoxides; (10) heavy metals and metal complexes; and (11) miscellaneous inducers [67]. While the biochemical mechanisms how these chemicals induce ARE-dependent gene expression are largely elusive, it is known that Michael acceptors promote Nrf2-dependent ARE-gene expression by forming a covalent adduct with the multiple sulfhydryl groups of cysteines in Keap1, an interesting hypothesis referred to as the “cysteine code hypothesis”. According to this hypothesis, Keap1, but not Nrf2, is a stress sensor. Michael acceptors are directly conjugated to cysteine (Cys) residues in Keap1 (Scheme 1) and these modifications lead to a conformational change of Keap1, resulting in the release of Nrf2 from the low affinity binding site (the latch) from poly-ubiquitination [68]. Among more than 20 cysteine residues found in Keap1, Cys-151. Cys-273, and Cys-288 were identified to serve as critical residues of stress sensors in vivo [69]. In addition, Hayes and colleagues have demonstrated using mass spectrometry that endogenous or exogenous chemical inducers of Nrf2 modified different Cys residues of Keap1 in cells [70]. Because a large number of Michael acceptors are found in naturally-occurring chemopreventive agents [curcumin, caffeic acid phenethyl ester (CAPE), flavonoids and sesquiterpenes as examples] all of which possess olefins conjugated with electron-withdrawing carbonyl groups in common, finding out whether and which Cys residues in Keap1 these natural compounds directly modify to exhibit Nrf2-dependent phase II cytoprotective gene activation is necessary to better clarify their chemopreventive mechanisms.
Scheme 1.
Adduct Formation of Keap1 with Michael Acceptor.
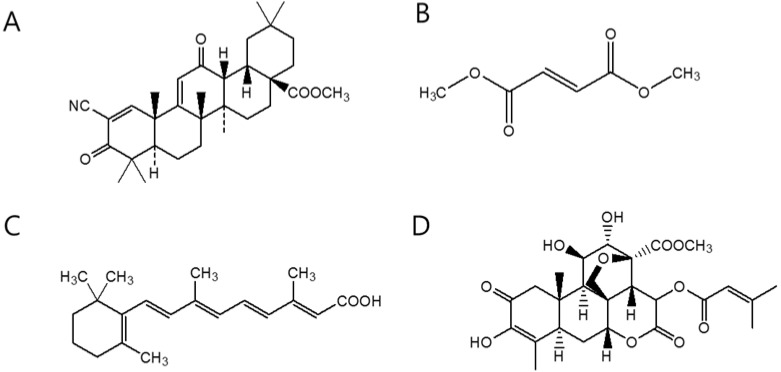
Considering that oxidative stress and inflammation are intimately correlated with each other, pharmaceutical companies have recently attempted to develop Nrf2 chemical activators as antioxidant inflammation modulators (AIMs). One of the most promising Nrf2 inducers was the methyl ester derivative of the synthetic triterpenoid, 2-cyano-3,12-dioxooleana-1,9(11)-dien-28-oic acid (CDDO-Me, Figure 4A). Because CDDO-Me induces Nrf2 at low nanomolar concentrations [71], it underwent development as a promising AIM under the generic name, bardoxolone methyl for treatment of advanced chronic kidney disease (CKD) and type 2 diabetes mellitus [72]. Unfortunately, the clinical trial of bardoxolone methyl was terminated in the phase III in October 2012 for safety concerns due to excess serious adverse events and mortality. Dimethyl fumarate (DMF, Figure 4B) is another synthetic Nrf2 activator that has been developed for potential AIM. Scannevin et al. have demonstrated that treatment of primary cultured central nervous system (CNS) cells or animals with DMF resulted in an increased nuclear level of Nrf2 and Phase II cytoprotective genes, thereby rendering a significant protection against oxidative challenges [73]. More importantly, an oral preparation of DMF successfully reduced the annual relapse rate in patients with multiple sclerosis (MS) in the phase III clinical trial [74] and the Food and Drug Administration (FDA) finally approved DMF for treatment of MS in March 2013 under the trade name of Tecfidera. Whether DMF possesses chemopreventive activities is unknown at present.
Figure 4.
Chemical Structures of (A) CDDO-Me; (B) DMF; (C) ATRA; and (D) brusatol.
On the other hand, the development of Nrf2 chemical inhibitors started to garner a significant interest in recent years due to the realization that a high level of Nrf2 is implicated in the chemoresistance and radioresistance [75]. Wolf and colleagues have previously reported that all-trans retinoic acid (ATRA, Figure 4C) is a chemical inhibitor of Nrf2. As such, they have demonstrated that treatment of ATRA or other retinoic acid receptor-α (RARα) agonists reduced the ability of Nrf2 binding to the ARE and suppressed the induction of ARE-driven genes by butylated hydroxyanisole (BHA) and tert-butyl hydroxyquinone (tBHQ). [76]. It is well known that retinoids, including ATRA exhibit significant chemopreventive effects against head and neck cancer [77]. The effects of retinoids are mediated by specific nuclear receptors, e.g., retinoic acid receptors (RAR-α, -β, and -γ) and retinoid X receptors (RXR-α, -β, and -γ), in which RAR and RXR form a heterodimer and exerts their biological effects by binding to retinoic acid response elements (RAREs) in the genome [78]. In addition to RARα, other nuclear receptors, such as peroxisome proliferator-activated receptor-γ (PPARγ), estrogen receptor-α (ERα), estrogen-related receptor-ß (ERRβ), and glucocorticoid receptor (GR) are reported to physically interact with Nrf2 and repress Nrf2-dependent gene expression [79], raising an intriguing possibility that Nrf2 can be implicated in hormone-related diseases. In addition to ATRA, several other natural chemicals have been reported to exert inhibitory effects on the Nrf2-ARE pathway. Zhang and colleagues have demonstrated that brusatol (Figure 4D), purified from the extract of plant Brucea javanica suppressed Nrf2-dependent gene expression and increased the sensitivity of cisplatin-mediated apoptosis of cancer cells in vitro and in vivo [80]. Similarly, recent studies have illustrated that several natural chemicals, such as luteolin [81], procyanidin [82], apigenin [83], and chrysin [84] can suppress the Nrf2 activity. The molecular mechanisms by which these natural compounds exhibit inhibitory effects on the Nrf2-ARE pathway are largely unclear. However, it should be noted that there exist other literatures illustrating that these compounds can act as Nrf2 activators in different experiment settings. Therefore, further studies will be necessary to establish the specificity and accurate mechanisms of action of these chemicals, if they are intended for development of chemotherapeutic adjuvants in the future.
5. Concluding Remarks
By now, we have discussed the regulatory mechanisms of the Keap1-Nrf2-ARE pathway. However, a growing evidence indicates that the regulation of the Keap1-Nrf2-ARE is more complex than thought and other molecular mechanisms are also involved. Among them, we believe that the epigenetic regulation of Nrf2 [85] and Keap1 [86] will particularly an interesting theme of future studies. Because Nrf2 exhibits protective effects against oxidants and electrophiles in normal cells but sustained Nrf2 activation favor the growth of tumors, it is possible to speculate that differential epigenetic regulation of Nrf2 and/or Keap1 might exist in normal and cancer cells. In addition, we are now beginning to appreciate that Nrf2 plays an important role in the regulation of hormonal outcomes and metabolism. Therefore, the implication of Nrf2 in hormonal and metabolic diseases warrants further investigations considering that the Keap1-Nrf2-ARE pathway is now moving into the realm of drug development.
Acknowledgments
Y.-S.K. was supported by the GRRC program of Gyeonggi province. [(GRRC-DONGGUK2014-B02), Development of novel recombinant enzymes and diagnostic platform], B.Y.C. was supported by Bio-Theme Cluster program of Korea Industrial Complex Corp (KICOX) [2013055-1].
Author Contributions
Y.-S.K. and B.Y.C wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Surh Y.J., Kundu J.K., Na H.K. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008;74:1526–1539. doi: 10.1055/s-0028-1088302. [DOI] [PubMed] [Google Scholar]
- 2.Circu M.L., Aw T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohen R., Nyska A. Oxidation of biological systems: Oxidative stress phenomena, antioxidants, redox reactions, and methods for their quantification. Toxicol. Pathol. 2002;30:620–650. doi: 10.1080/01926230290166724. [DOI] [PubMed] [Google Scholar]
- 4.Sykiotis G.P., Bohmann D. Stress-activated cap’n’collar transcription factors in aging and human disease. Sci. Signal. 2010 doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kundu J.K., Surh Y.J. Inflammation: Gearing the journey to cancer. Mutat. Res. 2008;659:15–30. doi: 10.1016/j.mrrev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Kensler T.W., Wakabayashi N., Biswal S. Cell survival responses to environmental stresses via the keap1-nrf2-are pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 7.Rushmore T.H., Morton M.R., Pickett C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 8.Ma Q. Transcriptional responses to oxidative stress: Pathological and toxicological implications. Pharmacol. Ther. 2010;125:376–393. doi: 10.1016/j.pharmthera.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Wakabayashi N., Slocum S.L., Skoko J.J., Shin S., Kensler T.W. When Nrf2 talks, who’s listening? Antioxid Redox Signal. 2010;13:1649–1663. doi: 10.1089/ars.2010.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sykiotis G.P., Habeos I.G., Samuelson A.V., Bohmann D. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr. Opin. Clin. Nutr. Metab. Care. 2011;14:41–48. doi: 10.1097/MCO.0b013e32834136f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plafker K.S., Nguyen L., Barneche M., Mirza S., Crawford D., Plafker S.M. The ubiquitin-conjugating enzyme UbcM2 can regulate the stability and activity of the antioxidant transcription factor Nrf2. J. Biol. Chem. 2010;285:23064–23074. doi: 10.1074/jbc.M110.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niture S.K., Khatri R., Jaiswal A.K. Regulation of Nrf2-an update. Free Radic. Biol. Med. 2014;66:36–44. doi: 10.1016/j.freeradbiomed.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nioi P., Nguyen T., Sherratt P.J., Pickett C.B. The carboxy-terminal Neh3 domain of Nrf2 is required for transcriptional activation. Mol. Cell. Biol. 2005;25:10895–10906. doi: 10.1128/MCB.25.24.10895-10906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh Y., Itoh K., Yoshida E., Miyagishi M., Fukamizu A., Yamamoto M. Two domains of Nrf2 cooperatively bind CBP, a creb binding protein, and synergistically activate transcription. Genes Cells. 2001;6:857–868. doi: 10.1046/j.1365-2443.2001.00469.x. [DOI] [PubMed] [Google Scholar]
- 15.McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J.D. Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 2004;279:31556–31567. doi: 10.1074/jbc.M403061200. [DOI] [PubMed] [Google Scholar]
- 16.Wang H., Liu K., Geng M., Gao P., Wu X., Hai Y., Li Y., Li Y., Luo L., Hayes J.D., et al. Rxralpha inhibits the Nrf2-are signaling pathway through a direct interaction with the Neh7 domain of Nrf2. Cancer Res. 2013;73:3097–3108. doi: 10.1158/0008-5472.CAN-12-3386. [DOI] [PubMed] [Google Scholar]
- 17.Itoh K., Wakabayashi N., Katoh Y., Ishii T., Igarashi K., Engel J.D., Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang M.I., Kobayashi A., Wakabayashi N., Kim S.G., Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc. Natl. Acad. Sci. USA. 2004;101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Padmanabhan B., Tong K.I., Kobayashi A., Yamamoto M., Yokoyama S. Structural insights into the similar modes of Nrf2 transcription factor recognition by the cytoplasmic repressor Keap1. J. Synchrotron Radiat. 2008;15:273–276. doi: 10.1107/S090904950705114X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong K.I., Kobayashi A., Katsuoka F., Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: A hinge and latch mechanism. Biol. Chem. 2006;387:1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 21.Tong K.I., Padmanabhan B., Kobayashi A., Shang C., Hirotsu Y., Yokoyama S., Yamamoto M. Different electrostatic potentials define etge and dlg motifs as hinge and latch in oxidative stress response. Mol. Cell. Biol. 2007;27:7511–7521. doi: 10.1128/MCB.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uruno A., Motohashi H. The Keap1-Nrf2 system as an in vivo sensor for electrophiles. Nitric Oxide. 2011;25:153–160. doi: 10.1016/j.niox.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Bassermann F., Eichner R., Pagano M. The ubiquitin proteasome system-Implications for cell cycle control and the targeted treatment of cancer. Biochim. Biophys. Acta. 2014;1843:150–162. doi: 10.1016/j.bbamcr.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickart C.M. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/S0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 25.Sumara I., Maerki S., Peter M. E3 ubiquitin ligases and mitosis: Embracing the complexity. Trends Cell Biol. 2008;18:84–94. doi: 10.1016/j.tcb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Chen C., Seth A.K., Aplin A.E. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol. Cancer Res. 2006;4:695–707. doi: 10.1158/1541-7786.MCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama K.I., Nakayama K. Ubiquitin ligases: Cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 28.Furukawa M., Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol. Cell. Biol. 2005;25:162–171. doi: 10.1128/MCB.25.1.162-171.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rada P., Rojo A.I., Chowdhry S., McMahon M., Hayes J.D., Cuadrado A. SCF/{beta}-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 2011;31:1121–1133. doi: 10.1128/MCB.01204-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chowdhry S., Zhang Y., McMahon M., Sutherland C., Cuadrado A., Hayes J.D. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene. 2013;32:3765–3781. doi: 10.1038/onc.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoki K., Kamioka Y., Matsuda M. Fluorescence resonance energy transfer imaging of cell signaling from in vitro to in vivo: Basis of biosensor construction, live imaging, and image processing. Dev. Growth Differ. 2013;55:515–522. doi: 10.1111/dgd.12039. [DOI] [PubMed] [Google Scholar]
- 32.Baird L., Lleres D., Swift S., Dinkova-Kostova A.T. Regulatory flexibility in the Nrf2-mediated stress response is conferred by conformational cycling of the Keap1-Nrf2 protein complex. Proc. Natl. Acad. Sci. USA. 2013;110:15259–15264. doi: 10.1073/pnas.1305687110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baird L., Swift S., Lleres D., Dinkova-Kostova A.T. Monitoring Keap1-Nrf2 interactions in single live cells. Biotechnol. Adv. 2014 doi: 10.1016/j.biotechadv.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Copple I.M. The Keap1-Nrf2 cell defense pathway-A promising therapeutic target? Adv. Pharmacol. 2012;63:43–79. doi: 10.1016/B978-0-12-398339-8.00002-1. [DOI] [PubMed] [Google Scholar]
- 35.Xu C., Huang M.T., Shen G., Yuan X., Lin W., Khor T.O., Conney A.H., Kong A.N. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 36.Ramos-Gomez M., Dolan P.M., Itoh K., Yamamoto M., Kensler T.W. Interactive effects of Nrf2 genotype and oltipraz on benzo[a]pyrene-DNA adducts and tumor yield in mice. Carcinogenesis. 2003;24:461–467. doi: 10.1093/carcin/24.3.461. [DOI] [PubMed] [Google Scholar]
- 37.Fahey J.W., Haristoy X., Dolan P.M., Kensler T.W., Scholtus I., Stephenson K.K., Talalay P., Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc. Natl. Acad. Sci. USA. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohkoshi A., Suzuki T., Ono M., Kobayashi T., Yamamoto M. Roles of Keap1-Nrf2 system in upper aerodigestive tract carcinogenesis. Cancer Prev. Res. 2013;6:149–159. doi: 10.1158/1940-6207.CAPR-12-0401-T. [DOI] [PubMed] [Google Scholar]
- 39.Vousden K.H., Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 40.Chen W., Sun Z., Wang X.J., Jiang T., Huang Z., Fang D., Zhang D.D. Direct interaction between Nrf2 and p21(Cip1/WAF1) upregulates the Nrf2-mediated antioxidant response. Mol. Cell. 2009;34:663–673. doi: 10.1016/j.molcel.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toledano M.B. The guardian recruits cops: The p53-p21 axis delegates prosurvival duties to the keap1-nrf2 stress pathway. Mol. Cell. 2009;34:637–639. doi: 10.1016/j.molcel.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Camp N.D., James R.G., Dawson D.W., Yan F., Davison J.M., Houck S.A., Tang X., Zheng N., Major M.B., Moon R.T. Wilms tumor gene on X chromosome (WTX) inhibits degradation of Nrf2 protein through competitive binding to Keap1 protein. J. Biol. Chem. 2012;287:6539–6550. doi: 10.1074/jbc.M111.316471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma J., Cai H., Wu T., Sobhian B., Huo Y., Alcivar A., Mehta M., Cheung K.L., Ganesan S., Kong A.N., et al. PALB2 interacts with Keap1 to promote Nrf2 nuclear accumulation and function. Mol. Cell. Biol. 2012;32:1506–1517. doi: 10.1128/MCB.06271-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hast B.E., Goldfarb D., Mulvaney K.M., Hast M.A., Siesser P.F., Yan F., Hayes D.N., Major M.B. Proteomic analysis of ubiquitin ligase Keap1 reveals associated proteins that inhibit Nrf2 ubiquitination. Cancer Res. 2013;73:2199–2210. doi: 10.1158/0008-5472.CAN-12-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitsuishi Y., Motohashi H., Yamamoto M. The Keap1-Nrf2 system in cancers: Stress response and anabolic metabolism. Front. Oncol. 2012;2:200. doi: 10.3389/fonc.2012.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taguchi K., Motohashi H., Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 47.Shibata T., Kokubu A., Gotoh M., Ojima H., Ohta T., Yamamoto M., Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 48.Shibata T., Ohta T., Tong K.I., Kokubu A., Odogawa R., Tsuta K., Asamura H., Yamamoto M., Hirohashi S. Cancer related mutations in Nrf2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc. Natl. Acad. Sci. USA. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solis L.M., Behrens C., Dong W., Suraokar M., Ozburn N.C., Moran C.A., Corvalan A.H., Biswal S., Swisher S.G., Bekele B.N., et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin. Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki T., Maher J., Yamamoto M. Select heterozygous Keap1 mutations have a dominant-negative effect on wild-type Keap1 in vivo. Cancer Res. 2011;71:1700–1709. doi: 10.1158/0008-5472.CAN-10-2939. [DOI] [PubMed] [Google Scholar]
- 51.DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taguchi K., Maher J.M., Suzuki T., Kawatani Y., Motohashi H., Yamamoto M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell. Biol. 2010;30:3016–3026. doi: 10.1128/MCB.01591-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satoh H., Moriguchi T., Takai J., Ebina M., Yamamoto M. Nrf2 prevents initiation but accelerates progression through the kras signaling pathway during lung carcinogenesis. Cancer Res. 2013;73:4158–4168. doi: 10.1158/0008-5472.CAN-12-4499. [DOI] [PubMed] [Google Scholar]
- 54.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 55.Mitsuishi Y., Taguchi K., Kawatani Y., Shibata T., Nukiwa T., Aburatani H., Yamamoto M., Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22:66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 56.Wu K.C., Cui J.Y., Klaassen C.D. Beneficial role of Nrf2 in regulating nadph generation and consumption. Toxicol. Sci. 2011;123:590–600. doi: 10.1093/toxsci/kfr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yates M.S., Tran Q.T., Dolan P.M., Osburn W.O., Shin S., McCulloch C.C., Silkworth J.B., Taguchi K., Yamamoto M., Williams C.R., et al. Genetic versus chemoprotective activation of Nrf2 signaling: Overlapping yet distinct gene expression profiles between Keap1 knockout and triterpenoid-treated mice. Carcinogenesis. 2009;30:1024–1031. doi: 10.1093/carcin/bgp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ooi A., Wong J.C., Petillo D., Roossien D., Perrier-Trudova V., Whitten D., Min B.W., Tan M.H., Zhang Z., Yang X.J., et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell. 2011;20:511–523. doi: 10.1016/j.ccr.2011.08.024. [DOI] [PubMed] [Google Scholar]
- 59.Adam J., Hatipoglu E., O’Flaherty L., Ternette N., Sahgal N., Lockstone H., Baban D., Nye E., Stamp G.W., Wolhuter K., et al. Renal cyst formation in fh1-deficient mice is independent of the hif/phd pathway: Roles for fumarate in Keap1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. doi: 10.1016/j.ccr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gottlieb E., Tomlinson I.P. Mitochondrial tumour suppressors: A genetic and biochemical update. Nat. Rev. Cancer. 2005;5:857–866. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 61.Lippman S.M., Hawk E.T. Cancer prevention: From 1727 to milestones of the past 100 years. Cancer Res. 2009;69:5269–5284. doi: 10.1158/0008-5472.CAN-09-1750. [DOI] [PubMed] [Google Scholar]
- 62.Jeong W.S., Keum Y.S., Chen C., Jain M.R., Shen G., Kim J.H., Li W., Kong A.N. Differential expression and stability of endogenous nuclear factor E2-related factor 2 (Nrf2) by natural chemopreventive compounds in HepG2 human hepatoma cells. J. Biochem. Mol. Biol. 2005;38:167–176. doi: 10.5483/bmbrep.2005.38.2.167. [DOI] [PubMed] [Google Scholar]
- 63.Keum Y.S., Jeong W.S., Kong A.N. Chemoprevention by isothiocyanates and their underlying molecular signaling mechanisms. Mutat. Res. 2004;555:191–202. doi: 10.1016/j.mrfmmm.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 64.Keum Y.S. Regulation of Nrf2-mediated phase ii detoxification and anti-oxidant genes. Biomol. Ther. (Seoul) 2012;20:144–151. doi: 10.4062/biomolther.2012.20.2.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niture S.K., Jain A.K., Jaiswal A.K. Antioxidant-induced modification of INrf2 cysteine 151 and PKC-delta-mediated phosphorylation of Nrf2 serine 40 are both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. J. Cell Sci. 2009;122:4452–4464. doi: 10.1242/jcs.058537. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 66.Jain A.K., Jaiswal A.K. Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J. Biol. Chem. 2006;281:12132–12142. doi: 10.1074/jbc.M511198200. [DOI] [PubMed] [Google Scholar]
- 67.Magesh S., Chen Y., Hu L. Small molecule modulators of Keap1-Nrf2-are pathway as potential preventive and therapeutic agents. Med. Res. Rev. 2012;32:687–726. doi: 10.1002/med.21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bryan H.K., Olayanju A., Goldring C.E., Park B.K. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 2013;85:705–717. doi: 10.1016/j.bcp.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto T., Suzuki T., Kobayashi A., Wakabayashi J., Maher J., Motohashi H., Yamamoto M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 2008;28:2758–2770. doi: 10.1128/MCB.01704-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McMahon M., Lamont D.J., Beattie K.A., Hayes J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liby K.T., Sporn M.B. Synthetic oleanane triterpenoids: Multifunctional drugs with a broad range of applications for prevention and treatment of chronic disease. Pharmacol. Rev. 2012;64:972–1003. doi: 10.1124/pr.111.004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crunkhorn S. Deal watch: Abbott boosts investment in Nrf2 activators for reducing oxidative stress. Nat. Rev. Drug Discov. 2012;11:96. doi: 10.1038/nrd3655. [DOI] [PubMed] [Google Scholar]
- 73.Scannevin R.H., Chollate S., Jung M.Y., Shackett M., Patel H., Bista P., Zeng W., Ryan S., Yamamoto M., Lukashev M., et al. Fumarates promote cytoprotection of central nervous system cells against oxidative stress via the nuclear factor (erythroid-derived 2)-like 2 pathway. J. Pharmacol. Exp. Ther. 2012;341:274–284. doi: 10.1124/jpet.111.190132. [DOI] [PubMed] [Google Scholar]
- 74.Fox R.J., Miller D.H., Phillips J.T., Hutchinson M., Havrdova E., Kita M., Yang M., Raghupathi K., Novas M., Sweetser M.T., et al. Placebo-controlled phase 3 study of oral BG-12 or glatiramer in multiple sclerosis. N. Engl. J. Med. 2012;367:1087–1097. doi: 10.1056/NEJMoa1206328. [DOI] [PubMed] [Google Scholar]
- 75.Suzuki T., Motohashi H., Yamamoto M. Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol. Sci. 2013;34:340–346. doi: 10.1016/j.tips.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 76.Wang X.J., Hayes J.D., Henderson C.J., Wolf C.R. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc. Natl. Acad. Sci. USA. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khuri F.R., Lippman S.M., Spitz M.R., Lotan R., Hong W.K. Molecular epidemiology and retinoid chemoprevention of head and neck cancer. J. Natl. Cancer Inst. 1997;89:199–211. doi: 10.1093/jnci/89.3.199. [DOI] [PubMed] [Google Scholar]
- 78.Bushue N., Wan Y.J. Retinoid pathway and cancer therapeutics. Adv. Drug Deliv. Rev. 2010;62:1285–1298. doi: 10.1016/j.addr.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Namani A., Li Y., Wang X.J., Tang X. Modulation of Nrf2 signaling pathway by nuclear receptors: Implications for cancer. Biochim. Biophys. Acta. 2014;1843:1875–1885. doi: 10.1016/j.bbamcr.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 80.Ren D., Villeneuve N.F., Jiang T., Wu T., Lau A., Toppin H.A., Zhang D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA. 2011;108:1433–1438. doi: 10.1073/pnas.1014275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang X., Wang H., Fan L., Wu X., Xin A., Ren H., Wang X.J. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic. Biol. Med. 2011;50:1599–1609. doi: 10.1016/j.freeradbiomed.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 82.Ohnuma T., Matsumoto T., Itoi A., Kawana A., Nishiyama T., Ogura K., Hiratsuka A. Enhanced sensitivity of A549 cells to the cytotoxic action of anticancer drugs via suppression of Nrf2 by procyanidins from cinnamomi cortex extract. Biochem. Biophys. Res. Commun. 2011;413:623–629. doi: 10.1016/j.bbrc.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 83.Gao A.M., Ke Z.P., Wang J.N., Yang J.Y., Chen S.Y., Chen H. Apigenin sensitizes doxorubicin-resistant hepatocellular carcinoma BEL-7402/ADM cells to doxorubicin via inhibiting PI3K/Akt/Nrf2 pathway. Carcinogenesis. 2013;34:1806–1814. doi: 10.1093/carcin/bgt108. [DOI] [PubMed] [Google Scholar]
- 84.Gao A.M., Ke Z.P., Shi F., Sun G.C., Chen H. Chrysin enhances sensitivity of BEL-7402/ADM cells to doxorubicin by suppressing PI3K/Akt/Nrf2 and ERK/Nrf2 pathway. Chem. Biol. Interact. 2013;206:100–108. doi: 10.1016/j.cbi.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 85.Yu S., Khor T.O., Cheung K.L., Li W., Wu T.Y., Huang Y., Foster B.A., Kan Y.W., Kong A.N. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of tramp mice. PLoS One. 2010;5:e8579. doi: 10.1371/journal.pone.0008579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Muscarella L.A., Parrella P., D’Alessandro V., la Torre A., Barbano R., Fontana A., Tancredi A., Guarnieri V., Balsamo T., Coco M., et al. Frequent epigenetics inactivation of Keap1 gene in non-small cell lung cancer. Epigenetics. 2011;6:710–719. doi: 10.4161/epi.6.6.15773. [DOI] [PubMed] [Google Scholar]