Abstract
From ethyl acetate-methanol extracts of leaves and twigs of Pseuduvaria trimera a new aporphine alkaloid; 8-hydroxy-1,4,5-trimethoxy-7-oxoaporphine or 8-hydroxyartabonatine C (1) was isolated, together with the known 1,2,3-trimethoxy-4,5-dioxo-6a,7-dehydroaporphine (ouregidione, 2). Their structures were elucidated by a combination of spectral methods; mainly 2D NMR; IR and MS. Compounds 1 and 2 exhibited cytotoxic activity with IC50 values of 26.36 ± 5.18 μM and 12.88 ± 2.49 μM, respectively, for human hepatocellular carcinoma HepG2 cells, and 64.75 ± 4.45 and 67.06 ± 3.5 μM, respectively, for human breast cancer MDA-MB231 cells. Both compounds displayed anti-cancer activity but less than that of doxorubicin; a conventional chemotherapeutic drug, the IC50 levels of which were 2.21 ± 1.72 and 1.83 ± 0.09 μM for HepG2 and MDA-MB231 cells, respectively.
Keywords: aporphine alkaloid, cytotoxicity, anti-cancer, Pseuduvaria trimera, Annonaceae
1. Introduction
P. trimera belongs to the family Annonaceae, which includes about eight species in Thailand (P. gardneri, P. macrophylla, P. monticola, P. muitiovulata, P. reticulata, P. rugosa, P. setosa, P. trimera) [1]. It forms evergreen and deciduous broad forests on base lime-stone mountains. P. trimera is classified as a tree, 20 m tall, branches pale gray, densely puberulent when young, glabrescent [2].
Previous work on plants of this genus have revealed the presence of alkaloids in every Pseuduvaria species investigated such as aporphine, 1,2,3-trimethoxy-4,5-dioxo-6a,7-dehydroaporphine and O-methylmoschatoline [3], N-methylouregidione, liriodenine, oxostephanine [4], pseuduvarines A and B [5], oxoanolobine [6] and 1,2,3-trimethoxy-5-oxonoraporphine (ouregidione) [4,7]. However, no phytochemical investigation of this plant species has been reported to date. Pseuduvaria species are traditionally used to treat fever, nausea, headache and stomach ailment and have been mostly studied for alkaloids [8]. In our search for biologically active constituents from this plant, we have subjected its extracts to in vitro screening for anticancer activity against two cancer cell lines (human hepatocellular carcinoma HepG2 cells and human breast cancer MDA-MB231cells). The goal of this study was to describe the isolation, structural elucidation and cytotoxicity of aporphine alkaloids from P. trimera.
2. Results and Discussion
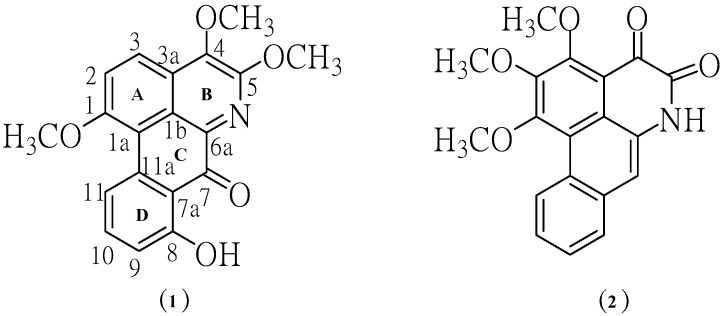
Phytochemical investigation of the ethyl acetate-methanol extract obtained from the mixture of leaves and twigs of P. trimera led to the isolation of two alkaloids, namely 8-hydroxy-1,4,5-trimethoxy-7-oxoaporphine (1) and 1,2,3-trimethoxy-4,5-dioxo-6a,7-dehydroaporphine (2). The structures of aporphine alkaloids 1 and 2 (Figure 1) were elucidated by spectroscopic methods, including 1H-NMR, 13C-NMR, UV, IR, MS.
Figure 1.
Structures of the isolated aporphine alkaloids.
Compound 1 crystallized as orange crystals, m.p. 251–252 °C and its molecular formula was determined to be C19H15NO5 by HR-ESIMS. Moreover, its UV spectrum exhibited absorption peaks at 210, 241, 317 and 417 nm indicating the presence of a highly conjugated system. The IR absorptions for OH (3249 cm−1), C=O (1705 cm−1), aromatic (1658, 1559, 1508, 1458 cm−1) and ether moieties (1281, 1211 cm−1) were also observed. From the UV and IR spectral data it was indicated that 1 was an oxoaporphine derivative [9]. The fragment ions at m/z 337 (M+) and 322 (M+-Me) in the mass spectrum of compound 1 indicated the presence of OMe group in the position next to N in the structure. The results from decarbonylation cleavage at m/z 294 indicated the presence of carbonyl group in ring C. The ion of m/z 91 supported that 1 contained one hydroxyl group in ring D (Figure 2).
Figure 2.
Most important observed fragmentations for compound 1.
The 1H-NMR spectrum of the major alkaloid, in addition to three methoxy (δ 4.10, s), (3.99, s) and 3.97, s) groups, had three obscured signals at δ 7.57, 7.60 and 7.62, and two multiplet signals at δ 7.88 and 9.34 in aromatic ring region, so adding together hydroxyl group (12.02, s), this accounts for all 15 protons. A downfield singlet proton at δ 12.02 was assigned to a hydroxyl group at the C-8 position due to the formation of hydrogen bonding with neighboring carbonyl group at C-7. Moreover, the sequential correlations of aromatic proton signals at C-9 (δ 7.57), C-10 (δ 7.88) and C-11 (δ 9.34) on ring D were clearly proven by the COSY spectrum. Additionally, the aromatic proton at C-2 (δ 7.60) and C-3 (δ 7.62) are also clearly supported by this COSY spectrum and HMBC correlation with C-1a ,C-3 (strong correlation) and C-1a (weak correlation), C-2, respectively (Figure 3). Moreover, the identification of H-9, 10, 11 in ring D was determined by NOE difference spectra. Irradiation of H-8 (OH) signal showed enhancement of the H-9, thus indicating that it definitely has one proton next to the OH group. In addition, irradiation at the methoxy signal of position 1 also showed enhancement of H-11. Accordingly, the H-10 was confirmed at this position since there was NOE enhancement with the H-9 (Figure 3). Correspondingly, the presence of a hydroxyl group in the molecule located in the ring D at C-8 was established on the basis of long-range 1H-13C correlation of the HO-8 at δ 12.02 with the carbon C-9 (δ 113.76), C-7 (δ 175.55). The 13C-NMR spectrum exhibited the presence of three methoxyl group, five methine, and eleven quaternary carbons (Table 1). In comparison with the literature data [10] the keto group at C-7 position usually resonance at δ 175 indicating the existence of this carbonyl at the peri-position.
Figure 3.
NOE experiment, significant correlations in the COSY and HMBC spectra.
Table 1.
1H-NMR (500 MHz), 13C-NMR (125 MHz) in DMSO-d6 1 and CDCl3 for 2.
| Position | 8-Hydroxy-1,4,5-trimethoxy-7-oxoaporphine (1) | 1,2,3-Trimethoxy-4,5-dioxo-6a,7-dehydroaporphine (2) | ||||
|---|---|---|---|---|---|---|
| * δ 1H m (J Hz) | δ 13C(DEPT) | * δ 1H m (J Hz) | δ 13C(DEPT) | |||
| 1 | - | 159.15 (C) | - | 158.66 (C) | ||
| 1a | - | 131.58 (C) | - | 116.26 (C) | ||
| 1b | - | 120.21 (C) | - | 120.31 (C) | ||
| 2 | 7.60 obsc. | 127.51 (CH) | - | 147.54 (C) | ||
| 3 | 7.62 obsc. | 126.87 (CH) | - | 160.44 (C) | ||
| 3a | - | 117.65 (C) | - | 117.62 (C) | ||
| 4 | - | 147.02 (C) | - | 175.41 (C) | ||
| 5 | - | 157.49 (C) | - | 157.63 (C) | ||
| 6a | - | 129.82 (C) | - | 128.38 (C) | ||
| 7 | - | 175.55 (C) | 7.83 s | 116.0 (CH) | ||
| 7a | - | 119.21 (C) | - | 131.77 (C) | ||
| 8 | - | 155.80 (C) | 7.98 m | 128.48 (CH) | ||
| 9 | 7.57 obsc. | 113.76 (CH) | 7.65 obsc. | 127.57 (CH) | ||
| 10 | 7.88 m | 128.17 (CH) | 7.66 obsc. | 127.44 (CH) | ||
| 11 | 9.34 m | 126.81 (CH) | 9.5 m | 127.24 (CH) | ||
| 11a | - | 125.91 (C) | - | 121.23 (C) | ||
| 1-OMe | 4.10 s | 61.09 (CH3) | 4.17 s | 62.08 (CH3) | ||
| 2-OMe | - | - | 4.10 s | 61.74 (CH3) | ||
| 3-OMe | - | - | 4.21 s | 61.17 (CH3) | ||
| 4-OMe | 3.99 s | 61.59 (CH3) | - | - | ||
| 5-OMe | 3.97 s | 61.65 (CH3) | - | - | ||
| 8-OH | 12.02 s | - | - | - | ||
| N-H | - | - | 11.77 s | - | ||
* δ in ppm from TMS [coupling constants (J) in Hz are given in parentheses]; obsc. = obscure signal.
In addition, the HMBC spectra gave further support for carbonyl group by correlations with hydroxy proton. The methoxy group at C-1 position of ring A was confirmed by NOE enhancement with H-11. In addition, the methoxy group at C-4 and 5 position of ring B confirmed by direct comparison of those chemical shifts with the 13C-NMR of artabonatine C, a compound isolated from Artabotrys uncinatus [10]. According to the 1H and 13C-NMR 1D/2D data this compound was identified as the oxoaporphine alkaloid. On the basis of the above data, the structure for 1 was formulated as 8-hydroxy-1,4,5-trimethoxy-7-oxoaporphine. It is a new alkaloid and does not appear to have been previously isolated from this species. Chromatographic separation of the ethyl acetate extract afforded compound 2. This compound is isomeric with compound 1. The structure was established by comparison of their UV, IR, EIMS, and 1D, 2D NMR data with the literature data [4,7,11].
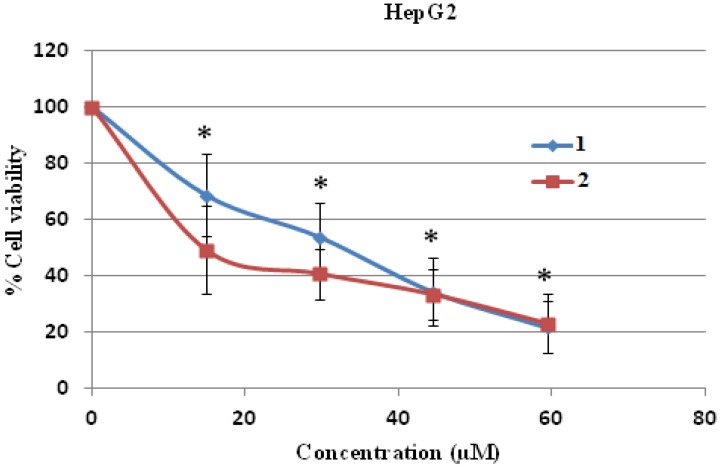
The cytotoxicity test was performed by the MTT assay and it was found that compounds 1 and 2 were toxic to both human hepatocellular carcinoma HepG2 and human breast cancer MDA-MB231 cell lines dose dependently, and these toxicities were statistically and significantly different at the IC10, IC20 and IC50 levels when compared to control (without treatment) (Figure 4 and Figure 5). Compound 2 was more cytotoxic to HepG2 cells than compound 1, with an IC50 concentration of 12.88 ± 2.49 μM compared to 26.36 ± 5.18 μM, respectively, and the IC50 values of both compounds were significantly different when compared to each other (Table 2). Both IC50 levels of compound 1 and 2 were statistically different compared to those of doxorubicin in both cells (Table 2 and Table 3).
Figure 4.
Cell cytotoxicity of 1 and 2 on human hepatocellular carcinoma HepG2 cells. HepG2 cells were treated with 1 and 2 at various concentrations for 24 h and the cell viability was determined by MTT assay. * p < 0.05, compared to control of both compound 1 and 2.
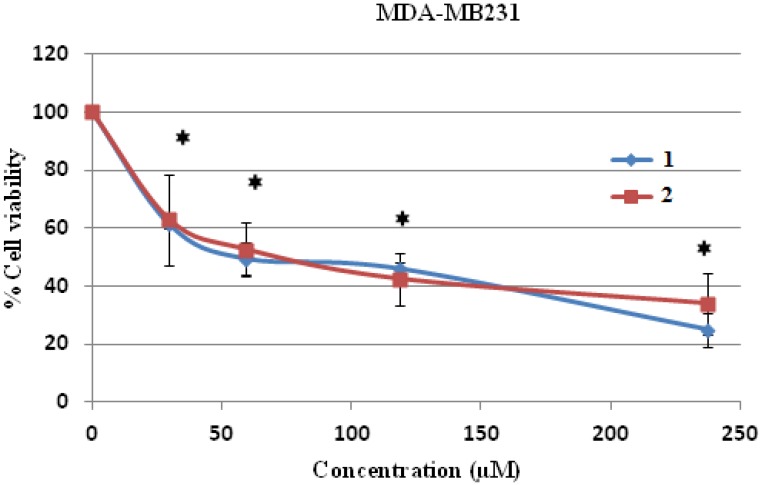
Figure 5.
Cell cytotoxicity of 1 and 2 on human breast cancer MDA-MB231cells. MDA-MB231 cells were treated with 1 and 2 at various concentrations for 24 h and the cell viability was determined by MTT assay. * p < 0.05, compared to control of both compound 1 and 2.
Table 2.
IC10, IC20 and IC50 levels of compound 1 and 2 in human hepatocellular carcinoma HepG2 cells.
| Compound | HepG2 | |||||
|---|---|---|---|---|---|---|
| IC10 (μM) | IC20 (μM) | IC50 (μM) * | ||||
| MEAN | SD | MEAN | SD | MEAN | SD | |
| 1 | 4.21 *a | ±1.60 | 8.74 *a | ±2.22 | 26.36 *a | ±5.18 |
| 2 | 1.74 *b | ±0.23 | 3.46 *b | ±1.10 | 12.88 *b,c | ±2.49 |
| Doxorubicin | - | - | - | - | 2.21 | ±1.72 |
* p < 0.05, a vs. control of compound 1, b vs. control of compound 2, c vs. IC50 level of compound 1.
Table 3.
IC10, IC20 and IC50 levels of compound 1 and 2 in human breast cancer MDA-MB231 cells.
| Compound | MDA-MB231 | ||||||
|---|---|---|---|---|---|---|---|
| IC10 (μM) | IC20 (μM) | IC50 (μM) * | |||||
| MEAN | SD | MEAN | SD | MEAN | SD | ||
| 1 | 5.02 *a | ±0.53 | 15.12 *a | ±1.24 | 64.75 *a,c | ±4.45 | |
| 2 | 3.37 *b | ±0.88 | 8.20 *b | ±2.45 | 67.06 *b | ±3.5 | |
| Doxorubicin | - | - | - | - | 1.83 | ±0.09 | |
* p < 0.05, a vs. control of compound 1, b vs. control of compound 2, c vs. IC50 level of compound 2.
However, when human breast cancer MDA-MB231 cells were treated with both compounds, it was shown that both compound 1 and 2 were cytotoxic to the MDA-MB231 cells up to the tested concentration of 240 μM (Figure 5). The IC50 levels of both compound 1 and 2 on human breast cancer MDA-MB231 cells were 64.75 ± 4.45 and 67.06 ± 3.5 μM, respectively (Table 3). It could be concluded that human hepatocellular carcinoma HepG2 cells were more sensitive to both compounds than human breast cancer MD-MB231 cells at the lower IC50 value level.
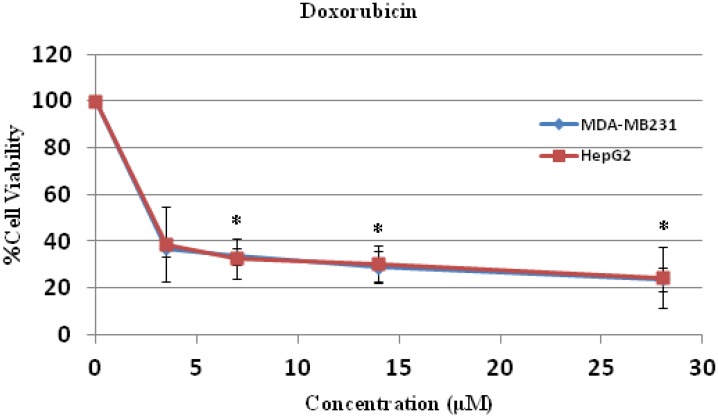
The anti-cancer activities of both compounds in both cell lines were less potent when compared to doxorubicin (Figure 6, Table 2 and Table 3). The mechanisms of action of doxorubicin are DNA intercalation [12], increased free radical production [13], and inhibition of topoisomerase II progression [14]. Doxorubicin is used for treatment of leukemia, lymphoma and several solid tumors, such as osteosarcoma. It also induces apoptosis and necrosis in healthy tissue of the brain, liver, kidney and heart. The drug influences Bcl-2/Bax apoptosis pathway and caspase activation [15]. However, further study of the mode and mechanism of cell death of compounds 1 and 2 will provide more information of anti-cancer activities of these two compounds and an in vivo assay in animal model is required before safe application for cancer treatment in human-beings.
Figure 6.
Cell cytotoxicity of doxorubicin on human hepatocellular carcinoma HepG2 and breast cancer MDA-MB231 cells. HepG2 and MDA-MB231 cells were treated with doxorubicin at various concentrations for 24 h and the cell viability were determined by MTT assay. * p < 0.05, compared to control.
3. Experimental
3.1. General Information
Melting points were recorded in degree Celsius (°C) and were measured on a digital Electrothermal melting apparatus. UV spectrum was measured on a Shimadzu 1601 spectrophotometer (Shimadzu, Kyoto, Japan). 1H and 13C-NMR, 1H-1H COSY, HMQC and HMBC spectra were recorded with a Unity plus 500 spectrometer (Varian Inc, Palo Alto, CA, USA) operating at 500 MHz for 1H, and 125 MHz for 13C. Low resolution mass spectra were recorded on a Thermo Finnegan Polaris Q mass spectrometer at 70 eV (probe) for EIMS. HRESIMS was obtained by using a Finnigan LC-Q Advantage Thermoquest spectrometer equipped with Xcalibur software (both intruments from Thermo Finnigan, Waltham, MA, USA). IR spectra in KBr disk were recorded on Shimadzu 8900 FTIR spectrophotometer. Column chromatography was conducted on silica gel 60 (Merck 7734, 70–230 mesh). TLC was performed on aluminium backed pre-coated silica gel 60 PF254 sheets and detection with using UV detector.
3.2. Plant Material
The mixture leaves and twigs of P. trimera (Annonaceae) were collected in August 2013 from Chiang Rai Province, the North Thailand. The species was identified by Mr. Narong Nantasean, from The Forest Herbarium, Department of National Park, Wildlife and Plant Conservation, Ministry of Natural Resources and Environment, Bangkok, Thailand. A voucher specimen (BKF.158070) was deposited in the herbarium of this institute.
3.3. Extraction and Isolation
The milled dried mixture of leaves and twigs (500 g) was extracted exhaustively in turn with hexane and ethyl acetate-methanol (1:2). The ethyl acetate-methanol extract, on removal of solvent under reduced pressure, gave a black residue (100 g). The extract of the ethyl acetate-methanol extract was fractionated by CC, silica gel, Merck No. 7734, Mesh 70–230 ASTM (600 g, 11 × 20 cm, hexane, hexane-ethyl acetate, ethyl acetate, ethyl acetate-methanol, methanol in order of increasing polarity) to give five fractions (F1–F5). Fraction F4 (31.2 g) was further subjected to CC (silica gel, 200 g, 5 × 16 cm, hexane-ethyl acetate in order of increasing polarity). Eight fractions (A1–A8) were ultimately obtained on combining the eluates on the basis of TLC. Further, the fraction A6 (3.7g) was rechromatographed over silica gel, eluting with dichloromethane, dichloromethane-methanol. The solvents were evaporated to dryness to afford seven subfractions (B1–B7). The subfraction B5 (0.61 g) yielded yellow solid which was recrystallized from methanol/dichlormethane (2:1) to afford orange crystals (26 mg) and identified as 6a,7-dehydro-1,4,5-trimethoxy-7-oxoaporphine (1). Further, the mother liquor of B5 was recrystallized from dichloromethane (2:1) to give yellow crystals (6.3 mg) of ouregidione (2, 6.3 mg).
8-Hydroxy-1,4,5-trimethoxy-7-oxoaporphine (1): Orange crystals, m.p. 251–252 °C. UV (MeOH) λmax (log ε): 417(4.58), 317(4.76), 241(5.12), 210(5.08) nm. IR (KBr) vmax 3384, 3249, 2936, 1705, 1658, 1559, 1508, 1458, 1281, 1211 cm−1. 1H-NMR (DMSO-d6, 500 MHz) data see Table 1, 13C-NMR (DMSO-d6, 125 MHz) data see Table 1. EI-MS m/z: 337 [M]+(26), 322(100), 307(6), 294(21), 297(16), 264(17), 91(15). COSY correlations H/H: 2/3, 3/2, 9/10, 10/9, 11, 11/10. HMBC correlations H/C:2/1a, 3; 3/1a, 2; 9/7a, 10, 10a; 10/9, 11, 11a; 11/1a, 7a, 11a; 1OCH3/1; 4OCH3/4; 5OCH3/5; 8OH/ 8. HR-ESI-MS (pos.) m/z: 360.0846 ([M+Na]+, C19H15NO5Na. calcd. 360.0848.
1,2,3-Trimethoxy-4,5-dioxo-6a,7-dehydroaporphine (2): Yellow crystals, m.p. 242–243 °C. UV (MeOH) λmax (log ε): 416(4.15), 317(4.34), 214(4.70), 210(4.66) nm. IR (KBr) vmax 3420, 2853, 1689, 1658, 1559, 1458, 1211 cm−1. 1H-NMR (CDCl3, 500 MHz) data see Table 1, 13C-NMR (CDCl3, 125 MHz) data see Table 1. EI-MS m/z: 337 [M]+(48), 322(100), 306(3), 294(29), 279(23), 264(15), 251(52), 236(21), 180(25), 152(23). COSY correlations H/H: 8/9, 9/10, 10/11. HMBC correlations H/C:7/1b, 8; 8/7, 10; 9/11; 10/9; 11/7a, 9; 1OCH3/1; 2OCH3/2; 3OCH3/3; NH/1b, 4. HR-ESI-MS (pos.) m/z: 360.0846 ([M+Na]+, C19H15NO5Na. calcd. 360.0849.
3.4. Cell Culture
The human hepatocellular carcinoma HepG2 and breast cancer MDA-MB231 cells were cultured in Leibovitz’s L-15 and Dulbecco’s modified eagle media (DMEM), respectively, with 25 mM NaHCO3, 20 mM HEPES, 100 units/mL penicillin, 100 μg/mL streptomycin and supplemented with 10% foetal bovine serum. The cell lines were grown at 37 °C in a 5% CO2 atmosphere. The cells (5 × 103) were treated with each compound at indicated concentrations for 24 h. The compound 1 and 2 was dissolved in dimethyl sulfoxide (DMSO) as a vehicle and the maximum volume used did not exceed 10 μL/mL of media to avoid the cytotoxicity of DMSO.
3.5. MTT Assay
Cell viability was measured by MTT assay (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) [16]. In brief, human hepatocellular carcinoma HepG2 and human breast cancer MDA-MB231 cells were seeded in 96-well plates at density of 5,000 cells/well. After 24 h incubation, HepG2 cells and MDA-MB231 cells were treated with or without compound 1 and 2 at various concentrations for 24 h at 37 °C in a humidified 5% CO2 atmosphere. Doxorubicin, a chemotherapeutic drug, was used as positive control. Non-treated cells were used as negative control. Then 15 μL (sterile stock solution of 5 mg/mL MTT dye) were added to each well and the solution was incubated for 4 h at 37 °C in a humidified 5% CO2 atmosphere. The medium was removed and 100 μL of DMSO were added to each well, and mixed to dissolve the blue crystal formazan. The plate was read at 540 nm, with a reference wavelength of 630 nm, using a microplate reader (Biotek, Winooski, VT, USA) The percentage of cell viability was determined as follows:
 |
4. Conclusions
Compound 1 and 2 displayed anti-cancer activity and have potential for use as anti-cancer drugs in human liver and breast cancer. but the anti-cancer activities of both compounds were less than those of doxorubicin in both cell lines. However, further research in an in vivo model is needed before any clinical usage.
Acknowledgments
The authors thank the Center for Innovation in Chemistry (PERCH-CIC), Commission on Higher Education, Ministry of Education, the grant of Thailand’s Office of Higher Education Commission for the Project on Higher Education and Research Promotion (HERP) and Lampang Rajabhat University Research Grant (SC07/2557) for financial support. Moreover, National Research Council of Thailand (NRCT), Thailand Research Fund and the Commission of Higher Education, grant No. RMU5080003 are grateful for the financial support to RB and BW.
Author Contributions
The contributions of the respective authors are as follows: Pompimon, W. is the principal investigator did the isolation, purification, identification and prepared the manuscript, hence the corresponding author. Sesang, W. contributed in the confirming of the isolation procedures, especially interpretation of the spectra and also part of the preparation of the manuscript. Punyanitya, S. is part of the grant that supported this work. Pitchuanchom, S and Udomputtimekakul, P. are elucidated and proved the structures of compounds 1 and 2. This plants are corrected and identified by Nuntasaen, N. Banjerdpongchai, R. and Wudtiwai, B. are carried out the bioassay and cotributed in the preparing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest
Footnotes
Sample Availability: Samples of the compounds 1–2 are not available from the authors.
References
- 1.Su Y.C.F., Chaowasku T., Saunders M.K. An extend phylogeny of Pseuduvaria. (Annonaceae) with descriptions of three new species and a reassessment of the gerneric status of oreomitra. Syst. Bot. 2010;36:1–11. [Google Scholar]
- 2.Su Y.C.F., Saunders R.M.K. Pseuduvaria. trimera (Craib.) Syst. Bot. Monogr. 2006;79:174. [Google Scholar]
- 3.Mahmood K., Chan K.C., Park M.H., Han Y.N., Han B.H. An aporphinoid alkaloid from Pseuduvaria. macrophylla. Phytochemistry. 1986;25:1509–1510. doi: 10.1016/S0031-9422(00)81328-X. [DOI] [Google Scholar]
- 4.Wirasathien L., Boonarkart C., Pengsuparp T., Suttisri R. Biological activities of alkaloids from Pseuduvaria setosa. Pharm. Biol. 2006;44:274–278. doi: 10.1080/13880200600714111. [DOI] [Google Scholar]
- 5.Taha H., Hadi A.H.A., Nordin N., Najmuldeen I.A., Mohamad K., Shirota O., Nugroho A.E., Piow W.C., Kaneda T., Morita H. Pseuduvarines A and B, two new cytotoxic dioxoaporphine alkaloids from Pseuduvaria rugosa. Chem. Pharm. Bull. 2011;59:896–897. doi: 10.1248/cpb.59.896. [DOI] [PubMed] [Google Scholar]
- 6.Ming Z.S., Shun Z.S., Ning X. Alkaloids from Pseuduvaria indochinensis. Phytochemistry. 1988;27:4004–4005. doi: 10.1016/0031-9422(88)83076-0. [DOI] [Google Scholar]
- 7.Uadkla O., Yodkeeree S., Buayairaksa M., Meepowpan P., Nuntasaen N., Limtrakul P., Pompimon W. Antiproliferative effect of alkaloids via cell cycle arrest from Pseuduvaria rugosa. Pharm. Biol. 2013;51:400–404. doi: 10.3109/13880209.2012.734314. [DOI] [PubMed] [Google Scholar]
- 8.Taha H., Arya A., Paydar M., Looi C.Y., Wong W.F., Murthy C.R.V., Noordin M.I., Ali H.M., Mustafa A.M., Hadi A.H.A. Upregulation of insulin secretion and downregulation of pro-inflammatory cytokines, oxidative stress and hyperglycemia in STZ-nicotinamide-induced type 2 diabetic rats by Pseuduvaria monticola bark extract. Food Chem. Toxicol. 2014;66:295–306. doi: 10.1016/j.fct.2014.01.054. [DOI] [PubMed] [Google Scholar]
- 9.Ge Y.W., Zhu S., Shang M.Y., Zang X.Y., Wang X., Bai Y.J., Li L., Komatsu K., Cai S.Q. Aristololactams and aporphines from the stems of Fissistigma oldhamii (Annonaceae) Phytochemistry. 2013;86:201–207. doi: 10.1016/j.phytochem.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh T.J., Chang F.R., Chia Y.C., Chen C.Y., Lin H.C., Chiu H.F., Wu Y.C. The alkaloids of Artabotrys uncinatus. J. Nat. Prod. 2001;64:1157–1161. doi: 10.1021/np010036k. [DOI] [PubMed] [Google Scholar]
- 11.Wueratne E.M.K., Hatanaka Y., Kikuchi T., Tezuka Y., Gunatilaka A.A.L. A dioxoaporphine and other alkaloids of two annonaceous plants of sri lanka. Phytochemistry. 1996;42:1703–1706. doi: 10.1016/0031-9422(96)00181-1. [DOI] [Google Scholar]
- 12.Momparler R.L., Karon M., Siegel S.E., Avila F. Effect of adriamycin on DNA, RNA and protein synthesis in cell-free systems and intact cells. Cancer Res. 1976;36:2891–2895. [PubMed] [Google Scholar]
- 13.Keizer H.G., Pinedo H.M., Schuurhuis G.J., Joenje H. Doxorubicin (Adriamycin): A critical review of free radical-dependent mechanisms of cytotoxicity. Pharmacol. Ther. 1990;47:219–231. doi: 10.1016/0163-7258(90)90088-J. [DOI] [PubMed] [Google Scholar]
- 14.Fornari F.A., Randolph J.K., Yalowich J.C., Ritke M.K., Gewirtz D.A. Interference by doxorubicin with DNA unwinding in MCF-7 breast tumor cells. Mol. Pharmacol. 1994;45:649–656. [PubMed] [Google Scholar]
- 15.Tacar O., Sriamornsak P., Dass C.R. Doxorubicin: An update on anticancer molecular action, toxicity and novel drug delivery systems. J. Pharm. Pharmacol. 2013;65:157–170. doi: 10.1111/j.2042-7158.2012.01567.x. [DOI] [PubMed] [Google Scholar]
- 16.Wudtiwai B., Sripanidkulchai B., Kongtawelert P., Banjerdpongchai R. Methoxyflavone derivatives modulate the effect of TRAIL-induced apoptosis in human leukemic cell lines. J. Hematol. Oncol. 2011;4:52–63. doi: 10.1186/1756-8722-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]