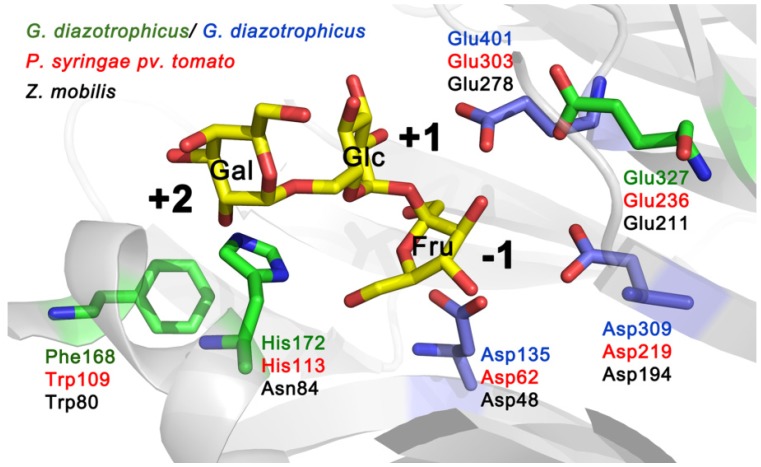
Figure 2.
Catalytic centre of G. diazotrophicus levansucrase LsdA (PDB ID: 1W18; [5]) with side chains of catalytic triad residues (Asp135, Asp309 and Glu401) indicated in blue. The structure of the protein was visualized using PyMol [27]. Raffinose (consisting of galactose, glucose and fructose residues) shown in substrate-binding pocket originates from the structure of the B. subtilis levansucrase SacB in complex with raffinose (PDB ID: 3BYN) that was superimposed with the LsdA structure. Positions of LsdA equivalent to Trp109, His113 and Glu236 of Lsc3 of P. syringae pv. tomato are shown on the model. Respective amino acids of levansucrases of different bacteria are shown in different colour: LevU of Z. mobilis (black), Lsc3 of P. syringae pv. tomato (red) and LsdA of G. diazotrophicus (green and blue).