Abstract
Spin labelling is a chemical technique that enables the integration of a molecule containing an unpaired electron into another framework for study. Given the need to understand the structure, dynamics, and conformational changes of biomacromolecules, spin labelling provides a relatively non-intrusive technique and has certain advantages over X-ray crystallography; which requires high quality crystals. The technique relies on the design of binding probes that target a functional group, for example, the thiol group of a cysteine residue within a protein. The unpaired electron is typically supplied through a nitroxide radical and sterically shielded to preserve stability. Pulsed electron paramagnetic resonance (EPR) techniques allow small magnetic couplings to be measured (e.g., <50 MHz) providing information on single label probes or the dipolar coupling between multiple labels. In particular, distances between spin labels pairs can be derived which has led to many protein/enzymes and nucleotides being studied. Here, we summarise recent examples of spin labels used for pulse EPR that serve to illustrate the contribution of chemistry to advancing discoveries in this field.
Keywords: electron paramagnetic resonance, spin label, DEER, PELDOR
1. Introduction
The use of pulsed electron paramagnetic resonance (EPR) to extract magnetic couplings hidden within the linewidth of conventional continuous-wave (CW) studies has led to a resurgence in the applications of EPR [1]. Typically, the couplings are electron-nuclear (hyperfine) or electron-electron interactions, and yield important information about the environment around the unpaired electron. Diamagnetic molecules can be studied by attaching a molecule containing an unpaired electron; known as “spin labelling”. This has led to a significant use in biology through site directed spin labelling (SDSL), which uses mutagenesis to attach labels to specific amino acids. To be able to study the molecular structure of biomacromolecules, techniques such as X-ray diffraction, require crystals from highly purified and monodisperse samples. Likewise, NMR; although able to study polydisperse samples, is limited by the size of the system. EPR in contrast is able to detect the paramagnetic species and ignore diamagnetic background. This makes EPR a very attractive technique for the study of biomacromolecules, possible in membranes and in whole cells.
There is a continuously growing array of pulse sequences [2] used to measure dipolar electron-electron interactions, which have been previously reviewed in detail [3,4]. The dipolar interaction is both distance and orientation dependent, and in principal both parameters can be extracted from pulse measurements. Typically, measurements are made on a frozen solution or in the solid state which causes all orientations of the macromolecule to be observed with respect to the pole faces of the magnet. The most widely applied technique is double electron electron resonance (DEER) or pulsed electron double resonance (PELDOR) which can measure the dipolar interaction between spin pairs and is typically used to extract distances in the range 17–100 Å [5,6,7,8]. In brief, this is a double resonance technique requiring two sets of microwave pulses at different frequencies; an inversion/pump pulse flips one of the spins and inverts the local dipolar field induced by that spin, and an observation echo sequence leads to a refocusing of all other interactions, except the dipolar interaction, which leads to modulation of the echo signal [9]. The overall intensity of the echo signal depends on a characteristic spin relaxation time called the phase memory time (Tm). It is advantageous to maximise Tm for a longer measurement window needed to measure distances > 40 Å. Longer distances can be measured through deuteration of the protein and buffer due to favourable relaxation [8]. In the case of broad EPR spectra, often found in metals, it is possible to excite specific orientations [10,11] with respect to the magnetic field and obtain structural information [12].
Another powerful technique used to extract the dipolar frequency is called double quantum coherence (DQC) developed by the Freed group [13,14,15]. The DQC method detects a double quantum coherence generated by spin-spin interactions. The methods of DEER and DQC are contrasted in the detailed review of Borbat and Freed [16].
Pulsed dipolar EPR has allowed numerous studies on the structure and conformational change of proteins [9] and can harness biological paramagnetic centres such as semiquinones [17], iron sulphur clusters [18] and tyrosyl radicals [19,20]. The techniques have also allowed applications to materials science [21,22].
There are many good reviews that focus on how spin labels can provide information about the structure and dynamics of proteins [23,24]. The design of spin labels needs to consider the labels influence on the protein structure as well as EPR spectral properties. The purpose of this article is to describe the general chemical scope of current labels used for pulsed dipolar EPR and to encourage future development.
2. Nitroxides
Incorporation of nitroxides by SDSL has, by large, become the dominant technique for DEER distance measurements on proteins [23,24,25]. There is considerable breadth to the understanding of the synthetic chemistry of nitroxides, being by far the most studied and synthesised spin labels [26]. The numerous uses of nitroxide radicals have probably played a part in their success as spin labels. Nitroxides are commonly used as co-oxidants [27], mediators for polymerisation [28], detectors of nitrogen monoxide [29] and for the determination of oxygen concentrations [30]. The considerable stability of nitroxides to organic synthetic conditions, allows their use in organic synthesis, creating an incredible library of spin labels, flexible to the needs of the experimental conditions [26,31,32,33].
For pulsed distance measurements, interpretation of EPR data using SDSL requires knowledge of the rotamers and internal dynamics of the respective side chains [24,34]. Different strategies have been used in the past to account for the local dynamics of a spin label in an arbitrary site of a protein. This information is best known for the methanethiosulfonate spin label (MTSL, 1); also known as R1. Rotamer libraries have been created for α helixes and β sheets [35,36,37], and molecular dynamics (MD) simulations and Monte Carlo conformational searches have been established [38,39], which are commonly used to extrapolate distance measurements between MTSL labels on proteins. There have been several attempts at MD simulations based on DEER constraints [40,41].
2.1. Nitroxides in the Literature
By far the most popular nitroxide spin label is MTSL (1, Figure 1). MTSL is highly selective for thiol groups, and is commonly attached to cysteine residues in proteins. The MTSL side chain (commonly known as R1) also has a minimum impact on the secondary and tertiary structure of proteins. There are, however, disadvantages to R1; the rotational dynamics of the spin label can be complicated and can lead to broad distance distributions being obtained, and both the N-O moiety and disulphide bonds to cysteine residues are susceptible to reducing conditions, such as those that are present inside live cells. Alternative spin labels are available for conditions that are reducing or for in-cell EPR (and will be discussed, in detail, later in this review).
Figure 1.
Structures of radical spin labels commonly used for SDSL of proteins.
In order to overcome the complexities associated with the flexibility of the R1 cysteine linker, alternative spin label side chains have been created, with significant reductions in the internal motions of the spin label. The 4-pyridyl substituted spin label 2, gives rise to the R1p side chain, and has been shown to have a considerable reduction in the flexibility of the side chain when in a solvent exposed α-helix [42]. This dramatically simplifies the interpretation of rotamer dynamics, producing narrower distance distributions, leading to greater accuracy. Other spin label side chains with constrained geometry include, RX 3 [43], a bi-functional analogue of R1, which has been shown to tether to two cysteines at positions i and i + 3 or i + 4 on an exposed α-helix. RX can also be positioned at i and i + 1 in a β strand [44].
2.1.1. Spin Labelling of Amino Acids
Non-native amino acids have been incorporated into peptides using solid-phase synthesis using traditional peptide coupling reagents [45,46]. Amino acid spin labels, such as 2,2,6,6-tetramethyl-piperidine-1-oxyl-4-amino-4-carboxylic acid (TOAC, 4) [46] and 3-amino-1-oxyl-2,2,5,5-tetramethyl pyrrolidine-4-carboxylic acid (POAC) [47] provide a fixed position for a spin label, with no internal rotamers, however, currently can only be incorporated into small peptides and proteins by total synthesis. Incorporation of TOAC via peptide coupling, however, does not occur in good yield [47]; often requiring multiple rounds of peptide coupling, due to the low nucleophilicity of the sterically hindered amino group. TOAC has a very limited range of backbone dihedral angles, creating a significant distortion of the secondary structure of proteins [46]. Despite this, TOAC is commonly used; for example, in studies of the Aib rich peptide alamethicin [48]. In order to combat the distortion of the secondary structure; the 4-(3,3,5,5-tetramethyl-2,6-dioxo-4-oxylpiperazin-1-yl)-l‑phenylglycine (TOPP, 5) spin label has been introduced, allowing greater flexibility of the backbone chain [49].
2.1.2. Spin Labels for DNA and RNA
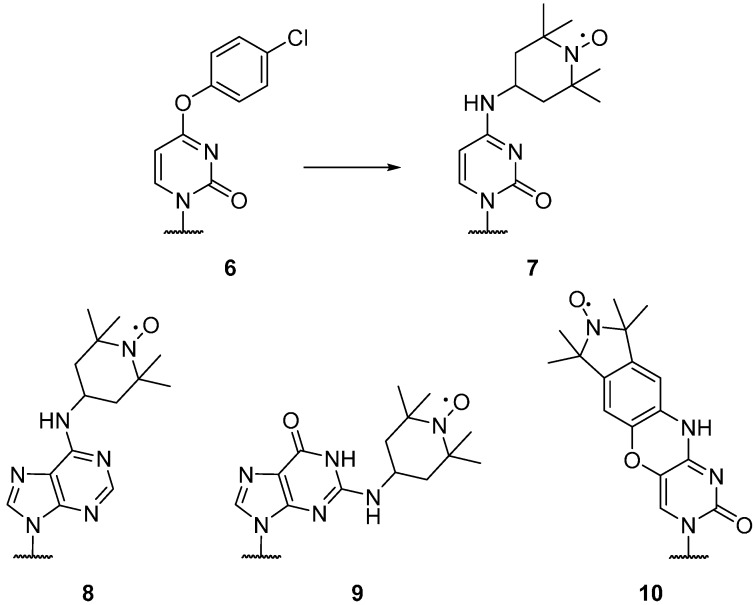
Nitroxides have been utilised for the specific spin labelling of deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), and can be attached to the bases, phosphate, and sugar backbone of individual nucleotides [50]. DEER measurements on nucleic acids are often carried out using rigid spin labels, which leads to significant orientation selection even at 9 GHz [12]. Commercially available “convertible” nucleotides [51]; O4-(4-chlorophenyl)-uridine (6), O6-(4-chlorophenyl)-inosine, and 2-fluoroinosine, incorporated into RNA sequences via solid-state synthesis, can be spin labelled using 4-amino-TEMPO; giving the spin labelled RNA bases cytosine (7), adenine (8) and guanine (9), respectively (Figure 2) [52].
Figure 2.
Spin labelled RNA bases.
An alternative rigid, profluorescent spin labelled cytosine nucleotide 10 has been reported by Barhate et al. [53], creating a rigid, tricyclic nitroxide labelled base for DNA via a seven step synthesis. This label has also been attached to RNA by Höbartner et al. [54] The RNA base uracil can be labelled 13 using alkyne substituted spin labels 11 (Scheme 1), which can be coupled via Sonogashira palladium coupling to 5-iodouridine (12) [55,56]. Both five [57] and six [58] membered nitroxideshavebeencoupledto 5-iodouridine, with five membered rings showing greater stability under reducing conditions [59]. The 3-(2-iodoacetamido)-proxyl spin label can be coupled to a 4-thiouridine residue in RNA [60,61]. Nucleotides with alkyne moieties have also been created [62,63], allowing the use of “click chemistry” for post-modification of DNA in solution with azido functionalised nitroxides.
Scheme 1.
Spin labelling of uracil (12).
The 2' positions on the ribose moiety of RNA have been spin labelled 16 (Scheme 2), from the 2'-amino RNA derivatives 14, using an isocyanate TEMPO derivative 15 [64,65]. Schiemann et al. have demonstrated that this spin label can be used to determine both distance and relative orientation of two spin-labelled nucleosides in DNA at 9 GHz [65].
Scheme 2.
Spin labelling of the 2' position on the sugar backbone of RNA.
The phosphate backbone of RNA/DNA can be spin labelled 19 by reacting the iodo-nitroxide 18 with a commercially available phosphorothioate modified nucleotides 17, with minimal perturbation of the RNA/DNA complex [66,67] (Scheme 3). Spin labelling of the phosphate backbone is more cost-effective than other nucleotide labelling procedures, as the cost of introducing phosphorothioates is significantly less than incorporating modified nucleotides [67].
Scheme 3.
Spin labelling of phosphate backbone of RNA.
2.2. Design Philosophy
2.2.1. Parent Ring Structures
There are three main groups of nitroxides that have been commonly used historically for SDSL; six membered rings (piperidines) 20, five membered rings (pyrrolines 21 and pyrrolidines), and fused ring systems (isoindolines) 22; each of which has associated advantages and disadvantages (Figure 3). Nitroxides with a piperidine parent structure are easily derived from triacetoamine [68], with few chemical steps, making cheap and quick functionalised spin labels. Nitroxides with a pyrrole parent structure are synthesised from 4-oxo-TEMPO using a Favorskii rearrangement [26,69,70], requiring additional multi-step synthesis. The six membered piperidine nitroxides are the least stable to chemically reducing conditions, with the five membered pyrroline and pyrrolidine nitroxides having substantially greater resistance [71,72].
Figure 3.
Parent structures of nitroxides; (a) a piperidine, (b) a pyrroline, (c) an isoindoline, (d) an imidazoline.
Fused ring aromatic nitroxides; such as the isoindolines 22, have been shown to exhibit several advantages over the piperidine and pyrroline nitroxides. Isoindolines have greater structural rigidity, thermal and chemical stability [73], and narrower EPR linewidths [74,75,76] than piperidine nitroxides and can double as fluorescent probes [77], making them useful for in-cell EPR. They can also be highly functionalised, specifically tailoring the spin label to its intended environment [78]; recent advances include the spin labelling of graphene with aryl radical diazonium tetrafluoroborate salts [79]. Imidizolines 23 have been developed to give a largely pH dependent EPR spectrum, and are commonly used as pH probes, and for in-cell pH imaging [80].
2.2.2. Alternatives to Methanethiosulfonate Linkages
There are many ways of linking nitroxides to proteins and substrates (Figure 4). Methanethiosulfonate moieties 24 are by far the most common used, creating selective disulphide bonds with cysteine residues on proteins. However, there are limitations; it is important to balance the rigidity of a spin label and the distortion of the protein.
Figure 4.
Structures of common nitroxide linkers.
Alternative cysteine specific linkers have also been created, such as the iodocetamido 25 [81] and maleimido 26 [82] and indanedione 27 [83] moieties, which have greater pH stability than the disulphide bonds of the methanethiosulfonate moiety. However, there are unfortunate drawbacks to maleimides; they can react with amines at high pH and they can hydrolyse to the maleamic acid, which may react with other maleimides, reducing their shelf-life [26].
Spin labels designed to target different amino acids are also available; with linkers specific to serine (28) [84], tyrosine (29) [85] and arginine (30) [86] reported in the literature. In 2011 an alternative tyrosine linker was developed, utilising a Mannich-type reaction with 4-amino-2,2,5,5-tetramethyl-3-imidazoline-1-yloxy nitroxide [87]. However, the greater scarcity of cysteine residues has meant that the methanethiosulfonate moiety dominates.
“Click chemistry” was developed by Kolb, Finn and Sharpless, as a way to create highly selective, modular carbon-heteroatom bonds, using 1,3-dipolar cycloadditions between organic azides and alkynes in the presence of Cu(I) catalysts [88]. Development of nitroxides containing organic azides 31 and alkynes 32 by Kálai, Hubbell, and Hideg [89], provided a site selective method of incorporating nitroxides into proteins, that was based on chemistry unused by cellular processes. Alkynes 32 may also be coupled to aromatic ring systems via Sonogashira palladium coupling [55,76], and boron esters 33 of pyrrolines [90] have been created, opening opportunities for the creation of increasingly complex radical spin labels.
Carboxylic acids 34 and amines 35 provide simple and reliable linkers for attaching spin labels. Often they are used in combination with each other, creating synthetic amino acids [47,91]. Carboxylic acids can be activated with peptide coupling reagents [92,93], or converted into acyl chlorides [94], facilitating nucleophilic attack via amines. Amine functionality allows the coupling of spin labels to a large array of carbonyls; such as, carboxylic acids, acid chlorides [95] and esters [96].
2.2.3. Steric Groups
Most nitroxides contain two gem-dimethyl substituted quaternary carbons (Figure 5), sterically shielding the nitroxide moiety, and thus providing kinetic stability. This steric stabilisation of the radical, leads to a long lasting, stable nitroxide, but also leads to a decrease of the relaxation time Tm. For standard 2,2,6,6-tetramethyl nitroxides 36, at temperatures above 70 K, the rotation of the methyl groups is fast enough to average in-equivalent electron-nuclear couplings, dramatically decreasing Tm [97,98]. When the two gem-dimethyl substituted quaternary carbons are replaced either with bulky aromatic groups; such as 1,1,3,3-tetraphenylisoindolin-2-yloxyl (TPHIO, 37) [99], or with spirocyclohexyl substituents 38 [91,100], the extra source of spin relaxation contributing to Tm is removed, leading to the low-temperature limit of Tm already being attained at 80–100 K, while 50–60 K are required for tetramethyl nitroxides.
Figure 5.
tructures of nitroxides with various steric groups on the 2, 6 positions.
Steric groups at the 4-position of pyrrolines have been shown to adopt a significantly restricted internal motion than the un-substituted R1 spin label side chain [42]. The presence of a rigid substituent, such as the 4-phenylpyrroline analogue 39 or the 4-pyridylpyrroline 40 (Figure 6), significantly restricts the motion of the side chain at a solvent exposed helical site. The 4‑phenyl substituent is, however, prone to aggregation, most likely due to the increased surface hydrophobicity of the spin label [42].
Figure 6.
Structures of pyrrolines with rigid substituents on the 4 position.
2.2.4. Isotope Effects
Isotopically labelled analogues of nitroxide spin labels, incorporating N15 (I = 1/2) into the N‑O moiety, have only two resonances rather than the three for N14 (I = 1) nitroxides; decreasing spectral width and therefore leading to an increase in signal intensity [101,102]. This is particularly useful to reduce detection limits of dilute systems, most notably for in-cell EPR (see below). Isotope spin labelling has enabled selective distance measurements between N14- N14 and N15- N15 spin labels [103]. This is particularly useful for heteropolymers and ensembles of macromolecules; where distance information can become complicated. The ability to distinguish different distance contributions between multiple spin labelled components, significantly decreases the complexity of the spectra.
3. Carbon Centred Spin Labels
Trityl Radicals
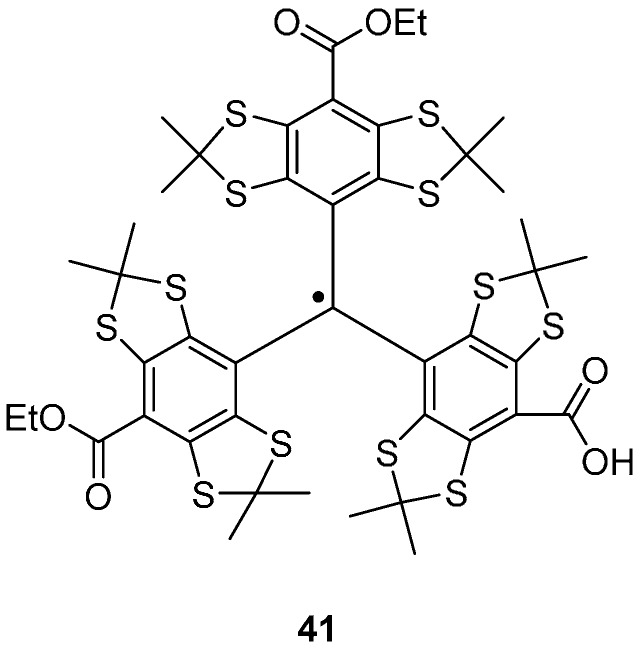
The tetrathiatriarylmethyl (trityl or TAM) radical (Figure 7), has been used as a spin label and offers certain advantages over other spin labels due to its narrow linewidth and relatively long Tm (microseconds at room temperature [104,105]), allowing a more physiological temperature range [106,107,108]. The synthesis of the tetrathiatriarylmethyl radical 41 (Figure 7) is described by Reddy et al. [109] and consists of five steps; the first four of which form the trityl alcohol from tetrachlorobenzene, further treatment with boron trifluoride diethyl etherate followed by tin(II) chloride forms the trityl radical. Trityl radicals are stabilised against dimerisation by substituted aryl groups and the in-cell survival time is in the range of hours [110].
Figure 7.
A tetrathiatriarylmethyl radical 41.
Yang et al. have attached TAM labels via disulfide linkages to cysteine residues in T4 lysozyme, using a dithiodipyridine TAM label 42 (Figure 8) [106]. When coupling to cysteine residues on a protein, 2-thiopyridone is also produced, the UV absorbance of which can be used to monitor the reaction [111]. Immobilisation on Sepharose, prevented averaging of the anisotropic dipolar interaction between spins induced by the rotational diffusion of the spin labelled protein. Distance measurements of ~2 nm were successfully performed using DQC in trityl-labelled protein system at 277 K [106].
Figure 8.
A cysteine specific TAM spin label CT02-TP 42.
Further properties of the trityl radical have been investigated by Schiemann group [108,112] who synthesised trityl-trityl and trityl-nitroxide model compounds with well-defined interspin distances in the range of 17 to 49 Å. They studied the nature of the dipolar coupling and observed deep DEER modulations and improved sensitivity in trityl-trityl DQC experiments (of a factor of two) compared to a nitroxide-nitroxide system under the same conditions. The authors note the increased steric bulk compared to nitroxide labels may lead to structural distortions in biological systems, but this will depend on the specific structure, and will (as is the case with nitroxides) have to be checked for structural perturbations caused by the spin label.
Recently, Shevelev et al. [107] demonstrated distance measurements of ~4.6 nm in nucleic acid duplex systems at physiological temperatures (310 K) using DQC and at 80 K using DQC and DEER. Distance distributions obtained from DQC measurements at 80 K and at 310 K correlated with the data obtained from DEER measurements at 80 K. The spin labelled nucleic acids, shown in Figure 9a,b, were prepared by coupling the acid chloride TAM spin label to a piperazine activated 5' terminus of two complementary 10-mer oligonucleotides; forming a double stranded spin-labelled nucleic acid duplex, that was subsequently immobilised electrostatically on common ion-exchange sorbent NucleosilDMA.
Figure 9.
(a) TAM-labelled oligonucleotide 43, (b) structure of the doubly-labelled nucleic acid duplex.
4. Photo-Excited Triplet States
Porphyrin Rings
A recent development in orthogonal labelling is to use a photo-excited triplet state (S = 1), localised on a porphyrin moiety coupled to the nitroxide radical (S = 1/2) [113]. Among organic chromophores, porphyrins have been widely studied by EPR spectroscopy, due to their high triplet yields, strong spin polarisation, reasonable relaxation rates, and moderate spectral anisotropy caused by the zero-field splitting (ZFS) interaction [114]. The triplet state has distinctive properties compared to metal centres; the large anisotropy due to the ZFS is accompanied by a strong spin polarisation of the spectrum, resulting from a non-Boltzmann population of the triplet-state sublevels by intersystem crossing from the corresponding excited singlet state [113,115]. The strong anisotropy of the triplet state ZFS can be potentially exploited to perform orientation selection; taking advantage of the compensating effect of the spin polarisation signal enhancement. A porphyrin-nitroxide system (bis-labelled peptide TPP-(Ala-Aib)4-Ala-TOAC-Ala-(Aib-Ala)2-OH) 44 (Figure 10) was synthesised by Di Valentin et al. [113] providing proof of the feasibility of using DEER spectroscopy to determine the inter-spin distance. The 15-residue peptide was labelled at the N-terminus with 5-(4-carboxyphenyl)-10,15,20-triphenylporphyrin (TPP) and at position 10 with TOAC. The peptide bridge connecting the paramagnetic probes consists of alternating L-alanine (Ala) and α‑aminoisobutyric acid (Aib) residues; known to promote an α-helix conformation and consequently a well-defined geometry in terms of distance, relative orientation, and restricted conformational flexibility.
Figure 10.
Chemical structure of the bis-labelled peptide TPP-(Ala-Aib)4-Ala-TOAC-Ala-(Aib-Ala)2-OH 44.
The DEER spectrum revealed a well-resolved and pronounced dipolar modulation. The significant extent of orientation selection expected in the relatively broad porphyrin triplet spectrum was not visible on the DEER spectrum and attributed to an observer position exciting almost all the possible molecular orientations with the tetrapyrrole plane parallel to the magnetic field, the lack of co-linearity between the zero field splitting principal axes and the spin−spin distance vector, and to a lesser extent by the small degree of rotational freedom of the para-substituted benzoyl group on the porphyrin. It is envisaged that this type of orthogonal approach can be extended to chromophores whose triplet state is highly polarised and well-characterised by EPR spectroscopy, such as porphyrin derivatives, fullerenes, and flavins [113,114,115]. It is also thought that chlorophylls and flavins could be exploited as endogenous photo-probes because of their presence in several classes of proteins.
5. Transition Metals
Many efforts have been recently devoted to the development of alternative spin labels with more attractive properties than conventional nitroxide radicals. Metals, such as copper (II), gadolinium (III), manganese (II) and nickel (II) have been utilised [116]. These systems present several EPR features, differing from nitroxide spin labels; firstly, they can have large hyperfine splitting and g-tensor anisotropy, which results in broad EPR spectra of which microwave pulses may only excite a fraction, resulting in strong orientation selection [117]. Secondly, relaxation rates are normally fast and limit the detection window and the accessible distance range. And thirdly, spin density can be distributed into the ligands, leading to the breakdown of the simple point-dipole model and the onset of an exchange coupling J [118].
5.1. Gadolinium (III)
Gadolinium (III) complexes have been utilised as spin labels for distance measurements in rigid and flexible model systems [119], for proteins [120,121,122] and for oligonucleotides [123], and are summarised in a recent review by Goldfarb [124].
There are several advantages in using Gd(III) ions (S = 7/2); firstly, the narrowing of the spin transition from ms = −1/2 to ms = 1/2 leads to increased sensitivity when using high magnetic fields. Secondly, the high transition probability of the S = 7/2 gadolinium spin with respect to the S = 1/2 nitroxide spin allows shorter microwave pump pulses for an equivalent microwave magnetic field. Lastly, the short spin-lattice relaxation time of Gd(III) complex allows the use of shorter repetition times, and the isotropic excitation resulting from the broadly distributed strengths and orientations of the ZFS typically suppresses orientation selection effects [21]. Gadolinium DEER measurements are usually performed at 25 K at a single magnetic field position, neglecting the orientation.
These advantages were demonstrated by Yagi et al. using Gd(III)-DOTA spin label 45 through reaction with a cysteine thiol groups to measure with outstanding accuracy distances of 6 nm between Gd(III) pairs at 95 GHz [122]. Furthermore, Matalon et al. [125] used gadolinium spin labels (Figure 11) to study transmembrane helical WALP peptides (composed of tryptophan (W), alanine (A) and leucine (L) amino acids) in model 1,2-dioleoyl-sn-glycero-3 phosphocholine (DOPC) vesicles systems. AWALP23 peptide was labelled at N and C termini cysteine residues with two different Gd-DOTA derivatives, DOTA 45 and C1 46. DEER measurements were performed at 95 GHz, showing distances of 3.7 nm and 4.3 nm with WALP23-DOTA 45 and WALP23-C1 46, respectively. The synthesis of C1 is described by Graham et al. [126].
Figure 11.
Gadolinium (III) spin labels DOTA chelate 45 and C1 46 with phenylethylamine substituents.
Gd(III) centres have also been used in combination with nitroxide radicals [127,128,129]. In particular, a combination of selective distance measurements in nitroxide-nitroxide, Gd-nitroxide and Gd-Gd pairs is of potential interest for structural studies of macromolecules and other nano-objects. In the Gd-nitroxide orthogonal spin pair; each type of paramagnetic centre opens the possibility to characterise the local environment of each spin probe, and macromolecular aggregation, without the need of additional singly labelled samples. This approach demonstrates good performance in distance measurements, and possessed high sensitivity using high-field EPR in protein systems [127,128,130,131]. Garbuio et al. [127] have used the Gd‑nitroxide orthogonal approach; to study bacteriophage T4-lysozyme using MTSL and the Gd(III) spin label 47 (Figure 12).
Figure 12.
The Gd(III) spin label 47; chelated by maleimido mono amide 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA), which can attach to the thiol group of cysteine residues.
5.2. Copper (II)/Nickel (II) Porphyrin/Nitroxide Systems
Copper (II) centres often occur in biological systems and play an important role in the macromolecular architecture as well as the structure and function of proteins and oligonucleotides. At 9 GHz the Cu(II) EPR spectrum extends over 700 G due to its g‑anisotropy and hyperfine coupling (I = 3/2 nucleus, 63Cu and 65Cu isotopes) so that the microwave pulses used in DEER typically excite only a small part of the spectrum, usually less than 50 MHz [12]. In DEER studies on Cu-nitroxide and Cu-Cu model systems [132,133], excitation in the g⊥ region of the copper spectrum led to negligible orientation selection and allowed a reasonable analysis of a distance distribution.
In order to resolve orientation effects, high quality experimental DEER data is required due to the small modulation depths and subtle changes with orientation [12]. Yang et al. have studied the optimal EPR conditions for data acquisition in the case of Cu-nitroxide and Cu-Cu systems with regard to commercial instrumentation [134]. Bode et al. have studied the influence of orientation selection, spin-density distribution, conformational flexibility, and exchange coupling on DEER measurements at 9 GHz for two structurally related nitroxide-labelled Cu(II) and Ni(II) porphyrin systems [118]. Cu(II)-Cu(II) distances have also been measured in proteins [116], such as the homotrimeric Cu-containing nitrite reductase [135], and the DNA modifying enzyme EcoRI endonuclease homodimer bound to its specific DNA recognition site [136].
6. Expression in Cells
6.1. Introducing Spin Labels via Unnatural Amino Acids
The complexity of measuring multiple spin pair interactions in proteins is usually avoided through mutations so that a single cysteine pair can be introduced in the desired positions. In the case of larger proteins when there may be a large number of cysteine residues, several rounds of mutagenesis will be required which is both time consuming and has an increased chance of perturbing the important structural and functional role played by these residues. Therefore, alternative labelling strategies are required to increase the scope of SDSL distance measurements to more complex problems.
One approach for spin labelling proteins is to use endogenous expression of unnatural amino acids [137,138]. These can be coded into the DNA by an amber stop codon (three letter code TAG) allowing the unnatural amino acids to be inserted into the sequence or to substitute a native residue, leaving all other amino acid residues untouched. This method works by introducing a suppressor tRNA which recognises amber stop codons in DNA and introduces the unnatural amino acid for coupling to the growing peptide. This technique is technically challenging because as well as synthesising an unnatural amino acid, a mutated tRNA-aminoacyl synthetase enzyme has to be evolved which can catalyse the esterification of the unnatural amino acid to the suppressor tRNA. Nonetheless over thirty different unnatural amino acids have been incorporated into proteins this way [138].
For introducing spin labels to proteins, this method has been used in two different ways: by introducing amino acids with paramagnetic side chains; or, by introducing unnatural amino acids with side chains of orthogonal chemical reactivity to the twenty natural amino acids, allowing subsequent spin labelling.
6.1.1. Unnatural Amino Acids with Paramagnetic Centres
As well as being able to retain native cysteines, expressing unnatural amino acids with paramagnetic centres has the advantage over other spin labels that the conformational degrees of freedom of the side chain are limited, as with short spin labelled peptides produced by solid phase synthesis. Since the intracellular environment is strongly reducing, the main synthetic challenge is to design an amino acid with a paramagnetic centre which can persist in cells without being reduced to the corresponding hydroxylamine over the several hours necessary for protein expression.
The first example of an endogenously expressed paramagnetic amino acid was demonstrated in 1994 using T4 lysozyme, singly labelled with the nitroxide linked amino acid 48 (Figure 13). CW EPR was used to measure the label in the purified protein [139]. Distance measurements require at least two labels but introducing more than one unnatural amino acid requires having multiple amber stop codons in the DNA sequence which can be recognised by the cellular machinery for termination of the growing peptide, leading to lower yields of expression. Recent developments in molecular biology have allowed proteins to be expressed in E. coli where this mechanism has been removed, rapidly expanding the applicability of this method [140,141,142,143]. The first example of DEER on protein with unnatural paramagnetic amino acids was reported earlier this year with the unnatural nitroxide amino acid 49 [144].
Figure 13.
Paramagnetic unnatural amino acids 48 and 49.
6.1.2. Unnatural Amino Acids with Orthogonal Chemical Reactivity
Introducing unnatural amino acids with orthogonal reactivity to the natural twenty amino acids allows selective labelling after expression. This allows a much greater scope for labelling strategies, however, the side chains are generally longer than when the paramagnetic centre is endogenously expressed.
The unnatural amino acid p-acetyl phenylalanine 50 [145] contains a ketone group which can be reacted with the hydroxylamine 51 to give the ketoxime linked nitroxide side chain K1 52 (Figure 14) and distances have been determined by DEER [146]. However, this reaction was demonstrated at pH 4 and required several hours at 37 °C; conditions which would not be tolerated by many proteins.
Figure 14.
Spin labelling of unnatural amino acids (a) p-acetyl phenylalanine 52 and (b) p-azidophenylalanine 55.
Click chemistry offers an opportunity for pH neutral labelling, although the use of Cu(I) as a catalyst is undesirable due to cytotoxicity and the need to maintain reducing conditions unfavourable with nitroxides. Copper-free click chemistry has been used for fluorescent labelling of unnatural amino acids [147] and subsequently for spin labelling [148]. The unnatural amino acid p-acetyl phenylalanine 53 with a cyclo-octyne label 54 gives a triazole linked nitroxide side chain 55 [148] (also referred to as T1 [24] in a metal-free synthesis. However, dipolar EPR distance measurements have not yet been published using a pair of these spin labels.
7. In-Cell EPR
Unnatural amino acid mutagenesis allows opportunities for detection of EPR signals in-cell without purification of the protein and continuous-wave EPR measurements have been recorded using unnatural amino acid 49 [144]. But the concentration of proteins expressed is limited and no in-cell dipolar EPR measurements have currently been reported using this method.
In-cell EPR has largely been approached by the microinjection of spin labelled samples into cells; using oocytes of the African frog Xenopus laevis. A potential problem was the reducing cellular environment; which could convert a nitroxide into its corresponding hydroxylamine. Therefore, methods involving microinjection require freeze-quenching the sample after a short incubation period.
The first in-cell DEER was reported in 2010 on the protein ubiquitin labelled with 3-maleimido-proxyl 56 (Figure 15), microinjected into Xenopus laevis, then freeze-quenched to prevent nitroxide reduction [149]. The maleimido linker group was chosen for its resistance to reductive cleavage.
Figure 15.
Spin labels used for in-cell DEER. The cysteine binding 3-maleimido-proxyl 9, and nucleic acid base binding labels TEMPA 57 and TPA 11.
In-cell DEER has also been carried out on nucleic acids using the spin labels TEMPA 57 [58] and TPA 11 [57,150] (Figure 15). It has been observed that TEMPA 57 injected cells had to be freeze‑quenched in as little as ten seconds [58], whereas cells containing TPA 11 linked nucleic acids could be incubated for up to seventy minutes because 5-membered ring nitroxides are significantly more stable than 6-membered ring nitroxides to reducing cellular conditions [59,71,72]. These longer incubation times of TPA 11 were preferable as this allowed nucleic acid conformations to reach equilibrium in the intracellular environment [150].
In Situ His Tag Labelling
Poly-histidine tags of recombinant proteins have been labelled in situ in the presence of nickel using a proxyl-trisNTA label 58 (Figure 16), allowing selective labelling from crude cell lysates and EPR measurement without purification [151].
Figure 16.
Histidine tag spin label proxyl-trisNTA 58. X = histidine coordination site.
The proxyl-trisNTA spin label 58 contains a proxyl nitroxide reporter group along with three nitrilotriacetic acid (NTA) groups which exploit the binding affinity of polyhistidine tags for nickel loaded NTA, which is commonly used as a protein purification strategy for polyhistidine tagged proteins. The short Tm relaxation time of the nickel bound label meant that it was not practical to measure the distance between a pair of these labels by DEER; but this label was usefully demonstrated as a pump spin with a separate MTSL as an observer spin.
8. Conclusions
Spin labelling techniques combined with pulse EPR techniques have allowed greater insight into biological structures particularly enriching our conformational mobile view of many proteins. The most widely used label is MTSL comprising a nitroxyl radical attached to a five-membered ring with a methanethiosulfonate moiety. In the case of nucleotides, TOAC has readily allowed the determination of orientation as well as distance. Gd(III) and trityl labels have also been validated for accurate dipolar measurements. Importantly, orthogonal spin labelling strategies have permitted discriminatory structural information. The need to measure inside live cells has resulted in promising experiments using nitroxides and this area is of active research. The use of Gd(III) and trityl labels have great potential for measurements within cells because of their favourable reduction properties. The novel idea to use photo-EPR sensitive labels has potential under well-chosen conditions. The advent of methods to incorporate unnatural amino acids into proteins which have multitudes of cysteine residues allows rich future biological targets. It is envisaged that further advances incorporating unnatural amino acids combined with novel labels, as well as developments in instrumentation [152], will lead to increased growth in EPR as a biochemical tool. Further applications of dipolar spectroscopy using labels to study materials are on-going in many laboratories. The design of future labels needs to consider EPR spectral features; resonance width, spin delocalisation and intensity, as well as chemical factors; reduction potential and molecular size. Greater rigidity of labels is also advantageous to allow orientation determination.
Acknowledgments
We are grateful for the EPSRC National EPR Research Facility & Service. AJF and MGC also thank Bruker for funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Schweiger A. Principles of pulse electron paramagnetic resonance. Oxford University Press; Oxford, UK: 2001. [Google Scholar]
- 2.Bowman M.K. Pulsed Electron Paramagnetic Resonance. In: Brustolon M., Giamello E., editors. Electron Paramagnetic Resonance: A Practitioner’s Toolkit. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2009. pp. 159–194. [Google Scholar]
- 3.Berliner L.J., Eaton G.R., Eaton S.S. Distance Measurements in Biological Systems by EPR. Kluwer Academic; New York, NY, USA: 2002. [Google Scholar]
- 4.Borbat P.P., Freed J.H. Pulse Dipolar Electron Spin Resonance: Distance Measurements. In: Timmel C.R., Harmer J.R., editors. Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences. Springer; Berlin, Germany: 2013. pp. 1–82. Structure and Bonding. [Google Scholar]
- 5.Milov A.D., Salikhov K.M., Shchirov M.D. Application of ENDOR in electron-spin echo for paramagnetic center space distribution in solids. Fiz. Tverd. Tela. 1981;23:975–982. [Google Scholar]
- 6.Milov A.D., Ponomarev A.B., Tsvetkov Y.D. Electron-electron double resonance in electron spin echo: Model biradical systems and the sensitized photolysis of decalin. Chem. Phys. Lett. 1984;110:67–72. doi: 10.1016/0009-2614(84)80148-7. [DOI] [Google Scholar]
- 7.Pannier M., Veit S., Godt A., Jeschke G., Spiess H.W. Dead-time free measurement of dipole–dipole interactions between electron spins. J. Magn. Reson. 2011;213:316–325. doi: 10.1016/j.jmr.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 8.Ward R., Bowman A., Sozudogru E., El-Mkami H., Owen-Hughes T., Norman D.G. EPR distance measurements in deuterated proteins. J. Magn. Reson. San Diego Calif. 1997. 2010;207:164–167. doi: 10.1016/j.jmr.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeschke G. DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 10.Larsen R.G., Singel D.J. Double electron—electron resonance spin-echo modulation—spectroscopic measurement of electron-spin pair separations in orientationally disordered solids. J. Chem. Phys. 1993;98:5134–5146. doi: 10.1063/1.464916. [DOI] [Google Scholar]
- 11.Denysenkov V.P., Prisner T.F., Stubbe J., Bennati M. High-field pulsed electron-electron double resonance spectroscopy to determine the orientation of the tyrosyl radicals in ribonucleotide reductase. Proc. Natl. Acad. Sci. USA. 2006;103:13386–13390. doi: 10.1073/pnas.0605851103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen A.M., Tait C.E., Timmel C.R., Harmer J.R. Orientation-Selective DEER Using Rigid Spin Labels, Cofactors, Metals, and Clusters. In: Timmel C.R., Harmer J.R., editors. Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences. Volume 152. Springer; Berlin, Germany: 2013. pp. 283–327. Structure and Bonding. [Google Scholar]
- 13.Saxena S., Freed J.H. Double quantum two-dimensional Fourier transform electron spin resonance: Distance measurements. Chem. Phys. Lett. 1996;251:102–110. doi: 10.1016/0009-2614(96)00075-9. [DOI] [Google Scholar]
- 14.Saxena S., Freed J.H. Theory of double quantum two-dimensional electron spin resonance with application to distance measurements. J. Chem. Phys. 1997;107:1317–1340. doi: 10.1063/1.474490. [DOI] [Google Scholar]
- 15.Borbat P.P., Freed J.H. Multiple-quantum ESR and distance measurements. Chem. Phys. Lett. 1999;313:145–154. doi: 10.1016/S0009-2614(99)00972-0. [DOI] [Google Scholar]
- 16.Borbat P.P., Freed J.H. Pros and cons of pulse dipolar ESR: DQC and DEER. EPR Newsl. 2007:21–33. [Google Scholar]
- 17.Kay C.W.M., Elsässer C., Bittl R., Farrell S.R., Thorpe C. Determination of the distance between the two neutral flavin radicals in augmenter of liver regeneration by pulsed ELDOR. J. Am. Chem. Soc. 2006;128:76–77. doi: 10.1021/ja057308g. [DOI] [PubMed] [Google Scholar]
- 18.Elsässer C., Brecht M., Bittl R. Pulsed electron-electron double resonance on multinuclear metal clusters: Assignment of spin projection factors based on the dipolar interaction. J. Am. Chem. Soc. 2002;124:12606–12611. doi: 10.1021/ja027348+. [DOI] [PubMed] [Google Scholar]
- 19.Bennati M., Weber A., Antonic J., Perlstein D.L., Robblee J., Stubbe J. Pulsed ELDOR spectroscopy measures the distance between the two tyrosyl radicals in the R2 subunit of the E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2003;125:14988–14989. doi: 10.1021/ja0362095. [DOI] [PubMed] [Google Scholar]
- 20.Fielding A.J., Brodhun F., Koch C., Pievo R., Denysenkov V., Feussner I., Bennati M. Multifrequency electron paramagnetic resonance characterization of PpoA, a CYP450 fusion protein that catalyzes fatty acid dioxygenation. J. Am. Chem. Soc. 2011;133:9052–9062. doi: 10.1021/ja202207t. [DOI] [PubMed] [Google Scholar]
- 21.Yulikov M., Lueders P., Warsi M.F., Chechik V., Jeschke G. Distance measurements in Au nanoparticles functionalized with nitroxide radicals and Gd(3+)-DTPA chelate complexes. Phys. Chem. Chem. Phys. PCCP. 2012;14:10732–10746. doi: 10.1039/c2cp40282c. [DOI] [PubMed] [Google Scholar]
- 22.Jeschke G. Determination of the nanostructure of polymer materials by electron paramagnetic resonance spectroscopy. Macromol. Rapid Commun. 2002;23:227–246. doi: 10.1002/1521-3927(20020301)23:4<227::AID-MARC227>3.0.CO;2-D. [DOI] [Google Scholar]
- 23.Bordignon E., Steinhoff H.-J. ESR Spectroscopy in Membrane Biophysics. Volume 27. Springer; New York, NY, USA: 2007. Membrane Protein Structure and Dynamics Studied by Site-Directed Spin-Labeling ESR; pp. 129–164. Biological Magnetic Resonance. [Google Scholar]
- 24.Hubbell W.L., López C.J., Altenbach C., Yang Z. Technological advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 2013;23:725–733. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeschke G. Conformational dynamics and distribution of nitroxide spin labels. Prog. Nucl. Magn. Reson. Spectrosc. 2013;72:42–60. doi: 10.1016/j.pnmrs.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Hideg K., Kalai T., Sar C.P. Recent results in chemistry and biology of nitroxides. J. Heterocycl. Chem. 2005;42:437–450. doi: 10.1002/jhet.5570420311. [DOI] [Google Scholar]
- 27.Merbouh N., Bobbitt J.M., Brückner C. Preperation of tetramethylpiperdine-1-oxoammonium salts and their use as oxidants in organic chemistry, a review. Org. Prep. Proced. Int. 2004;36:1–31. doi: 10.1080/00304940409355369. [DOI] [Google Scholar]
- 28.Hawker C.J., Bosman A.W., Harth E. New polymer synthesis by nitroxide mediated living radical polymerizations. Chem. Rev. 2001;101:3661–3688. doi: 10.1021/cr990119u. [DOI] [PubMed] [Google Scholar]
- 29.Joseph J., Kalyanaraman B., Hyde J.S. Trapping of nitric oxide by nitronyl nitroxides: An electron spin resonance investigation. Biochem. Biophys. Res. Commun. 1993;192:926–934. doi: 10.1006/bbrc.1993.1504. [DOI] [PubMed] [Google Scholar]
- 30.Shen J., Bottle S., Khan N., Grinberg O., Reid D., Micallef A., Swartz H. Development of isoindoline nitroxides for EPR oximetry in viable systems. Appl. Magn. Reson. 2002;22:357–368. doi: 10.1007/BF03166117. [DOI] [Google Scholar]
- 31.Rozantsev E.G., Ulrich H. Reactions of Radicals Not Involving the Free Valences. In: Ulrich H., editor. Free Nitroxyl Radicals. Springer; New York, NY, USA: 1970. pp. 53–66. [Google Scholar]
- 32.Brik M.-E. Chemistry of persistent free bi- and polyradicals. Heterocycles. 1995;41:2827–2873. doi: 10.3987/REV-95-469. [DOI] [Google Scholar]
- 33.Naik N., Braslau R. Synthesis and applications of optically active nitroxides. Tetrahedron. 1998;54:667–696. doi: 10.1016/S0040-4020(97)10061-8. [DOI] [Google Scholar]
- 34.Sale K., Song L., Liu Y.-S., Perozo E., Fajer P. Explicit treatment of spin labels in modeling of distance constraints from dipolar EPR and DEER. J. Am. Chem. Soc. 2005;127:9334–9335. doi: 10.1021/ja051652w. [DOI] [PubMed] [Google Scholar]
- 35.Cunningham T.F., McGoff M.S., Sengupta I., Jaroniec C.P., Horne W.S., Saxena S. High-resolution structure of a protein spin-label in a solvent-exposed β-sheet and comparison with DEER spectroscopy. Biochemistry. 2012;51:6350–6359. doi: 10.1021/bi300328w. [DOI] [PubMed] [Google Scholar]
- 36.Polyhach Y., Bordignon E., Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- 37.Sarver J.L., Townsend J.E., Rajapakse G., Jen-Jacobson L., Saxena S. Simulating the dynamics and orientations of spin-labeled side chains in a protein-DNA complex. J. Phys. Chem. B. 2012;116:4024–4033. doi: 10.1021/jp211094n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klose D., Klare J.P., Grohmann D., Kay C.W.M., Werner F., Steinhoff H.-J. Simulation vs. Reality: A Comparison of In Silico Distance Predictions with DEER and FRET Measurements. PLoS One. 2012;7:e39492. doi: 10.1371/journal.pone.0039492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fajer P.G., Brown L., Song L. Practical Pulsed Dipolar ESR (DEER) In: Hemminga M.A., Berliner L.J., editors. ESR Spectroscopy in Membrane Biophysics. Volume 27. Springer; New York, NY, USA: 2007. pp. 95–128. Biological Magnetic Resonance. [Google Scholar]
- 40.Islam S.M., Stein R.A., Mchaourab H.S., Roux B. Structural refinement from restrained-ensemble simulations based on EPR/DEER data: Application to T4 lysozyme. J. Phys. Chem. B. 2013;117:4740–4754. doi: 10.1021/jp311723a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hubbell W.L., Gross A., Langen R., Lietzow M.A. Recent advances in site-directed spin labeling of proteins. Curr. Opin. Struct. Biol. 1998;8:649–656. doi: 10.1016/S0959-440X(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 42.Fawzi N.L., Fleissner M.R., Anthis N.J., Kálai T., Hideg K., Hubbell W.L., Clore G.M. A rigid disulfide-linked nitroxide side chain simplifies the quantitative analysis of PRE data. J. Biomol. NMR. 2011;51:105–114. doi: 10.1007/s10858-011-9545-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kálai T. Synthesis and reactions of a symmetric paramagnetic pyrrolidine diene. Synthesis. 1999;1999:973–980. doi: 10.1055/s-1999-3502. [DOI] [Google Scholar]
- 44.Fleissner M.R., Bridges M.D., Brooks E.K., Cascio D., Kálai T., Hideg K., Hubbell W.L. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proc. Natl. Acad. Sci. USA. 2011;108:16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shelke S.A., Sigurdsson S.T. Site-Directed Nitroxide Spin Labeling of Biopolymers. In: Timmel C.R., Harmer J.R., editors. Structural Information from Spin-Labels and Intrinsic Paramagnetic Centres in the Biosciences. Volume 152. Springer; Berlin/Heidelberg, Germany: 2013. pp. 121–162. Structure and Bonding. [Google Scholar]
- 46.Schreier S., Bozelli J.C., Marín N., Vieira R.F.F., Nakaie C.R. The spin label amino acid TOAC and its uses in studies of peptides: Chemical, physicochemical, spectroscopic, and conformational aspects. Biophys. Rev. 2012;4:45–66. doi: 10.1007/s12551-011-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tominaga M., Barbosa S.R., Poletti E.F., Zukerman-Schpector J., Marchetto R., Schreier S., Paiva A.C., Nakaie C.R. Fmoc-POAC: [(9-fluorenylmethyloxycarbonyl)-2,2,5,5-tetramethylpyrrolidine-N-oxyl-3-amino-4-carboxylic acid]: A novel protected spin labeled β-amino acid for peptide and protein chemistry. Chem. Pharm. Bull. 2001;49:1027–1029. doi: 10.1248/cpb.49.1027. [DOI] [PubMed] [Google Scholar]
- 48.Marsh D., Jost M., Peggion C., Toniolo C. TOAC spin labels in the backbone of alamethicin: EPR studies in lipid membranes. Biophys. J. 2007;92:473–481. doi: 10.1529/biophysj.106.092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoller S., Sicoli G., Baranova T.Y., Bennati M., Diederichsen U. TOPP: A novel nitroxide-labeled amino acid for EPR distance measurements. Angew. Chem. Int. Ed. 2011;50:9743–9746. doi: 10.1002/anie.201103315. [DOI] [PubMed] [Google Scholar]
- 50.Reginsson G.W., Schiemann O. Spin labeling of DNA and RNA. In: Roberts G.C.K., editor. Encyclopedia of Biophysics. Springer; Berlin/Heidelberg, Germany: 2013. pp. 2429–2431. [Google Scholar]
- 51.Allerson C.R., Chen S.L., Verdine G.L. A chemical method for site-specific modification of RNA: The convertible nucleoside approach. J. Am. Chem. Soc. 1997;119:7423–7433. doi: 10.1021/ja962858n. [DOI] [Google Scholar]
- 52.Sicoli G., Wachowius F., Bennati M., Höbartner C. Probing secondary structures of spin-labeled RNA by pulsed EPR spectroscopy. Angew. Chem. Int. Ed. 2010;49:6443–6447. doi: 10.1002/anie.201000713. [DOI] [PubMed] [Google Scholar]
- 53.Barhate N., Cekan P., Massey A.P., Sigurdsson S.T. A nucleoside that contains a rigid nitroxide spin label: A fluorophore in disguise. Angew. Chem. Int. Ed. 2007;46:2655–2658. doi: 10.1002/anie.200603993. [DOI] [PubMed] [Google Scholar]
- 54.Höbartner C., Sicoli G., Wachowius F., Gophane D.B., Sigurdsson S.T. Synthesis and Characterization of RNA Containing a Rigid and Nonperturbing Cytidine-Derived Spin Label. J. Org. Chem. 2012;77:7749–7754. doi: 10.1021/jo301227w. [DOI] [PubMed] [Google Scholar]
- 55.Gannett P.M., Darian E., Powell J.H., Johnson E.M. A short procedure for synthesis of 4-ethynyl-2,2,6,6-tetramethyl-3,4-dehydro-piperidine-1-oxyl nitroxide. Synth. Commun. 2001;31:2137–2141. doi: 10.1081/SCC-100104464. [DOI] [Google Scholar]
- 56.Piton N., Mu Y., Stock G., Prisner T.F., Schiemann O., Engels J.W. Base-specific spin-labeling of RNA for structure determination. Nucleic Acids Res. 2007;35:3128–3143. doi: 10.1093/nar/gkm169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krstić I., Hänsel R., Romainczyk O., Engels J.W., Dötsch V., Prisner T.F. Long-range distance measurements on nucleic acids in cells by pulsed EPR spectroscopy. Angew. Chem. 2011;123:5176–5180. doi: 10.1002/ange.201100886. [DOI] [PubMed] [Google Scholar]
- 58.Azarkh M., Okle O., Singh V., Seemann I.T., Hartig J.S., Dietrich D.R., Drescher M. Long-range distance determination in a DNA model system inside Xenopus laevis oocytes by in-cell spin-label EPR. ChemBioChem. 2011;12:1992–1995. doi: 10.1002/cbic.201100281. [DOI] [PubMed] [Google Scholar]
- 59.Azarkh M., Okle O., Eyring P., Dietrich D.R., Drescher M. Evaluation of spin labels for in-cell EPR by analysis of nitroxide reduction in cell extract of Xenopus laevis oocytes. J. Magn. Reson. 2011;212:450–454. doi: 10.1016/j.jmr.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 60.Ramos A., Varani G. A new method to detect long-range protein-RNA contacts: NMR detection of electron-proton relaxation induced by nitroxide spin-labeled RNA. J. Am. Chem. Soc. 1998;120:10992–10993. doi: 10.1021/ja982496e. [DOI] [Google Scholar]
- 61.Duss O., Yulikov M., Jeschke G., Allain F.H.-T. EPR-aided approach for solution structure determination of large RNAs or protein-RNA complexes. Nat. Commun. 2014 doi: 10.1038/ncomms4669. [DOI] [PubMed] [Google Scholar]
- 62.Ding P., Wunnicke D., Steinhoff H.-J., Seela F. Site-directed spin-labeling of DNA by the azide-alkyne “click” reaction: nanometer distance measurements on 7-deaza-2'-deoxyadenosine and 2'-deoxyuridine nitroxide conjugates spatially separated or linked to a “dA-dT” base pair. Chem. Weinh. Bergstr. Ger. 2010;16:14385–14396. doi: 10.1002/chem.201001572. [DOI] [PubMed] [Google Scholar]
- 63.Jakobsen U., Shelke S.A., Vogel S., Sigurdsson S.T. Site-directed spin-labeling of nucleic acids by click chemistry: Detection of abasic sites in duplex DNA by EPR spectroscopy. J. Am. Chem. Soc. 2010;132:10424–10428. doi: 10.1021/ja102797k. [DOI] [PubMed] [Google Scholar]
- 64.Edwards T.E., Sigurdsson S.T. Site-specific incorporation of nitroxide spin-labels into 2'-positions of nucleic acids. Nat. Protoc. 2007;2:1954–1962. doi: 10.1038/nprot.2007.273. [DOI] [PubMed] [Google Scholar]
- 65.Schiemann O., Weber A., Edwards T.E., Prisner T.F., Sigurdsson S.T. Nanometer distance measurements on RNA using PELDOR. J. Am. Chem. Soc. 2003;125:3434–3435. doi: 10.1021/ja0274610. [DOI] [PubMed] [Google Scholar]
- 66.Qin P.Z., Haworth I.S., Cai Q., Kusnetzow A.K., Grant G.P.G., Price E.A., Sowa G.Z., Popova A., Herreros B., He H. Measuring nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nat. Protoc. 2007;2:2354–2365. doi: 10.1038/nprot.2007.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cai Q., Kusnetzow A.K., Hubbell W.L., Haworth I.S., Gacho G.P.C., van Eps N., Hideg K., Chambers E.J., Qin P.Z. Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res. 2006;34:4722–4730. doi: 10.1093/nar/gkl546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pauly H., Rossbach J. Ueber die bildung von pyrrolin- und pyrrolidin-derivaten aus triacetonamin. Berichte Dtsch. Chem. Ges. 1899;32:2000–2014. doi: 10.1002/cber.189903202110. [DOI] [Google Scholar]
- 69.Sosnovsky G., Cai Z. A study of the Favorskii rearrangement with 3-bromo-4-oxo-2,2,6,6-tetramethylpiperidine-1-oxyl. J. Org. Chem. 1995;60:3414–3418. doi: 10.1021/jo00116a029. [DOI] [Google Scholar]
- 70.Zhdanov R.I. Nitroxyl Radicals and Non-Radical Reactions of Free Radicals. In: Zhdanov R.I., editor. Bioactive Spin Labels. Springer; Berlin, Germany: 1992. pp. 23–82. [Google Scholar]
- 71.Couet W.R., Brasch R.C., Sosnovsky C., Lukszo J., Prakash I., Gnewech C.T., Tozer T.N. Influerce of chemical structure of nitroxyl spin labels on their reduction by ascorbic acid. Tetrahedron. 1985;41:1165–1172. doi: 10.1016/S0040-4020(01)96516-0. [DOI] [Google Scholar]
- 72.Morris S., Sosnovsky G., Hui B., Huber C.O., Rao N.U.M., Swartz H.M. Chemical and electrochemical reduction rates of cyclic nitroxides (nitroxyls) J. Pharm. Sci. 1991;80:149–152. doi: 10.1002/jps.2600800212. [DOI] [PubMed] [Google Scholar]
- 73.Bottle S.E., Chand U., Micallef A.S. Hydrogen abstraction from unactivated hydrocarbons using a photochemically excited isoindoline nitroxide. Chem. Lett. 1997;26:857–858. doi: 10.1246/cl.1997.857. [DOI] [Google Scholar]
- 74.Gillies D.G., Sutcliffe L.H., Wu X. NMR determination of EPR hyperfine coupling constants of some 5-(n-alkyl)-1,1,3,3-tetrakis(trideuteriomethyl)isoindolin-2-yloxyls. J. Chem. Soc. Faraday Trans. 1994;90:2345–2349. doi: 10.1039/ft9949002345. [DOI] [Google Scholar]
- 75.Bottle S.E., Gillies D.G., Micallef A.S., Reid D.A., Sutcliffe L.H. ESR measurements of the partitioning of some new spin probes in n-octanol-water. Magn. Reson. Chem. 1999;37:730–734. doi: 10.1002/(SICI)1097-458X(199910)37:10<730::AID-MRC530>3.0.CO;2-E. [DOI] [Google Scholar]
- 76.Keddie D.J., Fairfull-Smith K.E., Bottle S.E. The palladium-catalysed copper-free Sonogashira coupling of isoindoline nitroxides: A convenient route to robust profluorescent carbon-carbon frameworks. Org. Biomol. Chem. 2008;6:3135–3143. doi: 10.1039/b806963h. [DOI] [PubMed] [Google Scholar]
- 77.Blinco J.P., McMurtrie J.C., Bottle S.E. The first example of an azaphenalene profluorescent nitroxide. Eur. J. Org. Chem. 2007;2007:4638–4641. doi: 10.1002/ejoc.200700545. [DOI] [Google Scholar]
- 78.Reid D.A., Bottle S.E. The synthesis of water soluble isoindoline nitroxides and a pronitroxide hydroxylamine hydrochloride UV-Vis probe for free radicals. Chem. Commun. 1998:1907–1908. [Google Scholar]
- 79.Blinco J.P., Chalmers B.A., Chou A., Fairfull-Smith K.E., Bottle S.E. Spin-coated carbon. Chem. Sci. 2013;4:3411–3415. doi: 10.1039/c3sc51396c. [DOI] [Google Scholar]
- 80.Khramtsov V.V. Functional EPR Spectroscopy and Imaging of Nitroxides. In: Pifat-Mrzljak G., editor. Supramolecular Structure and Function 9. Springer; Dordrecht, The Netherlands: 2007. pp. 181–208. [Google Scholar]
- 81.Gruian C., Vulpoi A., Steinhoff H.-J., Simon S. Structural changes of methemoglobin after adsorption on bioactive glass, as a function of surface functionalization and salt concentration. J. Mol. Struct. 2012;1015:20–26. doi: 10.1016/j.molstruc.2012.01.045. [DOI] [Google Scholar]
- 82.Griffith O.H., McConnell H.M. A Nitroxide-maleimide Spin Label. Proc. Natl. Acad. Sci. USA. 1966;55:8–11. doi: 10.1073/pnas.55.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hankovszky H.O., Hideg K., Jerkovich G. Synthesis of 3-substituted 2,5-dihydro-2,2,5,5-tetramethyl-1H-pyrrol-1-yloxyl radicals, useful for spin-labelling of biomolecules. Synthesis. 1989;1989:526–529. doi: 10.1055/s-1989-27306. [DOI] [Google Scholar]
- 84.Berliner L.J., McConnell H.M. A spin-labeled substrate for alpha-chymotrypsin. Proc. Natl. Acad. Sci. USA. 1966;55:708–712. doi: 10.1073/pnas.55.4.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adackaparayil M., Smith J.H. Preparation and reactivity of a new spin label reagent. J. Org. Chem. 1977;42:1655–1656. doi: 10.1021/jo00429a042. [DOI] [Google Scholar]
- 86.Hankovszky O.H., Hideg K., Goldammer E.V., Matuszak E., Kolkenbrock H., Tschesche H., Wenzel H.R. New nitroxide reagents for the selective spin-labelling at the guanidino moiety of arginine residues in peptides and proteins. BBA-Protein Struct. Mol. Enzymol. 1987;916:152–155. doi: 10.1016/0167-4838(87)90223-8. [DOI] [Google Scholar]
- 87.Lorenzi M., Puppo C., Lebrun R., Lignon S., Roubaud V., Martinho M., Mileo E., Tordo P., Marque S.R.A., Gontero B., et al. Tyrosine-targeted spin labeling and EPR spectroscopy: An alternative strategy for studying structural transitions in proteins. Angew. Chem. Int. Ed. 2011;50:9108–9111. doi: 10.1002/anie.201102539. [DOI] [PubMed] [Google Scholar]
- 88.Kolb H.C., Finn M.G., Sharpless K.B. Click chemistry: Diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 89.Kálai T., Hubbell W., Hideg K. Click reactions with nitroxides. Synthesis. 2009;2009:1336–1340. doi: 10.1055/s-0028-1088018. [DOI] [Google Scholar]
- 90.Kálai T., Jekő J., Hideg K. Synthesis of a paramagnetic boronic acid as a useful synthetic building block and carbohydrate affinity spin probe. Tetrahedron Lett. 2004;45:8395–8398. doi: 10.1016/j.tetlet.2004.09.044. [DOI] [Google Scholar]
- 91.Rajca A., Kathirvelu V., Roy S.K., Pink M., Rajca S., Sarkar S., Eaton S.S., Eaton G.R. A spirocyclohexyl nitroxide amino acid spin label for pulsed EPR spectroscopy distance measurements. Chem. Weinh. Bergstr. Ger. 2010;16:5778–5782. doi: 10.1002/chem.200903102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McNulty J.C., Thompson D.A., Carrasco M.R., Millhauser G.L. Dap-SL: A new site-directed nitroxide spin labeling approach for determining structure and motions in synthesized peptides and proteins. FEBS Lett. 2002;529:243–248. doi: 10.1016/S0014-5793(02)03352-5. [DOI] [PubMed] [Google Scholar]
- 93.Kim N.-K., Murali A., DeRose V.J. A distance ruler for RNA using EPR and site-directed spin labeling. Chem. Biol. 2004;11:939–948. doi: 10.1016/j.chembiol.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 94.Kálai T., Kuppusamy M.L., Balog M., Selvendiran K., Rivera B.K., Kuppusamy P., Hideg K. Synthesis of N-substituted 3,5-bis(arylidene)-4-piperidones with high antitumor and antioxidant activity. J. Med. Chem. 2011;54:5414–5421. doi: 10.1021/jm200353f. [DOI] [PubMed] [Google Scholar]
- 95.Kósa C., Danko M., Hrdlovič P. Preparation and spectral characterization of fluorescence probes based on 4-N,N-dimethylamino benzoic acid and sterically hindered amines. J. Fluoresc. 2012;22:1371–1381. doi: 10.1007/s10895-012-1076-7. [DOI] [PubMed] [Google Scholar]
- 96.Mravljak J., Ojsteršek T., Pajk S., Sollner Dolenc M. Coumarin-based dual fluorescent spin-probes. Tetrahedron Lett. 2013;54:5236–5238. doi: 10.1016/j.tetlet.2013.07.084. [DOI] [Google Scholar]
- 97.Dzuba S.A., Maryasov A.G., Salikhov K.M., Tsvetkov Y.D. Superslow rotations of nitroxide radicals studied by pulse EPR spectroscopy. J. Magn. Reson. 1984;58:95–117. [Google Scholar]
- 98.Lindgren M., Eaton G.R., Eaton S.S., Jonsson B.-H., Hammarström P., Svensson M., Carlsson U. Electron spin echo decay as a probe of aminoxyl environment in spin-labeled mutants of human carbonic anhydrase II†. J. Chem. Soc. Perkin Trans. 2. 1997:2549–2554. [Google Scholar]
- 99.Sato H., Kathirvelu V., Fielding A., Blinco J.P., Micallef A.S., Bottle S.E., Eaton S.S., Eaton G.R. Impact of molecular size on electron spin relaxation rates of nitroxyl radicals in glassy solvents between 100 and 300 K. Mol. Phys. 2007;105:2137–2151. doi: 10.1080/00268970701724966. [DOI] [Google Scholar]
- 100.Kathirvelu V., Smith C., Parks C., Mannan M.A., Miura Y., Takeshita K., Eaton S.S., Eaton G.R. Relaxation rates for spirocyclohexyl nitroxyl radicals are suitable for interspin distance measurements at temperatures up to about 125 K. Chem. Commun. 2009:454–456. doi: 10.1039/b817758a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Khan N., Blinco J.P., Bottle S.E., Hosokawa K., Swartz H.M., Micallef A.S. The evaluation of new and isotopically labeled isoindoline nitroxides and an azaphenalene nitroxide for EPR oximetry. J. Magn. Reson. 2011;211:170–177. doi: 10.1016/j.jmr.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abe J., Ueki S., Arata T., Nakazawa S., Yamauchi S., Ohba Y. Improved sensitivity by isotopic substitution in distance measurements based on double quantum coherence EPR. Appl. Magn. Reson. 2012;42:473–485. doi: 10.1007/s00723-012-0317-x. [DOI] [Google Scholar]
- 103.Jeschke G., Zimmermann H., Godt A. Isotope selection in distance measurements between nitroxides. J. Magn. Reson. 2006;180:137–146. doi: 10.1016/j.jmr.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 104.Fielding A.J., Carl P.J., Eaton G.R., Eaton S.S. Multifrequency EPR of four triarylmethyl radicals. Appl. Magn. Reson. 2005;28:231–238. doi: 10.1007/BF03166758. [DOI] [Google Scholar]
- 105.Owenius R., Eaton G.R., Eaton S.S. Frequency (250 MHz to 9.2 GHz) and viscosity dependence of electron spin relaxation of triarylmethyl radicals at room temperature. J. Magn. Reson. 2005;172:168–175. doi: 10.1016/j.jmr.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 106.Yang Z., Liu Y., Borbat P., Zweier J.L., Freed J.H., Hubbell W.L. Pulsed ESR dipolar spectroscopy for distance measurements in immobilized spin labeled proteins in liquid solution. J. Am. Chem. Soc. 2012;134:9950–9952. doi: 10.1021/ja303791p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shevelev G.Y., Krumkacheva O.A., Lomzov A.A., Kuzhelev A.A., Rogozhnikova O.Y., Trukhin D.V., Troitskaya T.I., Tormyshev V.M., Fedin M.V., Pyshnyi D.V., et al. Physiological-temperature distance measurement in nucleic acid using triarylmethyl-based spin labels and pulsed dipolar EPR spectroscopy. J. Am. Chem. Soc. 2014;136:9874–9877. doi: 10.1021/ja505122n. [DOI] [PubMed] [Google Scholar]
- 108.Reginsson G.W., Kunjir N.C., Sigurdsson S.T., Schiemann O. Trityl radicals: Spin labels for nanometer-distance measurements. Chem.-Eur. J. 2012;18:13580–13584. doi: 10.1002/chem.201203014. [DOI] [PubMed] [Google Scholar]
- 109.Reddy T.J., Iwama T., Halpern H.J., Rawal V.H. General synthesis of persistent trityl radicals for EPR imaging of biological systems. J. Org. Chem. 2002;67:4635–4639. doi: 10.1021/jo011068f. [DOI] [PubMed] [Google Scholar]
- 110.Bobko A.A., Dhimitruka I., Zweier J.L., Khramtsov V.V. Trityl radicals as persistent dual function pH and oxygen probes for in vivo electron paramagnetic resonance spectroscopy and imaging: Concept and experiment. J. Am. Chem. Soc. 2007;129:7240–7241. doi: 10.1021/ja071515u. [DOI] [PubMed] [Google Scholar]
- 111.Grassetti D.R., Murray J.F. Determination of sulfhydryl groups with 2,2'- or 4,4'-dithiodipyridine. Arch. Biochem. Biophys. 1967;119:41–49. doi: 10.1016/0003-9861(67)90426-2. [DOI] [PubMed] [Google Scholar]
- 112.Kunjir N.C., Reginsson G.W., Schiemann O., Sigurdsson S.T. Measurements of short distances between trityl spin labels with CW EPR, DQC and PELDOR. Phys. Chem. Chem. Phys. PCCP. 2013;15:19673–19685. doi: 10.1039/c3cp52789a. [DOI] [PubMed] [Google Scholar]
- 113.Di Valentin M., Albertini M., Zurlo E., Gobbo M., Carbonera D. Porphyrin triplet state as a potential spin label for nanometer distance measurements by PELDOR spectroscopy. J. Am. Chem. Soc. 2014;136:6582–6585. doi: 10.1021/ja502615n. [DOI] [PubMed] [Google Scholar]
- 114.Dolphin D. The Porphyrins. Volume IV. Physical Chemistry, Part B. Academic Press; New York, NY, USA: 1978. [Google Scholar]
- 115.Lubitz W., Lendzian F., Bittl R. Radicals, radical pairs and triplet states in photosynthesis. Acc. Chem. Res. 2002;35:313–320. doi: 10.1021/ar000084g. [DOI] [PubMed] [Google Scholar]
- 116.Ji M., Ruthstein S., Saxena S. Paramagnetic metal ions in pulsed ESR distance distribution measurements. Acc. Chem. Res. 2014;47:688–695. doi: 10.1021/ar400245z. [DOI] [PubMed] [Google Scholar]
- 117.Roessler M.M., King M.S., Robinson A.J., Armstrong F.A., Harmer J., Hirst J. Direct assignment of EPR spectra to structurally defined iron-sulfur clusters in complex I by double electron-electron resonance. Proc. Natl. Acad. Sci. USA. 2010;107:1930–1935. doi: 10.1073/pnas.0908050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bode B.E., Plackmeyer J., Bolte M., Prisner T.F., Schiemann O. PELDOR on an exchange coupled nitroxide copper(II) spin pair. J. Organomet. Chem. 2009;694:1172–1179. doi: 10.1016/j.jorganchem.2008.11.029. [DOI] [Google Scholar]
- 119.Potapov A., Song Y., Meade T.J., Goldfarb D., Astashkin A.V., Raitsimring A. Distance measurements in model bis-Gd(III) complexes with flexible “bridge”. Emulation of biological molecules having flexible structure with Gd(III) labels attached. J. Magn. Reson. 2010;205:38–49. doi: 10.1016/j.jmr.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Potapov A., Yagi H., Huber T., Jergic S., Dixon N.E., Otting G., Goldfarb D. Nanometer-scale distance measurements in proteins using Gd3+ spin labeling. J. Am. Chem. Soc. 2010;132:9040–9048. doi: 10.1021/ja1015662. [DOI] [PubMed] [Google Scholar]
- 121.Gordon-Grossman M., Kaminker I., Gofman Y., Shai Y., Goldfarb D. W-Band pulse EPR distance measurements in peptides using Gd3+-dipicolinic acid derivatives as spin labels. Phys. Chem. Chem. Phys. 2011;13:10771–10780. doi: 10.1039/c1cp00011j. [DOI] [PubMed] [Google Scholar]
- 122.Yagi H., Banerjee D., Graham B., Huber T., Goldfarb D., Otting G. Gadolinium tagging for high-precision measurements of 6 nm distances in protein assemblies by EPR. J. Am. Chem. Soc. 2011;133:10418–10421. doi: 10.1021/ja204415w. [DOI] [PubMed] [Google Scholar]
- 123.Song Y., Meade T.J., Astashkin A.V., Klein E.L., Enemark J.H., Raitsimring A. Pulsed dipolar spectroscopy distance measurements in biomacromolecules labeled with Gd(III) markers. J. Magn. Reson. 2011;210:59–68. doi: 10.1016/j.jmr.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goldfarb D. Gd3+ spin labeling for distance measurements by pulse EPR spectroscopy. Phys. Chem. Chem. Phys. PCCP. 2014;16:9685–9699. doi: 10.1039/c3cp53822b. [DOI] [PubMed] [Google Scholar]
- 125.Matalon E., Huber T., Hagelueken G., Graham B., Frydman V., Feintuch A., Otting G., Goldfarb D. Gadolinium(III) spin labels for high-sensitivity distance measurements in transmembrane helices. Angew. Chem. Int. Ed. 2013;52:11831–11834. doi: 10.1002/anie.201305574. [DOI] [PubMed] [Google Scholar]
- 126.Graham B., Loh C.T., Swarbrick J.D., Ung P., Shin J., Yagi H., Jia X., Chhabra S., Barlow N., Pintacuda G., et al. DOTA-amide lanthanide tag for reliable generation of pseudocontact shifts in protein NMR spectra. Bioconjug. Chem. 2011;22:2118–2125. doi: 10.1021/bc200353c. [DOI] [PubMed] [Google Scholar]
- 127.Garbuio L., Bordignon E., Brooks E.K., Hubbell W.L., Jeschke G., Yulikov M. Orthogonal Spin Labeling and Gd(III)—Nitroxide Distance Measurements on Bacteriophage T4-Lysozyme. J. Phys. Chem. B. 2013;117:3145–3153. doi: 10.1021/jp401806g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kaminker I., Yagi H., Huber T., Feintuch A., Otting G., Goldfarb D. Spectroscopic selection of distance measurements in a protein dimer with mixed nitroxide and Gd3+ spin labels. Phys. Chem. Chem. Phys. PCCP. 2012;14:4355–4358. doi: 10.1039/c2cp40219j. [DOI] [PubMed] [Google Scholar]
- 129.Joseph B., Korkhov V.M., Yulikov M., Jeschke G., Bordignon E. Conformational cycle of the vitamin B12 ABC importer in liposomes detected by double electron-electron resonance (DEER) J. Biol. Chem. 2014;289:3176–3185. doi: 10.1074/jbc.M113.512178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kaminker I., Tkach I., Manukovsky N., Huber T., Yagi H., Otting G., Bennati M., Goldfarb D. W-band orientation selective DEER measurements on a Gd3+/nitroxide mixed-labeled protein dimer with a dual mode cavity. J. Magn. Reson. 2013;227:66–71. doi: 10.1016/j.jmr.2012.11.028. [DOI] [PubMed] [Google Scholar]
- 131.Lueders P., Jäger H., Hemminga M.A., Jeschke G., Yulikov M. Distance measurements on orthogonally spin-labeled membrane spanning WALP23 polypeptides. J. Phys. Chem. B. 2013;117:2061–2068. doi: 10.1021/jp311287t. [DOI] [PubMed] [Google Scholar]
- 132.Narr E., Godt A., Jeschke G. Selective measurements of a nitroxide-nitroxide separation of 5 nm and a nitroxide-copper separation of 2.5 nm in a terpyridine-based copper(II) complex by pulse EPR spectroscopy. Angew. Chem. Int. Ed. 2002;41:3907–3910. doi: 10.1002/1521-3773(20021018)41:20<3907::AID-ANIE3907>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 133.Becker J.S., Saxena S. Double quantum coherence electron spin resonance on coupled Cu(II)—Cu(II) electron spins. Chem. Phys. Lett. 2005;414:248–252. doi: 10.1016/j.cplett.2005.08.072. [DOI] [Google Scholar]
- 134.Yang Z., Ji M., Saxena S. Practical aspects of copper ion-based double electron electron resonance distance measurements. Appl. Magn. Reson. 2010;39:487–500. doi: 10.1007/s00723-010-0181-5. [DOI] [Google Scholar]
- 135.Van Wonderen J.H., Kostrz D.N., Dennison C., MacMillan F. Refined distances between paramagnetic centers of a multi-copper nitrite reductase determined by pulsed EPR (i DEER) spectroscopy. Angew. Chem. Int. Ed. 2013;52:1990–1993. doi: 10.1002/anie.201208166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang Z., Kurpiewski M.R., Ji M., Townsend J.E., Mehta P., Jen-Jacobson L., Saxena S. ESR spectroscopy identifies inhibitory Cu2+ sites in a DNA-modifying enzyme to reveal determinants of catalytic specificity. Proc. Natl. Acad. Sci. USA. 2012;109:E993–E1000. doi: 10.1073/pnas.1200733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Noren C.J., Anthony-Cahill S.J., Griffith M.C., Schultz P.G. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- 138.Liu C.C., Schultz P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 139.Cornish V.W., Benson D.R., Altenbach C.A., Hideg K., Hubbell W.L., Schultz P.G. Site-specific incorporation of biophysical probes into proteins. Proc. Natl. Acad. Sci. USA. 1994;91:2910–2914. doi: 10.1073/pnas.91.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Johnson D.B.F., Xu J., Shen Z., Takimoto J.K., Schultz M.D., Schmitz R.J., Xiang Z., Ecker J.R., Briggs S.P., Wang L. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 2011;7:779–786. doi: 10.1038/nchembio.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Johnson D.B.F., Wang C., Xu J., Schultz M.D., Schmitz R.J., Ecker J.R., Wang L. Release factor one is nonessential in Escherichia coli. ACS Chem. Biol. 2012;7:1337–1344. doi: 10.1021/cb300229q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Loscha K.V., Herlt A.J., Qi R., Huber T., Ozawa K., Otting G. Multiple-site labeling of proteins with unnatural amino acids. Angew. Chem. Int. Ed. 2012;51:2243–2246. doi: 10.1002/anie.201108275. [DOI] [PubMed] [Google Scholar]
- 143.Chatterjee A., Sun S.B., Furman J.L., Xiao H., Schultz P.G. A versatile platform for single- and multiple-unnatural amino acid mutagenesis in Escherichia coli. Biochemistry. 2013;52:1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schmidt M.J., Borbas J., Drescher M., Summerer D. A genetically encoded spin label for electron paramagnetic resonance distance measurements. J. Am. Chem. Soc. 2014;136:1238–1241. doi: 10.1021/ja411535q. [DOI] [PubMed] [Google Scholar]
- 145.Wang L., Zhang Z., Brock A., Schultz P.G. Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2003;100:56–61. doi: 10.1073/pnas.0234824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fleissner M.R., Brustad E.M., Kálai T., Altenbach C., Cascio D., Peters F.B., Hideg K., Peuker S., Schultz P.G., Hubbell W.L. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc. Natl. Acad. Sci. USA. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Plass T., Milles S., Koehler C., Schultz C., Lemke E.A. Genetically encoded copper-free click chemistry. Angew. Chem. Int. Ed. 2011;50:3878–3881. doi: 10.1002/anie.201008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kálai T., Fleissner M.R., Jekő J., Hubbell W.L., Hideg K. Synthesis of new spin labels for Cu-free click conjugation. Tetrahedron Lett. 2011;52:2747–2749. doi: 10.1016/j.tetlet.2011.03.077. [DOI] [Google Scholar]
- 149.Igarashi R., Sakai T., Hara H., Tenno T., Tanaka T., Tochio H., Shirakawa M. Distance determination in proteins inside Xenopus laevis oocytes by double electron−electron resonance experiments. J. Am. Chem. Soc. 2010;132:8228–8229. doi: 10.1021/ja906104e. [DOI] [PubMed] [Google Scholar]
- 150.Azarkh M., Singh V., Okle O., Seemann I.T., Dietrich D.R., Hartig J.S., Drescher M. Site-directed spin-labeling of nucleotides and the use of in-cell EPR to determine long-range distances in a biologically relevant environment. Nat. Protoc. 2013;8:131–147. doi: 10.1038/nprot.2012.136. [DOI] [PubMed] [Google Scholar]
- 151.Baldauf C., Schulze K., Lueders P., Bordignon E., Tampé R. In-situ spin labeling of his-tagged proteins: Distance measurements under in-cell conditions. Chem.-Eur. J. 2013;19:13714–13719. doi: 10.1002/chem.201301921. [DOI] [PubMed] [Google Scholar]
- 152.Cruickshank P.A.S., Bolton D.R., Robertson D.A., Hunter R.I., Wylde R.J., Smith G.M. A kilowatt pulsed 94 GHz electron paramagnetic resonance spectrometer with high concentration sensitivity, high instantaneous bandwidth, and low dead time. Rev. Sci. Instrum. 2009;80:103102. doi: 10.1063/1.3239402. [DOI] [PubMed] [Google Scholar]