Abstract
EphB2 and EphA2 control stemness and differentiation in the intestinal mucosa, but the way they cooperate with the complex mechanisms underlying tumor heterogeneity and how they affect the therapeutic outcome in colorectal cancer (CRC) patients, remain unclear. MicroRNA (miRNA) expression profiling along with pathway analysis provide comprehensive information on the dysregulation of multiple crucial pathways in CRC.
Through a network-based approach founded on the characterization of progressive miRNAomes centered on EphA2/EphB2 signaling during tumor development in the AOM/DSS murine model, we found a miRNA-dependent orchestration of EphB2-specific stem-like properties in earlier phases of colorectal tumorigenesis and the EphA2-specific control of tumor progression in the latest CRC phases. Furthermore, two transcriptional signatures that are specifically dependent on the EphA2/EphB2 signaling pathways were identified, namely EphA2, miR-423-5p, CREB1, ADAMTS14, and EphB2, miR-31-5p, mir-31-3p, CRK, CXCL12, ARPC5, SRC.
EphA2- and EphB2-related signatures were validated for their expression and clinical value in 1663 CRC patients. In multivariate analysis, both signatures were predictive of survival and tumor progression.
The early dysregulation of miRs-31, as observed in the murine samples, was also confirmed on 49 human tissue samples including preneoplastic lesions and tumors. In light of these findings, miRs-31 emerged as novel potential drivers of CRC initiation.
Our study evidenced a miRNA-dependent orchestration of EphB2 stem-related networks at the onset and EphA2-related cancer-progression networks in advanced stages of CRC evolution, suggesting new predictive biomarkers and potential therapeutic targets.
Electronic supplementary material
The online version of this article (10.1186/s12943-018-0912-z) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, MicroRNA, EphA2 and EphB2, Cancer stem cells, Biomarkers
EphA2 and EphB2 are tyrosine kinase receptors that are involved in complex biochemical mechanisms underlying tumor heterogeneity primarily by guiding the positioning of intestinal cells with distinct stemness or differentiation properties [1]. EphA2high cells are localized in the upper differentiated crypt region, while EphB2 is expressed in a decreasing gradient from the crypt base toward the upper cell compartment and corresponds to an intestinal stem cell (ISC) marker [2]. At the onset of colorectal cancer (CRC), early events trigger continuous stem-like self-renewing state, giving rise to adenoma, while additional driver pathway alterations confer invasive behavior in advanced carcinoma [3]. EphA2 and EphB2 undergo progressive dysregulation in carcinogenesis that may resemble these different stages, so representing attractive druggable targets [4].
MicroRNA (miRNA) redundancy and the capacity of individual miRNAs to simultaneously regulate large cohorts of genes have shaped by evolution combinatorial miRNA-target networks that profoundly affect cellular properties, including the promotion of tumorigenic processes [5]. A network-based approach founded on the characterization of progressive miRNAomes centered on EphA2/EphB2 signaling during tumor development may unveil important regulatory networks and oncogenic targets.
Given that EphA2 and EphB2 have opposite distribution and roles along the intestinal crypt, we hypothesized that these receptors may influence alternative cell behaviors on initial or advanced CRC stages. Through an integrative translational approach (Additional File 1: Figure S1) and a miRNAome-guided pathway analysis, we defined two transcriptional signatures that are associated with EphB2 cells/early CRC phases and EphA2 cells/late CRC phases, with significant prognostic value.
Results and discussion
Distinct miRNAs characterize progressive stages of CRC development
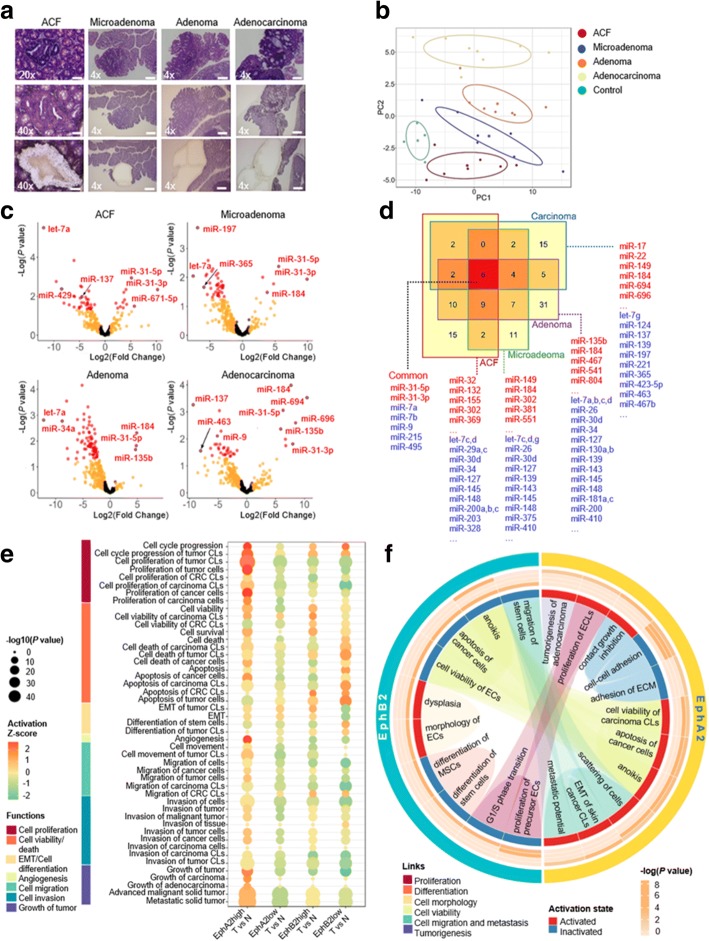
Given the poorly foreseeable multiphase pattern of human CRC (hCRC) development, we used the well-characterized azoxymethane (AOM)/dextran sodium sulfate (DSS) murine model of sporadic CRC to obtain a predictable neoplastic evolution through the colorectal “Aberrant crypt foci (ACF)-microadenoma-adenoma-carcinoma” sequence, where each phase was expected to most likely progress to the following one [6, 7] (Fig. 1a; Additional file 1: Figure S2).
Fig. 1.
MiRNA expression profiling and functional enrichment analysis in progressive stages of murine CRC development: a Laser capture microdissection (LCM) of pretumoral lesions and tumors. First row, standard hematoxylin-eosin staining; second row, samples before LCM; third row, samples after LCM. Original magnification × 4 (scale bar 200 μm), × 20 (scale bar 50 μm), or × 40 (scale bar 20 μm). b Clustering of phases and control samples based on the first two principal components (PC1 and PC2) in a p partial least square - discriminant analysis (sPLS-DA) model. Control: normal colon mucosa of untreated mice. c Volcano plots of differentially expressed (DE) miRNAs (P value < 0.05 and |fold change| > 2) (red dots), miRNAs with only |fold change| > 2 (orange dots), and other miRNAs (black dots). The results were obtained through comparisons between each phase and control samples. d Venn diagram representing the number of miRNAs altered in each phase and shared between different phases (numbers in Venn diagram) of CRC. Sets of well-known CRC-associated miRNAs are indicated for each CRC phase (blue: down-regulated; red: up-regulated). e Enriched biological functions regulated by the DE miRNAs for Tumor (T) vs. Normal (N) of EphA2- and EphB2-positive cell comparisons. EphA2high tumor cells are significantly enriched in activated functions related to cell proliferation, cell viability and cell death, angiogenesis, cell migration, cell invasion and growth of tumor (P < 0.0001). EphB2high tumor cells show instead a significant enrichment of functions related to cell viability and a decreased or no activation of cell death/apoptosis (P < 0.0001). EMT activation and cell differentiation inactivation are also observed in EphB2high tumor cells, a behavior coherently inverted in EphB2low tumor cells (P < 0.0001). Sizes of bubbles are proportional to the statistical significance of the enriched functions (Fisher’s exact test). Gradients of color vary from red (activation) to green (inhibition) according to the Z-scores calculated for each enriched function and for each comparison. Functions are grouped into macro-categories. f Biological functions regulated by the DE miRNAs (and their target genes) with concordant fold changes between EphB2high cells and tissues and EphA2high cells and tissues. The outer circle marks the biological functions that are enriched in the EphA2high and EphB2high cells. The heatmap represents -log(P value) for each biological function evaluated in the four tissue types of adenocarcinoma (outer), adenoma, microadenoma, and ACF (inner). Blue and red rectangles represent the predicted activation and inactivation, respectively, of biological functions. These rectangles are linked by colored edges according to the macro-categories they belong to (proliferation, differentiation, cell morphology, etc.). Statistical significance was calculated using Fisher’s exact test as implemented in IPA. Abbreviations: CLs, cell lines; CRC, colorectal cancer; ECs, epithelial cells; ECLs, EC lines; ECM, extracellular matrix; EMT, epithelial-mesenchymal transition; MSCs, mesenchymal stem cells
Of 641 miRNAs analyzed by TaqMan Low-Density Array (TLDA), 121 miRNAs were differentially expressed in the four murine CRC phases relative to normal mucosa (P < 0.05) (Fig. 1b, c, and d; Additional file 1: Figures S3 and S4). We found that the AOM/DSS model not only recapitulates several genetic modifications of hCRC [6], but also showed the aberrant expression of miRNAs belonging to hCRC-related pathways. In particular, miR-135b, targeting APC/β-catenin pathway [8], was up-regulated in murine adenoma (P < 0.05); let-7 family and miR-143, acting as tumor-suppressors in KRAS-driven CRC [9], were down-regulated already in ACF (let-7a, c, d, f P < 0.05) and in microadenoma (miR-143 P < 0.005); miR-9 was down-regulated in all the stages (P < 0.05), correlating with its tumor-suppressor function in hCRC by mediating invasion and metastasis [10] (Fig. 1c, d). More importantly, miR-31-5p and miR-31-3p were the most up-regulated in the earliest phases (P < 0.05 in ACF) (Fig. 1c, d), suggesting that these miRNAs may be involved in the CRC initiation.
Distinct EphA2/EphB2-specific miRNAomes orchestrate cell proliferation/stemness/migration in progressive murine CRC stages
Based on cytofluorimetric, immunohistochemical and molecular characterization (Additional file 2: Methods and Materials) of EphA2high/low and EphB2high/low cells isolated from murine adenocarcinoma and normal colon (Additional file 1: Figure S5), we ascertained that EphA2 and EphB2 mark murine tumor cell subpopulations bearing either a differentiated or a stem-like phenotype, respectively.
With this assumption, we performed a TLDA-based miRNA expression analysis to characterize miRNAomes related to EphA2high/low and EphB2high/low CRC cells (Additional file 3: Tables S1, S2).
A list of 180 miRNAs was associated with EphA2 and CRC. Moreover, according to the EphB2high CRC cells well-accepted identity of putative CSCs [2], we found they were enriched with miRNAs dysregulated in colon CSCs, such as miR-137, miR-34a, miR-215, miR-328 and miR-203 (P < 0.0001) [11].
Figure 1e describes a functional enrichment of the EphA2/EphB2 differential miRNAomes. All the functions analyzed were principally under control of miRNA targets listed in Additional file 3: Table S3.
When compared with the miRNAomes of murine ACF, microadenoma, adenoma and adenocarcinoma, the EphA2 and EphB2 tumor cells’ miRNAomes resulted in the alternate control of specific functions in different CRC stages (Fig. 1f; Additional file 3: Table S4).
It is worth noting that functions concerning cell morphology/cell differentiation and the inactivation of cell proliferation and cell survival/death were exclusively associated with EphB2 and enriched in ACF. In contrast, cell survival/death and cell proliferation, cell migration and metastasis and the reduction of cell adhesion were significantly associated with EphA2 and enriched in adenoma and adenocarcinoma.
These results indicate that while ACF might be enriched with EphB2high tumor-initiating cells, which exhibit driver-gene/miRNA influencing CRC initiation via stem-related functions, the latest stages of murine CRC show an enrichment with EphA2high cells orchestrating functions primarily related to tumor malignancy.
Establishment of two CRC EphA2/EphB2-specific signatures
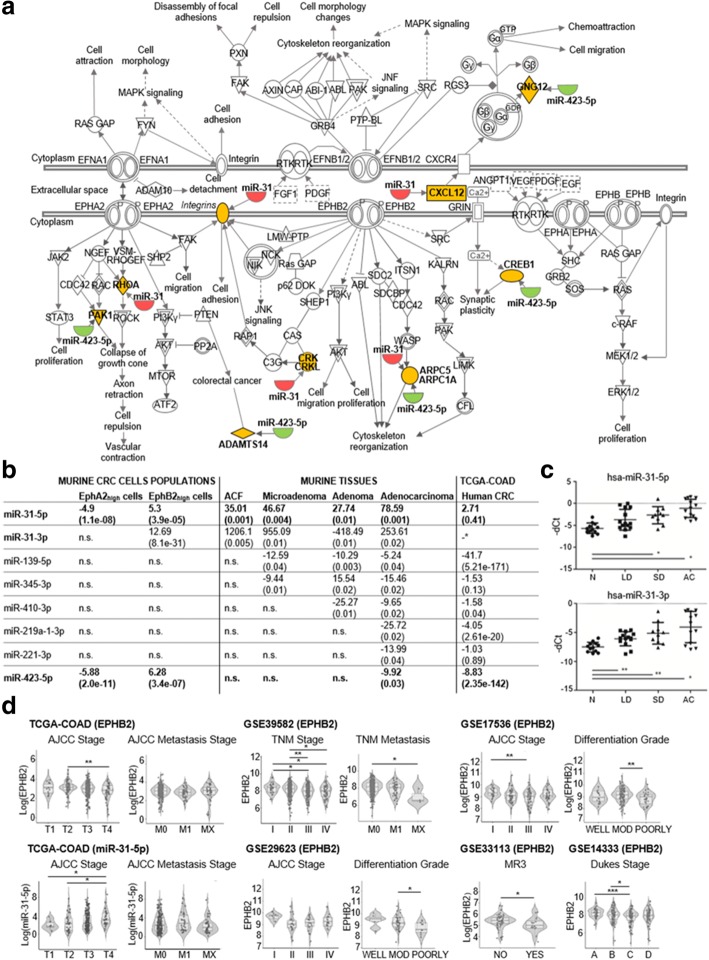
Considering the EphA2/EphB2 macro-pathway (Fig. 2a), we compared Eph pathway-related miRNAs included in our preclinical results to those dysregulated in CRC patients (The Cancer Genome Atlas - colon adenocarcinoma, TCGA-COAD). Eight miRNAs were concordantly dysregulated in both murine and human CRC (Fig. 2b). Strikingly, among them, miR-31-5p and miR-31-3p were up-regulated in all four murine CRC stages and in the EphB2high tumor cells. Conversely, miR-423-5p was down-regulated in the advanced murine CRC phases and in the EphA2high tumor cells. These miRNAs displayed inverse regulation in the two cell subpopulations (Fig. 2b), supporting the strong association of miRs-31 and miR-423-5p not only with specific CRC phases but also with specific Eph receptor-expressing cells.
Fig. 2.
a Scheme of a macro-pathway including both the EphA2 and EphB2 pathways with their cross-talk links. Down-regulated and up-regulated miRNAs (miRs-31 and miR-423-5p) and their mRNA targets are colored in green, red and yellow respectively. b Summary table of Fold Change data with correlated P values shown in brackets of 8 Eph pathway-related miRNAs concordantly DE in human CRC (TCGA-COAD) and in both EphA2 or EphB2 murine CRC cells. In bold the miRNAs concordantly DE in hCRC and in both murine EphA2/EphB2 cells and tissues. *no -3p and -5p specification in miRNA nomenclature. Abbreviations: n.s., not significant. c qPCR analysis of miR-31-5p and miR-31-3p expression in normal colon mucosa (N), colorectal adenoma with low dysplasia (LD), colorectal adenoma with high dysplasia (HD), and adenocarcinoma (AC) human samples. Data are represented as the means +/− standard deviation (SD). Data are normalized by geometric mean of RNU48 and RNU6B. Statistically significant differences were calculated using Student’s t-test. *P < 0.05, **P < 0.01. d EphB2 and miR-31-5p analysis on hCRC staging progression (AJCC stage, TNM stage, Dukes stage), metastasis, and differentiation grade. Dots indicate individual data points, and their distribution is represented by violin and box plots. The box plots show the interquartile range and median value of the distribution. Abbreviations: FC, fold change; MOD, moderately differentiated; MR3, metastasis or recurrence within 3 years. *P < 0.05; **P < 0.01 and ***P < 0.001; Mann-Whitney-U test
The pathway analysis of 617 genes target of miRs-31 and miR-423-5p (Additional file 3: Tables S5 and S6) resulted to significantly enrich CRC-related signaling pathways (P value < 0.05) (Additional file 1: Figures S6 and S7).
On that basis, we defined two EphA2-/EphB2-related signatures by the combination of miRs-31 and miR-423-5p with their experimentally validated target genes belonging to the EphA2/EphB2 macro-pathway. Therefore, the EphB2 signature comprehended miRs-31, ROHA, CRKL, CRK, ITGA5, CXCL12, ARPC5, RASA1 and SRC, while the EphA2 signature was composed of miR-423-5p, ARPC1A, PAK1, GNG12, CREB1 and ADAMTS14.
Up-regulation of miRs-31 occurs at very early stages of hCRC development
Given the importance of the early dysregulation of both miR-31-5p and miR-31-3p in the murine CRC, for their possible involvement in driving tumor initiation, we focused on the validation of their expression levels in the earliest stages of hCRC. We used a selected set of 49 human samples including low dysplasia (LD)-adenoma (n = 14), severe dysplasia (SD)-adenoma (n = 10), adenocarcinoma (n = 13) and normal mucosa (n = 12), that are not available in public datasets. qPCR analysis confirmed a significant overexpression of both miRs-31 in all the samples with an upward trend from LD-adenoma (preneoplastic) to SD-adenoma and adenocarcinoma, relative to normal mucosa (Student’s t-test, P < 0.05) (Fig. 2c). This is in line with recent demonstrations of miR-31-5p acting as a master modulator of the ISCs niche signaling during normal homeostasis, regeneration and tumorigenesis [12] or as potential driver of lung cancer initiation via KRAS pathway [13].
Prognostic value of EphA2/EphB2-specific signatures
Both EphA2 and EphB2 signatures were validated in 1663 CRC patients from nine cohorts of TCGA-COAD and Gene Expression Omnibus (GEO) databases, through a differential expression analysis between tumor and normal samples (Additional file 3: Table S7; Table 1a). EPHA2 was overexpressed in different datasets (P < 0.02) and miR-423-5p was concomitantly down-regulated (P < 0.01), whereas CREB1 and ADAMTS14 were overexpressed (P < 0.05). However, no significant changes in the expression levels of both EPHA2 and miR-423-5p were observed when they were analyzed in correlation with tumor staging progression (Additional file 1: Figure S8). EPHB2 was significantly overexpressed in TCGA-COAD (P = 0.03), GSE35834 cohort (P = 0.0008) and in patients with colorectal adenoma of GSE4183 cohort (P < 0.001); miR-31-5p was also overexpressed (P < 0.03) and its targets CRK, CXCL12, ARPC5 and SRC were down-regulated in three out of six cohorts. It is worth noting that in the analysis of the GSE4183 cohort, the expression level of EphB2 decreased in the transition from adenoma to carcinoma. In line with this observation, we found that EPHB2 expression levels gradually decreased with the progression of the TNM/AJCC stage and with metastasis (Fig. 2d). These results support the model that during CRC progression, stem EphB2-dependent functions are confined to a very small cell fraction of the tumor mass [7, 14], which likely represents the tumorigenic cell reservoir. Moreover, a gradual increase of miR-31-5p expression levels was observed with the progression of the TNM/AJCC stage (Fig. 2d), suggesting that it could be associated not only with particular miRNA patterns of stem-like EphB2high cells during tumorigenesis, but also with miRNA patterns of different cell populations replenishing the tumor mass in progressive CRC stages. Our findings are in line with functional studies showing that miR-31 has pleiotropic activity and is typically overexpressed with high expression correlating with advanced CRC disease [15].
Table 1.
Validation of the EphA2 and EphB2 signatures in GEO and TCGA-COAD public databases
| a. | ||||||
|
GSE35982
FC (P value) |
GSE35834
FC (P value) |
TCGA-COAD
FC (P value) |
GSE4183 | |||
|
Adenoma
FC (P value) |
Carcinoma
FC (P value) |
IBD
FC (P value) |
||||
| EPHA2 | -1.16 (0.35) |
1.18
(0.02) |
1.27 (0.40) |
1.7 (0.07) |
1.67 (0.08) |
2.16
(0.003) |
| miR-423-5p | -2.2 (0.30) |
-1.26 (0.09) |
-8.82
(2.35e-142) |
- | - | - |
| ARPC1A | -1.07 (0.20) |
1.12 (0.17) |
-1.09 (0.35) |
1.14 (0.32) |
1.16 (0.24) |
-1.13 (0.39) |
| PAK1 | -1.01 (0.94) |
-1.07 (0.25) |
-1.21 (0.01) |
-1.27 (0.02) |
-1.49 (0.0006) |
-1.63 (7.42e-05) |
| GNG12 | 1.11 (0.46) |
-1.63 (2.48e-08) |
-1 (0.128) |
-1.44 (0.005) |
-1.28 (0.04) |
-1.35 (0.02) |
| CREB1 | -1.06 (0.40) |
-1.04 (0.38) |
2.72
(2.85e-34) |
1.16 (0.15) |
1.36
(0.005) |
1.26
(0.04) |
| ADAMTS14 | -1.03 (0.67) |
1.03 (0.52) |
2.81
(0.0003) |
-1.05 (0.85) |
1.32 (0.21) |
1.52
(0.05) |
| EPHB2 | 1.19 (0.49) |
1.33
(0.0008) |
1.75
(0.03) |
3.39
(7.75e-06) |
1.26 (0.16) |
-1.13 (0.67) |
| miR-31-5p |
49.65
(0.02) |
3.17
(0.03) |
2.71 (0.41) |
- | - | - |
| miR-31-3p |
14.73
(0.005) |
- | - | - | - | - |
| RHOA | 1.10 (0.08) |
-1.01 (0.71) |
-1.08 (0.28) |
1.12 (0.06) |
-1.29 (0.21) |
1.09 (0.13) |
| CRKL | 1.07 (0.54) |
1.12 (0.002) |
1.18 (0.03) |
1.20 (0.139) |
1.13 (0.14) |
-1.19 (0.22) |
| CRK | -1.11 (0.36) |
-1.25
(0.0008) |
-1.12 (0.10) |
-1.22
(0.02) |
-1.15 (0.09) |
-1.13 (0.12) |
| ITGA5 | 1.78 (0.30) |
-1.08 (0.38) |
-1.13 (0.64) |
1.88 (0.59) |
4.50 (0.03) |
4.56 (0.03) |
| CXCL12 | -1.19 (0.69) |
-2.40
(9.44e-14) |
-3.00
(6.55e-18) |
-6.91
(9.80E-09) |
-2.90
(2.95E-06) |
-1.48
(0.01) |
| ARPC5 |
-1.18
(0.05) |
1.01 (0.62) |
1.09 (0.36) |
-1.33 (0.06) |
-1.36
(0.05) |
-1.37
(0.05) |
| RASA1 | -1.03 (0.61) |
1.19 (0.04) |
2.60 (1.92e-20) |
1.21 (0.02) |
1.11 (0.23) |
-1.10 (0.30) |
| SRC | 1.23 (0.16) |
1.20 (0.0008) |
-1.37
(0.02) |
-1.37 (0.10) |
-1.57
(0.03) |
1.79 (0.03) |
| b. | ||||||
| Multivariate analysis of EPHA2 genetic signature without hsa-miR-423-5p | ||||||
| Relative Risk | 95% CI | P value | ||||
| TCGA-COAD (OS) | ||||||
| ADAMTS14 | 1.07 | 0.97 to 1.19 | 0.17 | |||
| CREB1 | 0.97 | 0.87 to 1.09 | 0.65 | |||
| GSE33113 (MR3) | ||||||
| ADAMTS14 | 1.03 | 0.99 to 1.06 | 0.05 | |||
| CREB1 | 1.01 | 0.99 to 1.03 | 0.44 | |||
| GSE14333 (DFS) | ||||||
| ADAMTS14 | 0.76 | 0.65 to 0.90 | 0.001 | |||
| CREB1 | 0.99 | 0.54 to 1.84 | 0.99 | |||
| GSE17536 (OS) | ||||||
| ADAMTS14 | 0.79 | 0.26 to 2.45 | 0.69 | |||
| CREB1 | 1.43 | 0.59 to 3.48 | 0.44 | |||
| GSE29623 (OS) | ||||||
| ADAMTS14 | 0.36 | 0.04 to 3.06 | 0.36 | |||
| CREB1 | 0.82 | 0.13 to 5.1 | 0.83 | |||
| Multivariate analysis of EPHB2 genetic signature without hsa-miR-31-5p | ||||||
| Relative Risk | 95% CI | P value | ||||
| TCGA-COAD (OS) | ||||||
| CRK | 0.97 | 0.94 to 1.01 | 0.15 | |||
| CXCL12 | 1.03 | 0.99 to 1.07 | 0.17 | |||
| ARPC5 | 0.99 | 0.96 to 1.02 | 0.53 | |||
| SRC | 1.00 | 0.97 to 1.02 | 0.79 | |||
| GSE33113 (MR3) | ||||||
| CRK | 1.01 | 1.00 to 1.01 | 0.01 | |||
| CXCL12 | 1.00 | 1.00 to 1.00 | 0.02 | |||
| ARPC5 | 1.00 | 1.00 to 1.01 | 0.05 | |||
| SRC | 1.00 | 0.95 to 1.05 | 0.95 | |||
| GSE14333 (DFS) | ||||||
| CRK | 0.80 | 0.54 to 1.20 | 0.29 | |||
| CXCL12 | 0.74 | 0.60 to 0.92 | 0.01 | |||
| ARPC5 | 0.81 | 0.50 to 1.30 | 0.38 | |||
| SRC | 1.08 | 0.79 to 1.48 | 0.63 | |||
| GSE17536 (OS) | ||||||
| CRK | 1.14 | 0.49 to 2.66 | 0.75 | |||
| CXCL12 | 0.98 | 0.69 to 1.39 | 0.90 | |||
| ARPC5 | 1.27 | 0.37 to 4.35 | 0.70 | |||
| SRC | 0.10 | 0.02 to 0.54 | 0.01 | |||
| GSE29623 (OS) | ||||||
| CRK | 0.83 | 0.15 to 4.52 | 0.83 | |||
| CXCL12 | 1.02 | 0.45 to 2.32 | 0.96 | |||
| ARPC5 | 0.25 | 0.03 to 2.03 | 0.19 | |||
| SRC | 0.09 | 0.002 to 4.13 | 0.22 | |||
| Multivariate analysis of EPHA2 genetic signature with hsa-miR-423-5p | ||||||
| Relative Risk | 95% CI | P value | ||||
| TCGA-COAD (OS) | ||||||
| ADAMTS14 | 1.07 | 0.97 to 1.18 | 0.18 | |||
| CREB1 | 0.98 | 0.87 to 1.1009 | 0.67 | |||
| hsa-miR-423-5p | 1.00 | 0.99 to 1.00 | 0.69 | |||
| GSE 29623 (OS) | ||||||
| ADAMTS14 | 0.35 | 0.04 to 3.02 | 0.34 | |||
| CREB1 | 0.82 | 0.13 to 5.12 | 0.83 | |||
| hsa-miR-423-5p | 1.04 | 0.71 to 1.52 | 0.85 | |||
| Multivariate analysis of EPHB2 genetic signature with hsa-miR-31-5p | ||||||
| Relative Risk | 95% CI | P value | ||||
| TCGA-COAD (OS) | ||||||
| CRK | 0.97 | 0.93 to 1.01 | 0.11 | |||
| CXCL12 | 1.03 | 0.99 to 1.07 | 0.10 | |||
| ARPC5 | 0.99 | 0.97 to 1.02 | 0.65 | |||
| SRC | 1.00 | 0.98 to 1.03 | 0.90 | |||
| hsa-miR-31-5p | 1.00 | 1.00 to 1.01 | 0.001 | |||
| GSE29623 (OS) | ||||||
| CRK | 0.69 | 0.12 to 3.81 | 0.67 | |||
| CXCL12 | 1.31 | 0.53 to 3.21 | 0.56 | |||
| ARPC5 | 0.28 | 0.04 to 2.23 | 0.23 | |||
| SRC | 0.12 | 0.003 to 4.88 | 0.26 | |||
| hsa-miR-31-5p | 1.17 | 0.95 to 1.46 | 0.14 | |||
Validation of the EphA2 and EphB2 signatures in GEO and TCGA-COAD public databases; Fold Change data of miR-423-5p and miRs-31 coherent target genes (miRNA and target gene expression levels are inversely correlated) with significant P values (P < 0.05) are indicated in bold (a). Multivariate Cox proportional hazards regression analysis of EPHA2 and EPHB2 genetic signatures; significant P values (P < 0.05) are indicated in bold (b)
In addition, we evaluated the collective prognostic effect of the EphA2 and EphB2 signatures (multivariate Cox-regression analysis) (Table 1b). Regarding the EphA2 fingerprint, ADAMTS14 was a risk factor for disease-free survival (DFS) (P < 0.001) and metastasis or recurrence within 3 years (MR3) (P < 0.05). Concerning the EphB2 fingerprint, SRC was a risk factor for overall survival (OS) (P < 0.01), CXCL12 was significant for DFS (P < 0.01), CRK, CXCL12 and ARPC5 were significant for MR3 (P < 0.05). Furthermore, miR-31-5p was deemed a risk factor in TCGA-COAD (P < 0.001). Our results confirm the involvement of these molecules in a complex network affected by EphA2/EphB2 signaling pathways and their crosstalk.
Conclusions
EphA2 and EphB2 contribute, in a pleiotropic manner, to the CRC pathogenesis. We demonstrated that the early dysregulation of EphB2 at the CRC onset correlates with specific stem-like properties and that the late overexpression of EphA2 triggers tumor progression signals in the invasive CRC phase. These properties are orchestrated by distinct miRNAomes and are associated with miR-31-5p/miR-31-3p, miR-423-5p and their EphA2/EphB2-related target genes. These signatures have significant clinical value and may unveil clinical predictive biomarkers and potential therapeutic markers.
Additional files
Supplementary Figures. (DOCX 3766 kb)
Methods and Materials. (DOCX 24 kb)
Supplementary Tables. (XLSX 77 kb)
Acknowledgements
Not applicable.
Funding
This work was partially supported by grants from Italian Ministry of Education, University and Research (MIUR) national research program: MIUR RBAP10KJC5 to ALV, MIUR PON02_00576_3329762/3 AMIDERHA to VMF, and MIUR PON03PE_00232_Distretto H-Bio to VMF and GP, and Italian Association for Cancer Research: AIRC IG 14368 to ALV and AIRC MFAG 10520 to MLP. This work was also partially supported by the Italian Ministry of Health, IRCCS Ricerca Corrente RC1803LO36 to VMF. Bioinformatics analyses carried out by GP were performed using the infrastructures of the Italian node of ELIXIR funded by MIUR and Consiglio Nazionale delle Ricerche (CNR).
Availability of data and materials
The authors declare that the data supporting the findings of this study are available within the article and its supplementary information files.
Abbreviations
- ACF
Aberrant crypt foci
- AOM
Azoxymethane
- CRC
Colorectal cancer
- CSCs
Cancer stem cells
- DFS
Disease-free survival
- DSS
Dextran sodium sulfate
- GEO
Gene Expression Omnibus
- ISCs
Intestinal stem cells
- LD
Low dysplasia
- miRNAs
microRNAs
- MR3
Metastasis or recurrence within 3 years
- OS
Overall survival
- SD
Severe dysplasia
- TCGA-COAD
The Cancer Genome Atlas - colon adenocarcinoma
- TLDA
TaqMan Low-Density Array
Authors’ contributions
VMF conceived and oversaw the project. MDR designed and performed all the experiments, including the animal experiments, the laser capture microdissection, the immunohistochemistry and the molecular analyses. TM and CF performed all the bioinformatics and statistical analyses. GP contributed with IPA analysis. JGF oversaw laser capture microdissection, miRNA profiling and contributed to project conceiving. ES oversaw the animal experiments. MDR, LL, MLP and EM contributed to the molecular analyses. MS and GL performed the cell biology experiments and cytofluorimetric analysis. MGD and EP collected the tissue samples of CRC patients and performed histological analyses. VMF and MDR interpreted data and wrote the manuscript. TM, CF, MLP, GP, ES, JGF and ALV discussed the results and assisted in revisions. All authors read and approved the final manuscript
Ethics approval and consent to participate
Use and treatment of mice was approved by the Italian Ministry of Health with decree no. 336/2013-B.
The collection of clinical samples was approved by the Ethics Committee of the IRCCS “Regina Elena” National Cancer Institute and all patients signed an informed consent in accordance with the WMA Declaration of Helsinki 2013.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mariangela De Robertis, Phone: +393492609075, Email: mariangela.derobertis@gmail.com.
Tommaso Mazza, Email: t.mazza@css-mendel.it.
Caterina Fusilli, Email: c.fusilli@css-mendel.it.
Luisa Loiacono, Email: luisa.loiacono@gmail.com.
Maria Luana Poeta, Email: poetaluana@gmail.com.
Massimo Sanchez, Email: massimo.sanchez@iss.it.
Emanuela Massi, Email: e.massi@unicampus.it.
Giuseppe Lamorte, Email: g.lamorte@css-mendel.it.
Maria Grazia Diodoro, Email: maria.diodoro@ifo.gov.it.
Edoardo Pescarmona, Email: edoardo.pescarmona@ifo.gov.it.
Emanuela Signori, Email: emanuela.signori@ift.cnr.it.
Graziano Pesole, Email: graziano.pesole@uniba.it.
Angelo Luigi Vescovi, Email: a.vescovi@operapadrepio.it.
Jesus Garcia-Foncillas, Email: jesus.garciafoncillas@oncohealth.eu.
Vito Michele Fazio, Phone: +3906-22541-9180, Email: fazio@unicampus.it, Email: vmfazio@operapadrepio.it.
References
- 1.Kania A, Klein R. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat Rev Mol Cell Biol. 2016;17:240–256. doi: 10.1038/nrm.2015.16. [DOI] [PubMed] [Google Scholar]
- 2.Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 3.Tauriello DVF, Calon A, Lonardo E, Batlle E. Determinants of metastatic competency in colorectal cancer. Mol Oncol. 2017;11:97–119. doi: 10.1002/1878-0261.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lodola A, Giorgio C, Incerti M, Zanotti I, Tognolini M. Targeting Eph/ephrin system in cancer therapy. Eur J Med Chem. 2017;142:152–162. doi: 10.1016/j.ejmech.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 5.Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genetics. 2016;17:719–732. doi: 10.1038/nrg.2016.134. [DOI] [PubMed] [Google Scholar]
- 6.De Robertis M, Massi E, Poeta ML, Carotti S, Morini S, Cecchetelli L, et al. The AOM/DSS murine model for the study of colon carcinogenesis: from pathways to diagnosis and therapy studies. J Carcinog. 2011;10:9. doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Robertis M, Loiacono L, Fusilli C, Poeta ML, Mazza T, Sanchez M, et al. Dysregulation of EGFR pathway in EphA2 cell subpopulation significantly associates with poor prognosis in colorectal Cancer. Clin Cancer Res. 2017;23:159–170. doi: 10.1158/1078-0432.CCR-16-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagel R, le Sage C, Diosdado B, van der Waal M, Oude Vrielink JA, Bolijn A, et al. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 9.Kent OA, Chivukula RR, Mullendore M, et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park YR, Lee ST, Kim SL, Liu YC, Lee MR, Shin JH, et al. MicroRNA-9 suppresses cell migration and invasion through downregulation of TM4SF1 in colorectal cancer. Int J Oncol. 2016;48:2135–2143. doi: 10.3892/ijo.2016.3430. [DOI] [PubMed] [Google Scholar]
- 11.De Robertis M, Poeta ML, Signori E, Fazio VM. Current understanding and clinical utility of miRNAs regulation of colon cancer stem cells. Semin Cancer Biol. 2018. 10.1016/j.semcancer.2018.08.008. [Epub ahead of print]. [DOI] [PubMed]
- 12.Tian Y, Ma X, Lv C, Sheng X, Li X, Zhao R, et al. Stress responsive miR-31 is a major modulator of mouse intestinal stem cells during regeneration and tumorigenesis. Elife. 2017;6:e29538. 10.7554/eLife.29538. [DOI] [PMC free article] [PubMed]
- 13.Edmonds MD, Boyd KL, Moyo T, Mitra R, Duszynski R, Arrate MP, et al. MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer. J Clin Invest. 2016;126:349–364. doi: 10.1172/JCI82720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeuner A, Todaro M, Stassi G, De Maria R. Colorectal cancer stem cells: from the crypt to the clinic. Cell Stem Cell. 2014;15:692–705. doi: 10.1016/j.stem.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 15.Schee K, Boye K, Abrahamsen TW, Fodstad O, Flatmark K. Clinical relevance of microRNA miR-21, miR-31, miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC Cancer. 2012;12:505. doi: 10.1186/1471-2407-12-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures. (DOCX 3766 kb)
Methods and Materials. (DOCX 24 kb)
Supplementary Tables. (XLSX 77 kb)
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the article and its supplementary information files.