Abstract
Antimicrobial natural preparations involving cinnamon, storax and propolis have been long used topically for treating infections. Cinnamic acids and related molecules are partly responsible for the therapeutic effects observed in these preparations. Most of the cinnamic acids, their esters, amides, aldehydes and alcohols, show significant growth inhibition against one or several bacterial and fungal species. Of particular interest is the potent antitubercular activity observed for some of these cinnamic derivatives, which may be amenable as future drugs for treating tuberculosis. This review intends to summarize the literature data on the antimicrobial activity of the natural cinnamic acids and related derivatives. In addition, selected hybrids between cinnamic acids and biologically active scaffolds with antimicrobial activity were also included. A comprehensive literature search was performed collating the minimum inhibitory concentration (MIC) of each cinnamic acid or derivative against the reported microorganisms. The MIC data allows the relative comparison between series of molecules and the derivation of structure-activity relationships.
Keywords: cinnamic acid, coumaric acids, hybrids, antimicrobial, tuberculosis
1. Introduction
Cinnamic acids are a group of aromatic carboxylic acids (C6–C3) appearing naturally in the plant kingdom. They are formed in the biochemical route that yields lignin, the polymeric material that provides mechanical support to the plant cell wall [1]. Cinnamic acids occur in all green plants [2], although in minute quantities covalently bound to cell walls [3], but also in the reproductive organs of flowering plants [4]. Cinnamic acids are formed in the biosynthetic pathway leading to phenyl-propanoids, coumarins, lignans, isoflavonoids, flavonoids, stilbenes, aurones, anthocyanins, spermidines, and tannins [5]. These secondary metabolites play key physiological roles in plant growth, development, reproduction and disease resistance [6,7]. The first step of this pathway is catalyzed by the phenylalanine ammonia lyase (PAL), a widely distributed phenylpropanoid enzyme present in green plants, algae, fungi, and even in some prokaryotes [8]. This enzyme deaminates l-phenylalanine to yield (E)-cinnamic acid, which undergoes other enzymatic transformations, yielding a diversity of related products [5].
The term “cinnamic” derives from the spice cinnamon (Cinnamomum zeilanicum) which has been used since antiquity as a flavoring agent and for its stimulant, carminative, antiseptic and insecticide properties [9]. The bark of several species of Cinnamomum contain considerable amounts of (E)-cinnamaldehyde, a volatile aldehyde responsible for the pungent, sweet and hot flavor of cinnamon [10,11]. Cinnamaldehyde and the essential oils of the species of Cinnamomum have antimicrobial activity both against bacteria and fungi [12,13]. Cinnamic acids are also readily available from coffee beans, tea, mate, cocoa, apples and pears, berries, citrus, grape, brassicas vegetables, spinach, beetroot, artichoke, potato, tomato, celery, faba beans, and cereals [14]. Cinnamic acids often appear as ester conjugates with quinic acid, known as the chlorogenic acids, but they can also form esters with other acids, sugars or lipids, or form amides with aromatic and aliphatic amines.
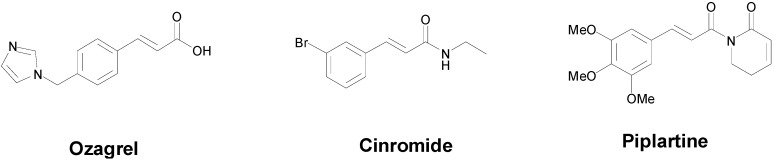
In the last ten years, the interest of researchers on the cinnamic acid moiety has notably increased. The number of published reports having the word “cinnamic” in the title, has almost doubled, from 341 in the years 1993–2003 to 633 in the period 2004–2014 according to the Scopus database (until mid-November 2014). If both “cinnamic” and “antimicrobial” keywords are used, the number of published articles increased from 1 in the period 1993–2003, to 7 in the period 2004–2014. There is no doubt that the cinnamic acids currently attracts the attention of chemists from different perspectives. Ozagrel (Figure 1), a thromboxane A2 synthase inhibitor, is in fact an imidazole para-substituted cinnamic acid that is employed therapeutically for treating acute ischemic stroke [15]. Cinromide (Figure 1) is an antiepileptic experimental drug studied in clinical trials during the 80 decade with a favorable profile to suppress generalized convulsions, but however displayed considerable toxicity [16]. Piplartine (Figure 1) is another cinnamic-related molecule showing an attractive biological horizon [17]. This cinnamic amide was first-time isolated from the roots of Piper tuberculatum [18], and later proved to be a promising anti-cancer scaffold [19,20].
Figure 1.
Chemical structures of some therapeutically important cinnamic acid-containing molecules: ozagrel, cinromide and piplartine.
Several reviews and studies have appeared in the literature focusing on a particular medicinal application of cinnamic-related molecules, for example on anticancer [21], antituberculosis [22], antimalarial [23], antifungal [24], antimicrobial [25], antiatherogenic [26] and antioxidant [25] activities. In addition a number of reviews directed towards the synthetic methods used to prepare cinnamic acids and related molecules have appeared in the literature [27,28,29]. Cinnamic acids have also been used by medicinal chemists to alter the potency, permeability, solubility or other parameters of a selected drug or pharmacophore.
Infectious diseases caused by bacteria, fungi and viruses are still a prominent global health problem, particularly in developing, low-income countries [30,31]. Every year in the whole planet, around 4 million persons die from acute respiratory infections, around 3 million individuals die from enteric infections, around 1.8 persons die from human immunodeficiency virus (HIV), around 1.3 million die from tuberculosis (TB), and 0.7 million die from malaria [32,33,34]. Other thousand people deeply suffer the consequences of being infected by neglected tropical pathogens Schistosoma mansoni, Onchocerca volvulus, Trypanosoma spp., Leishmania spp., Mycobacterium ulcerans, Mycobacterium leprae, Wucheria spp., and others. Although some of these diseases are caused by parasites, many are caused by bacteria and fungi. Anti-bacterial and anti-fungal drugs, which are broadly known as antimicrobials, started to be used in chemotherapy since the 1940 decade [35]. New antimicrobial classes were discovered in the 1940–1970 period, and were successfully introduced to clinical practice. However as soon as the new drugs were employed, the first drug-resistant strains started to appear [36,37]. In addition the widespread use of antibiotics for animal feed and human consumption during the last forty years, has fostered the emergence of resistance in several pathogenic organisms. Infection with drug-resistant strains is typically associated with longer treatment times, higher toxicity and higher costs. Nowadays some infections are extremely difficult to treat such as extensively drug-resistant tuberculosis (XDR-TB) [38], community-associated methicillin-resistant Staphylococcus aureus (MRSA) [39] and pan-resistant Klebsiella and Escherichia coli strains [40], and they pose an enormous challenge to clinicians. The presence of these “superbugs” call for drugs with novel mechanisms of action [41]. The cinnamic skeleton is considered an interesting scaffold for the development of novel antimicrobials, however little is known about its antimicrobial mechanism of action. A recent report proposed that cinnamic acids caused fungal growth inhibition by interacting with benzoate 4-hydroxylase, an enzyme responsible for aromatic detoxification [42]. However this enzyme occurs in fungi but not in prokaryotes, and because the cinnamic acids have proven anti-bacterialeffects [43,44,45,46], other targets may be implicated in their biological effects.
This review intends to bring the attention of scientists on the antimicrobial potential of the cinnamic acids. The focus is brought to the chemical entities containing the cinnamic skeleton, displaying fungal or bacterial growth inhibition. Emphasis is placed on whole-cell inhibitory potency, structure-activity relationships and mechanism of action studies. The review is organized in different sections according to the functional groups decorating the C6–C3 cinnamic skeleton. The first section deals with natural and synthetic cinnamic acids, the second section is concerned with cinnamic esters and amides, the third section deals with cinnamic aldehydes and alcohols, and finally the fourth section covers the hybrid covalently-bound molecules between a cinnamic acid and any other biologically-relevant molecule.
2. Natural and Synthetic Cinnamic Acids
Honey and propolis are both bee (Apis mellifera) products made from the nectar of the flowers, and they have long been used for their antimicrobial properties [47,48]. We should recall that a report from 1978, postulated the presence of cinnamic amides in the reproductive organs of flowers [4]. Propolis and honey contain hundreds of different organic compounds, and the cinnamic acids and their esters are typically present in these natural bees products [49,50,51,52]. Cinnamic acids with varied substitution on the aryl ring, and their esters have been identified in Iranian propolis showing minimum inhibitory concentration (MIC) values between 125 and 500 mg/L against bacteria and fungi [52]. Other studies have confirmed the antimicrobial potential of propolis [53,54]. Although secondary metabolites such as flavonoids, sesquiterpenoids present in propolis may have antimicrobial activity, cinnamic acids are likely to contribute to the observed effect.
The minimum inhibitory concentration (MIC) values for the natural cinnamic acids against different bacteria, as determined by different researchers using different methods, are shown in Table 1. It was surprising to find huge differences in the MIC values for the same compounds against the same species as reported by different authors. The existence of controversial results was already noted by Wen et al., in the seminal work of the antilisterial effect of natural phenolic acids [46]. The differences may be attributed to the diversity of experimental methods for MIC determination, often measuring distinct end-points, using different inoculum sizes, different culture media, and using particular strains with varying susceptibilities [55]. Although these discrepancies obscure the antimicrobial potential of the cinnamic acid class, there is clear tendency of the molecules to inhibit the growth of a wide variety of microorganisms by molecular mechanisms that are still unknown.
Table 1.
Minimum inhibitory concentration values of natural and synthetic cinnamic acids 1–24.
| Compound | Microbial Strain | MIC | Refs. |
|---|---|---|---|
| cinnamic acid (1) | Aeromonas hydrophila MTCC 646 | 7.7 mM | [59] |
| Aeromonas salmonicida MTCC 1522 | 5.6 mM | [59] | |
| Aspergillus flavus UBA294 | 1.7 mM | [65] | |
| Aspergillus niger ATCC 11394 | 844 µM | [65] | |
| Aspergillus terreus INM 031783 | 1.7 mM | [65] | |
| Candida albicans | 405 µM | [66] | |
| Edwardiella tarda MTCC 2400 | 7.0 mM | [59] | |
| Enterococcus faecalis | 6.75 mM | [12] | |
| Escherichia coli | 6.75 mM | [12] | |
| Escherichia coli | 5.0 mM | [56] | |
| Escherichia coli | >6.75 mM | [58] | |
| Escherichia coli ATCC 25922 | 9.0 mM | [57] | |
| Listeria monocytogenes | 13.5 mM | [46] | |
| Morganella morganni | >6.75 mM | [58] | |
| Mycobacterium tuberculosis H37Rv | 270 µM | [60] | |
| Mycobacterium tuberculosis H37Rv | 675 µM | [62] | |
| Neisseria gonorrhoeae | 6.75 mM | [58] | |
| Pasteurella multocida | >6.75 mM | [58] | |
| Proteus mirabilis | >6.75 mM | [58] | |
| Pseudomonas aeruginosa | 6.75 mM | [12] | |
| Salmonella sp. | 6.75 mM | [12] | |
| Salmonella typhimurium LT2 | 7.5 mM | [56] | |
| Staphylococcus aureus | 6.75 mM | [12] | |
| Staphylococcus epidermis | 6.75 mM | [12] | |
| Streptococcus pyogenes 10535 | 844 µM | [77] | |
| Vibrio parahaemolyticus | 6.75 mM | [12] | |
| 4-coumaric acid (3) | Aspergillus flavus UBA294 | 1.5 mM | [65] |
| Aspergillus niger ATCC 11394 | 761 µM | [65] | |
| Aspergillus terreus INM 031783 | 1.5 mM | [65] | |
| Bacillus cereus No-8 | 2.44 mM | [70] | |
| Bacillus subtilis NCIMB 8649 | 2.0 mM | [68] | |
| Bacillus subtilis 9372 | 122 µM | [67] | |
| Escherichia coli NCIMB 12210 | 2.0 mM | [68] | |
| Escherichia coli | 6.09 mM | [58] | |
| Escherichia coli #916 | 3.04 mM | [69] | |
| Escherichia coli O157:H7 | 2.74 mM | [70] | |
| Escherichia coli ATCC 25922 | 490 µM | [67] | |
| Fusarium oxysporum | 3.5 mM | [73] | |
| Fusarium verticillioides | 2.2. mM | [73] | |
| Lactobacillus brevis | 6.09 mM | [72] | |
| Lactobacillus collinoides | 6.09 mM | [72] | |
| Lactobacillus hilgardii IFI-CA 49 | 6.09 mM | [71] | |
| Lactobacillus rhamnosus #299 | 3.04 mM | [69] | |
| Listeria monocytogenes | 13.4 mM | [46] | |
| Morganella morganni | >6.09 mM | [58] | |
| Mycobacterium tuberculosis H37Rv | 244 µM | [60] | |
| Neisseria gonorrhoeae | 6.09 mM | [58] | |
| Pasteurella multocida | 6.09 mM | [58] | |
| Pediococcus pentosaceus IFI-CA 85 | 4.87 mM | [71] | |
| Proteus mirabilis | >6.09 mM | [58] | |
| Pseudomonas syringae NCIMB 649 | 2.0 mM | [68] | |
| Saccharomyces cerevisiae 019 391 | >8.0 mM | [68] | |
| Salmonella typhimurium #450 | 3.04 mM | [69] | |
| Salmonella typhimurium NRRL E4463 | 2.44 mM | [70] | |
| Salmonella typhimurium 50013 | 122 µM | [67] | |
| Schizosaccharomyces pombe 039 917 | 8.0 mM | [68] | |
| Shigella disenteriae 51302 | 61 µM | [67] | |
| Sporobolomyces roseus 043 529 | 8.0 mM | [68] | |
| Staphylococcus aureus # 917 | 761 µM | [69] | |
| Staphylococcus aureus NCTC 10657 | >3.65 mM | [70] | |
| Staphylococcus aureus 6538 | 122 µM | [67] | |
| Streptococcus pneumoniae ATCC 49619 | 122 µM | [67] | |
| Streptococcus pyogenes 10535 | 761 µM | [77] | |
| 3-coumaric acid (4) | Aspergillus flavus UBA294 | 1.5 mM | [65] |
| Aspergillus niger ATCC 11394 | 1.5 mM | [65] | |
| Aspergillus terreus INM 031783 | 1.5 mM | [65] | |
| Mycobacterium tuberculosis H37Rv | 366 µM | [60] | |
| 2-coumaric acid (5) | Bacillus cereus No-8 | 2.44 mM | [70] |
| Escherichia coli #916 | 1.5 mM | [69] | |
| Escherichia coli O157:H7 | 2.74 mM | [70] | |
| Lactobacillus rhamnosus #299 | 1.5 mM | [69] | |
| Morganella morganni | >6.09 mM | [58] | |
| Mycobacterium tuberculosis H37Rv | 122 µM | [60] | |
| Neisseria gonorrhoeae | >6.09 mM | [58] | |
| Pasteurella multocida | >6.09 mM | [58] | |
| Proteus mirabilis | >6.09 mM | [58] | |
| Salmonella typhimurium NRRL E4463 | 2.44 mM | [70] | |
| Salmonella typhimurium #450 | 1.5 mM | [69] | |
| Staphylococcus aureus NCTC 10657 | >3.65 mM | [70] | |
| Staphylococcus aureus # 917 | 760 µM | [69] | |
| caffeic acid (6) | Aspergillus flavus UBA294 | >1.39 mM | [65] |
| Aspergillus flavus | >5.5 mM | [73] | |
| Aspergillus fumigatus | >5.5 mM | [73] | |
| Aspergillus niger ATCC 11394 | >1.39 mM | [65] | |
| Aspergillus terreus INM 031783 | >1.39 mM | [65] | |
| Bacillus cereus No-8 | 1.94 mM | [70] | |
| Bacillus subtilis NCIMB 8649 | 4.0 mM | [68] | |
| Campylobacter jejuni KC40 | >1.0 mM | [93] | |
| Candida albicans 62 | 694 µM | [77] | |
| Candida albicans biofilm | 1.42 mM | [78] | |
| Candida albicans planktonic | 710 µM | [78] | |
| Escherichia coli | >5.5 mM | [58] | |
| Escherichia coli #916 | 2.78 mM | [69] | |
| Escherichia coli | 1.78 mM | [57] | |
| Escherichia coli NCIMB 12210 | 8.0 mM | [68] | |
| Escherichia coli O157:H7 | 1.94 mM | [70] | |
| Fusarium oxysporum | >5.5 mM | [73] | |
| Fusarium verticilioides | >5.5 mM | [73] | |
| Lactobacillus hilgardii IFI-CA 49 | 4.44 mM | [71] | |
| Lactobacillus rhamnosus #299 | <1.39 mM | [69] | |
| Listeria monocytogenes | 16.1 mM | [46] | |
| Morganella morganni | >5.5 mM | [58] | |
| Neisseria gonorrhoeae | >5.5 mM | [58] | |
| Pasteurella multocida | 5.5 mM | [58] | |
| Pediococcus pentosaceus IFI-CA 85 | 3.89 mM | [71] | |
| Penicillium brevicompactum | >5.5 mM | [73] | |
| Penicillium expansum | >5.5 mM | [73] | |
| Proteus mirabilis | >5.5 mM | [58] | |
| Pseudomonas syringae NCIMB 649 | 4.0 mM | [68] | |
| Saccharomyces cerevisiae 019 391 | >8.0 mM | [68] | |
| Salmonella typhimurium #450 | 2.78 mM | [69] | |
| Salmonella typhimurium NRRL E4463 | 1.94 mM | [70] | |
| Schizosaccharomyces pombe 039 917 | >8.0 mM | [68] | |
| Sporobolomyces roseus 043 529 | >8.0 mM | [68] | |
| Staphylococcus aureus 209 | 694 µM | [77] | |
| Staphylococcus aureus # 917 | 694 µM | [69] | |
| Staphylococcus aureus NCTC 10657 | 2.22 mM | [70] | |
| Streptococcus pyogenes 10535 | 694 µM | [77] | |
| ferulic acid (7) | Aspergillus flavus | >5.15 mM | [73] |
| Aspergillus flavus UBA294 | 161 µM | [65] | |
| Aspergillus fumigatus | >5.15 mM | [73] | |
| Aspergillus niger | >10 mM | [80] | |
| Aspergillus niger ATCC 11394 | 322 µM | [65] | |
| Aspergillus terreus INM 031783 | >1.3 mM | [65] | |
| Bacillus cereus No-8 | 2.06 mM | [70] | |
| Bacillus subtilis | 6.0 mM | [80] | |
| Bacillus subtilis NCIMB 8649 | 2.0 mM | [68] | |
| Candida albicans | >10 mM | [80] | |
| Candida albicans ATCC 10231 | 659 µM | [81] | |
| Candida krusei ATCC 6258 | 659 µM | [81] | |
| Enterococcus faecalis ATCC 29212 | 659 µM | [81] | |
| Escherichia coli | >5.15 mM | [58] | |
| Escherichia coli ATCC 25922 | 1.3 mM | [81] | |
| Escherichia coli O157:H7 | 2.32 mM | [70] | |
| Escherichia coli CECT 434 | 515 µM | [79] | |
| Escherichia coli IFO13275 | >5.0 mM | [94] | |
| Escherichia coli NCIMB 12210 | 2.0 mM | [68] | |
| Fusarium oxysporum | >5.15 mM | [73] | |
| Fusarium verticilioides | >5.15 mM | [73] | |
| Klebsiella pneumoniae RSKK 574 | 1.3 mM | [81] | |
| Listeria monocytogenes | 13.9 mM | [46] | |
| Listeria monocytogenes ATCC 15313 | 6.44 mM | [79] | |
| Morganella morganni | >5.15 mM | [58] | |
| Neisseria gonorrhoeae | 5.15 mM | [58] | |
| Pasteurella multocida | 5.15 mM | [58] | |
| Pediococcus pentosaceus IFI-CA 85 | 4.63 mM | [71] | |
| Penicillium brevicompactum | >5.15 mM | [73] | |
| Penicillium expansum | >5.15 mM | [73] | |
| Proteus mirabilis | >5.15 mM | [58] | |
| Pseudomonas aeruginosa ATCC 10145 | 515 µM | [79] | |
| Pseudomonas syringae NCIMB 649 | 2.0 mM | [68] | |
| Saccharomyces cerevisiae | 6.0 mM | [80] | |
| Saccharomyces cerevisiae 019 391 | 4.0 mM | [68] | |
| Salmonella enteriditis IFO3133 | >5.0 mM | [94] | |
| Salmonella typhimurium NRRL E4463 | 2.06 mM | [70] | |
| Schizosaccharomyces pombe 039 917 | 8.0 mM | [68] | |
| Sporobolomyces roseus 043 529 | 2.0 mM | [68] | |
| Staphylococcus aureus | 6.0 mM | [80] | |
| Staphylococcus aureus 209 | 644 µM | [77] | |
| Staphylococcus aureus ATCC 29213 | 1.3 mM | [81] | |
| Staphylococcus aureus CECT 976 | 5.7 mM | [79] | |
| Staphylococcus aureus IFO 12732 | 2.0 mM | [94] | |
| Staphylococcus aureus NCTC 10657 | 3.09 mM | [70] | |
| Streptococcus pyogenes 10535 | 644 µM | [77] | |
| sinapic acid (8) | Bacillus subtilis NCIMB 8649 | 2.0 mM | [68] |
| Bacillus subtilis FAD 110 | 1.3 mM | [83] | |
| Escherichia coli NCIMB 12210 | 2.0 mM | [68] | |
| Escherichia coli IFO13275 | 2.2 mM | [94] | |
| Escherichia coli AW 1.7 | 3.1 mM | [83] | |
| Listeria innocua ATCC 330909 | 1.3 mM | [83] | |
| Listeria monocytogenes ATCC 7644 | 900 µM | [83] | |
| Pseudomonas fluorescens ATCC 13525 | 2.7 mM | [83] | |
| Pseudomonas syringae NCIMB 649 | 4.0 mM | [68] | |
| Saccharomyces cerevisiae 019 391 | >8.0 mM | [68] | |
| Salmonella enteriditis IFO3133 | 2.0 mM | [94] | |
| Schizosaccharomyces pombe 039 917 | >8.0 mM | [68] | |
| Sporobolomyces roseus 043 529 | >8.0 mM | [68] | |
| Staphylococcus aureus 209 | 558 µM | [77] | |
| Staphylococcus aureus IFO 12732 | 1.8 mM | [94] | |
| Staphylococcus aureus ATCC 6538 | 1.3 mM | [83] | |
| Streptococcus pyogenes 10535 | 558 µM | [77] | |
| 4-methoxycinnamic acid (9) | Aspergillus niger | 50.4 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 50.4 µM | [44] | |
| Escherichia coli | 164 µM | [44] | |
| Escherichia coli | 281 µM | [84] | |
| Micrococcus luteus | 449 µM | [84] | |
| Salmonella enteriditis | 337 µM | [84] | |
| Staphylococcus aureus | 337 µM | [84] | |
| Staphylococcus aureus | 203 µM | [44] | |
| 3,4-methylenedioxy-cinnamic acid (10) | Mycobacterium tuberculosis H37Rv | 312 µM | [60] |
| Mycobacterium tuberculosis H37Rv | >520 µM | [85] | |
| 4-nitrocinnamic acid (11) | Bacillus subtilis IFO 3009 | 891 µM | [86] |
| Escherichia coli IFO 3301 | 794 µM | [86] | |
| 3-nitrocinnamic acid (12) | Aspergillus niger | 43.5 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 43.5 µM | [44] | |
| Escherichia coli | 252 µM | [44] | |
| Staphylococcus aureus | 252 µM | [44] | |
| 4-aminocinnamic acid (13) | Bacillus subtilis IFO 3009 | 602 µM | [86] |
| Escherichia coli IFO 3301 | 708 µM | [86] | |
| 4-chlorocinnamic acid (14) | Bacillus subtilis IFO 3009 | 708 µM | [86] |
| Escherichia coli IFO 3301 | 708 µM | [86] | |
| 4-O-prenylcoumaric acid (16) | Mycobacterium tuberculosis H37Rv | 86.1 µM | [60] |
| 4-O-geranylcoumaric acid (17) | Mycobacterium tuberculosis H37Rv | 66.8 µM | [60] |
| 3-O-prenylcoumaric acid (18) | Mycobacterium tuberculosis H37Rv | 172 µM | [60] |
| 2-O-prenylcoumaric acid (19) | Mycobacterium tuberculosis H37Rv | 258 µM | [60] |
| 3-prenyl-4-coumaric acid (= drupanin) (20) | Aspergillus fumigatus ATCC 26934 | >1.1 mM | [92] |
| Aspergillus flavus ATCC 9170 | >1.1 mM | [92] | |
| Aspergillus niger ATCC 9029 | >1.1 mM | [92] | |
| Candida albicans ATCC 10231 | 1.1 mM | [92] | |
| Candida tropicalis CEREMIC 131 | >1.1 mM | [92] | |
| Cryptococcus neoformans ATCC 32264 | 1.1 mM | [92] | |
| Epidermophyton floccosum C114 | 215 µM | [92] | |
| Escherichia coli ATCC 25922 | >1.1 mM | [92] | |
| Microsporum canis C112 | 431 µM | [92] | |
| Microsporum gypseum C115 | 431 µM | [92] | |
| Staphylococcus aureus LMS | >1.1 mM | [92] | |
| Methicillin-resistant Staphylococcus aureus | >1.1 mM | [92] | |
| Trichophyton mentagrophytes ATCC 9972 | 431 µM | [92] | |
| Trichophyton rubrum C113 | 431 µM | [92] | |
| 4-acetyl-3-prenyl-4-coumaric acid (21) | Aspergillus fumigatus ATCC 26934 | >912 µM | [92] |
| Aspergillus flavus ATCC 9170 | >912 µM | [92] | |
| Aspergillus niger ATCC 9029 | >912 µM | [92] | |
| Candida albicans ATCC 10231 | >912 µM | [92] | |
| Candida tropicalis CEREMIC 131 | >912 µM | [92] | |
| Cryptococcus neoformans ATCC 32264 | >912 µM | [92] | |
| Epidermophyton floccosum C114 | 364 µM | [92] | |
| Escherichia coli ATCC 25922 | >912 µM | [92] | |
| Microsporum canis C112 | >912 µM | [92] | |
| Microsporum gypseum C115 | 912 µM | [92] | |
| Staphylococcus aureus LMS | >912 µM | [92] | |
| Methicillin-resistant Staphylococcus aureus | >912 µM | [92] | |
| Trichophyton mentagrophytes ATCC 9972 | 456 µM | [92] | |
| Trichophyton rubrum C113 | 456 µM | [92] | |
| 3,5-diprenyl-4-coumaric acid (22) | Aspergillus fumigatus ATCC 26934 | >833 µM | [92] |
| Aspergillus flavus ATCC 9170 | >833 µM | [92] | |
| Aspergillus niger ATCC 9029 | >833 µM | [92] | |
| Candida albicans ATCC 10231 | 833 µM | [92] | |
| Candida tropicalis CEREMIC 131 | >833 µM | [92] | |
| Cryptococcus neoformans ATCC 32264 | >833 µM | [92] | |
| Epidermophyton floccosum C114 | 166 µM | [92] | |
| Escherichia coli ATCC 25922 | >833 µM | [92] | |
| Microsporum canis C112 | >833 µM | [92] | |
| Microsporum gypseum C115 | >833 µM | [92] | |
| Staphylococcus aureus LMS | 833 µM | [92] | |
| Methicillin-resistant Staphylococcus aureus | 833 µM | [92] | |
| Trichophyton mentagrophytes ATCC 9972 | >833 µM | [92] | |
| Trichophyton rubrum C113 | 416 µM | [92] | |
| 4-acetyl-3,5-diprenyl-4-coumaric acid (23) | Aspergillus fumigatus ATCC 26934 | >731 µM | [92] |
| Aspergillus flavus ATCC 9170 | >731 µM | [92] | |
| Aspergillus niger ATCC 9029 | >731 µM | [92] | |
| Candida albicans ATCC 10231 | >731 µM | [92] | |
| Candida tropicalis CEREMIC 131 | >731 µM | [92] | |
| Cryptococcus neoformans ATCC 32264 | >731 µM | [92] | |
| Epidermophyton floccosum C114 | 292 µM | [92] | |
| Escherichia coli ATCC 25922 | >731 µM | [92] | |
| Microsporum canis C112 | >731 µM | [92] | |
| Microsporum gypseum C115 | >731 µM | [92] | |
| Staphylococcus aureus LMS | >731 µM | [92] | |
| Methicillin-resistant Staphylococcus aureus | >731 µM | [92] | |
| Trichophyton mentagrophytes ATCC 9972 | 731 µM | [92] | |
| Trichophyton rubrum C113 | 365 µM | [92] | |
| O-acetylferulic acid (24) | Candida albicans ATCC 10231 | 540 µM | [81] |
| Candida krusei ATCC 6258 | 540 µM | [81] | |
| Enterococcus faecalis ATCC 29212 | 540 µM | [81] | |
| Escherichia coli ATCC 25922 | 1.1 mM | [81] | |
| Klebsiella pneumoniae RSKK 574 | 1.1 mM | [81] | |
| Staphylococcus aureus ATCC 29213 | 1.1 mM | [81] |
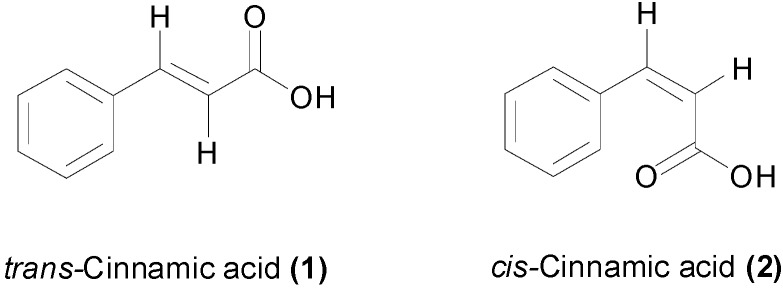
Cinnamic acid (1, Figure 2) showed a weak antibacterial effect against most of Gram-negative and Gram-positive species of bacteria, with MIC values higher than 5.0 mM [12,46,56,57,58] (Table 1). The same level of potency was observed against the fish pathogens Aeromonas hydrophila, Aeromonas salmonicida and Edwardiella tarda with MIC values between 5.6 and 7.7 mM [59]. However cinnamic acid was found to be much more active against the tuberculosis-causing bacteria, Mycobacterium tuberculosis H37Rv, with an MIC values of 270–675 µM using the SPOTi and the radiometric Bactec assays [60,61,62]. The free carboxylic acid and the presence of the α,β-unsaturation were both required for the anti-TB activity [60]. Rastogi et al., reported an MIC value of 675 µM against the H37Rv strain and varying values between 337 µM and 1.4 mM for multiple drug-resistant (MDR) clinical M. tuberculosis isolates [62]. The study found that 1 enhanced synergistically the effect of anti-TB drugs such as amikacin, ofloxacin and clofazimine. In addition its geometric isomer, cis-cinnamic acid (2) (Figure 2), was approximately 120 times more active than the trans isomer, with minimum bactericidal concentrations (MBC) values of 16.9 µM for 2, compared to 2.0 mM for 1, against an MDR M. tuberculosis strain [63]. The specific anti-TB effect of cinnamic acid may explain the traditional use of storax (Liquidambar orientalis) and cinnamon for treating TB in the 19th century [64]. Cinnamic acid also demonstrated anti-fungal activity with MIC values of 1.7 mM against Aspergillus terreus and Aspergillus flavus, being more active against Aspergillus niger with an MIC value of 844 µM [65]. Against Candida albicans, an MIC value of 405 µM has been found [66], which is comparable to the potency against M. tuberculosis.
Figure 2.
Chemical structures of trans- and cis-cinnamic acids
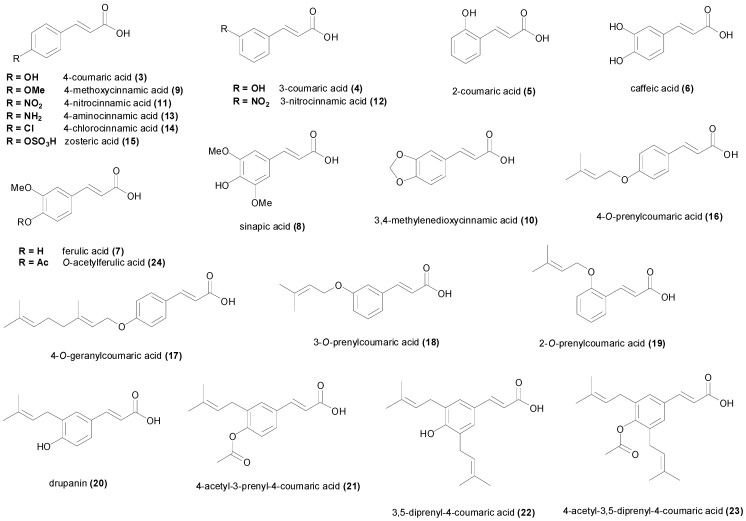
The widely distributed natural phenol 4-hydroxycinnamic acid (3, Figure 3), also known as 4-coumaric acid, has been found to be comparatively more potent bacterial growth inhibitor compared to cinnamic acid (1) (Table 1). However the reported MIC values of 4-coumaric acid vary to great extent from one species or strain to another. The MIC values against some Gram-negative bacteria such as Shigella disenteriae 51302 was low (MIC = 61 µM) [67], but however against Neisseria gonorrhoeae or Listeria monocytogenes or E. coli, the MIC was high (MIC > 6.0 mM) [46,58]. The same was true against Gram-positive species, with some studies reporting low MIC values for strains of Staphylococcus aureus and Bacillus subtilis whereas other reports have published higher MIC values [67,68,69,70]. A study showed that 3 disrupted the outer membrane of the Gram-negative bacteria S. disenteriae increasing the permeabilization, and in addition, the compound interacted with DNA and thus it may inhibit essential biochemical processes related to nucleic acids [67]. 4-Coumaric acid (3) completely inhibited M. tuberculosis H37Rv growth at 244 µM concentration [60], being therefore slightly more active than cinnamic acid. The compound had weak inhibition of lactic acid bacteria with MIC values of 6.09 mM [71,72] but was however relatively more active against the economically important phytopathogenic fungi Fusarium oxysporum and Fusarium verticillioides, with MIC values of 3.5 mM and 2.2 mM respectively [73]. Against Aspergillus spp MIC values have been reported between 1.5 mM and 760 µM [65].
Figure 3.
Chemical structures of differently substituted natural and synthetic cinnamic acids.
The structural isomers 3-coumaric acid (4) and 2-coumaric acid (5) (Figure 3 and Table 1) are less common in nature [74,75] compared to the 4-isomer, and have been less studied. In particular, reports of the antimicrobial activity of the 3-coumaric acid are scarce. 3-Coumaric acid (4) was less active (MIC = 366 µM) compared with the 4-isomer against M. tuberculosis H37Rv, while 2-coumaric acid (5) was the isomer with the highest activity (MIC = 122 µM) [60]. Similarly, 2-coumaric acid showed stronger antimicrobial activity against E. coli, S. aureus, Salmonella typhimurium and Lactobacillus rhamnosus in comparison with 4-coumaric acid, with MIC values between 1.5 mM and 760 µM [69]. However another report published the same MIC values against S. aureus (MIC > 3.6 mM), Bacillus cereus (MIC = 2.4 mM), E. coli (MIC = 2.7 mM) and S. typhimurium (MIC = 2.7 mM) [70]. The 3-isomer (4) displayed moderate antifungal activity (MIC > 1.5 mM) against A. terreus, A. flavus and A. niger [65].
The other abundant cinnamic acids in nature are caffeic acid (6), ferulic acid (7) and sinapic acid (8) (Figure 3), which have been studied for their antimicrobial activities [58,68,73]. A similar pattern was observed for these three natural cinnamic acids, showing a weak growth inhibition against Gram-negative bacteria compared to Gram-positive bacteria and fungi (Table 1). The pH of the media has been reported to exert influence on growth inhibition, with lower pH values increasing the activity of the acids [76] by probably favoring a greater proportion of un-dissociated acid. The lowest MIC values for caffeic acid (6) were found against some strains of S. aureus and Streptococcus pyogenes 10535 (MIC = 694 µM) [69,77]. Caffeic acid showed significant growth inhibition of planktonic C. albicans with MIC between 694 and 710 µM [77,78]. Other species of bacteria and fungi were less susceptible to caffeic acid. Ferulic acid (7) demonstrated significant antibacterial activity against S. aureus 209 and Streptococcus pyogenes 10535 with MIC values of 644 µM [77]. Pseudomonas aeruginosa ATCC 10145 and E. coli CECT 434 were also susceptible to ferulic acid (MIC = 515 µM) [79]. In addition the strain A. niger ATCC 11394 showed a low MIC value (MIC = 322 µM) [65], however the MIC value against another strain of A. niger was found to be higher than 10 mM [80]. Interestingly A. flavus UBA 294 was the microorganism with the lowest MIC value for ferulic acid (MIC = 161 µM) [65]. A similar pattern was noticed for C. albicans, one strain being reported as susceptible with an MIC value of 659 µM [81], whereas another report showed an MIC value higher than 10 mM [80]. Sinapic acid (8), which is the most substituted of the common naturally-occurring cinnamic acids, showed significant activity against S. aureus 209 and Streptococcus pyogenes 10535 (MIC = 558 µM) [77]. The acid also demonstrated anti-Campylobacter activity with MIC values ranging from 696 µM to 1.40 mM [82]. Sinapic acid was fairly active against L. monocytogenes ATCC 7644 with an MIC value of 900 µM [83]. Sinapic acid (8) was completely inactive at a concentration of 4.46 mM against the phytopathogenic fungi F. oxysporum, A. flavus, Penicillium brevicompactum and others [73], in contrast with caffeic and ferulic acid.
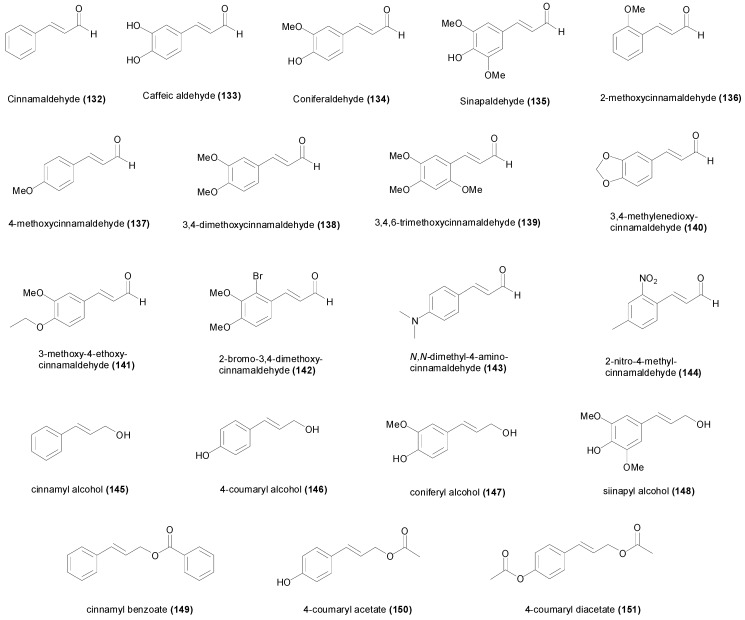
Some cinnamic acids with a particular substitution pattern on the aryl ring have been prepared and examined for their antimicrobial activity. The compound 4-methoxycinnamic acid (9) was isolated from the Argentinian medicinal plant Baccharis grisebachii and its antimicrobial activity was evaluated [84]. This acid showed a potent antibacterial and antifungal effect with MIC values ranging between 50.4 and 449 µM (Table 1) [44,84]. Interestingly, the acid (9) showed higher growth inhibition against fungal species compared to bacteria, and Gram-negative and Gram-positive bacteria were equally inhibited by the compound. The acid 3,4-methylenedioxycinnamic acid (10) has been reported to inhibit Mycobacterium tuberculosis H37Rv with one report displaying an MIC value of 312 µM [60] and the other an MIC value higher than 520 µM [85]. The effect of the position of the nitro group on antimicrobial activity suggest that 4-nitrocinnamic acid (11) is more active than 3-nitrocinnamic acid (12), however the comparison data results from two different studies [44,86]. The seminal report from 1940, found that none of the positional isomers of nitrocinnamic acid inhibited S. aureus or E. coli at the highest dilution tested [87]. A noteworthy MIC value was found for 12 against the fungal species A. niger and C. albicans (MIC = 43.5 µM) [44]. Although the nitro groups can be readily reduced to amino groups, only 4-aminocinnamic (13) acid has been evaluated for its antimicrobial properties. This acid showed inhibitory activity of B. subtilis and E. coli with respective MIC values of 602 and 708 µM [86]. No information on the antimicrobial properties of the positional isomers of 4-aminocinnamic acid could be found. Although most of the halogen derivatives of cinnamic acid have been prepared [88,89], no information about their antimicrobial profile of activity was found in literature, except for 4-chlorocinnamic acid (14). This acid showed MIC values of 708 µM against both E. coli and B. subtilis [86]. Zosteric acid (15) which is naturally present in the eelgrass Zostera marina, has been found to display powerful antifouling properties by preventing bacterial biofilm formation on the surface of water-submerged objects [90]. Zosteric acid did not show any growth inhibitory activity against M. tuberculosis H37Rv [60], however the compound inhibited biofilm formation of C. albicans at 41 µM [91], but no MIC values were found in the literature search.
Among the prenylated coumaric acids, 4-O-prenylcoumaric acid (16) showed potent inhibition of M. tuberculosis H37Rv with an MIC value of 86 µM [60]. The MIC values for 4-O-geranylcoumaric acid (17), 3-O-prenylcoumaric acid (18) and 2-O-prenylcoumaric acid (19) against the same bacterial species were found to be respectively 69, 172 and 258 µM [60]. If we compare the anti-TB activity of the coumaric acids with the O-prenylcoumaric acids, it is clear that O-prenylation increases the activity for the 4- and 3-isomers, whereas for the 2-isomer, O-prenylation reduces the anti-TB activity. The C-prenylated coumaric acid drupanin (20), also known as 3-prenyl-4-coumaric acid, showed fungal growth inhibition particularly against dermatophytes species such as Epidermophyton floccosum C114, Microsporum gypseum C115, Microsporum canis C112, Trichophyton mentagrophytes ATCC 9972 and Trichophyton rubrum C113 with respective MIC values of 215, 431, 431, 431 and 431 µM [92]. Against the bacterial species, drupanin was weakly active with MIC values higher than 1.1 mM. The O-acetyl derivative of drupanin, 4-acetyl-3-prenyl-4-coumaric acid (21) showed a similar pattern of activity, being able to inhibit the dermatophyte E. floccosum C114 with an MIC value of 364 µM [92]. It was therefore slightly less active than the non-acetylated compound.
The compound 3,5-diprenyl-4-coumaric acid (22) was comparatively more active growth inhibitor of E. floccosum C114 achieving an MIC values of 166 µM [92]. Its O-acetyl derivative (23) was again slightly less active than the non-acetylated compound (Table 1), suggesting that the free phenolic OH is essential for potent activity of this class of molecules. The 4-O-acetyl derivative of ferulic acid, namely 4-O-acetylferulic acid (24), prepared by synthesis, inhibited the growth of C. albicans, Candida krusei and Enterococcus faecalis at 540 µM, but was less active against S. aureus, E. coli and Klebsiella pneumoniae [81]. Its activity was found to be comparatively similar to the antimicrobial effect of ferulic acid.
3. Natural and Synthetic Cinnamic Esters and Amides
3.1. Esters
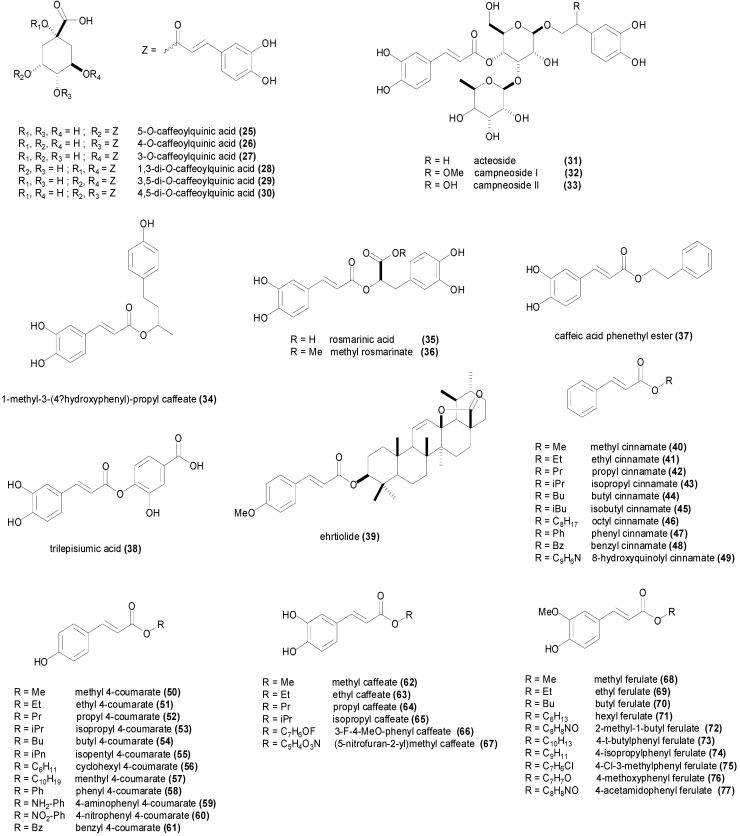
The chlorogenic acids are a family of natural esters of hydroxycinnamic acids (coumaric, caffeic, ferulic and sinapic acids) with (−)-quinic acid [14]. The most common chlorogenic acid is 5-O-caffeoylquinic acid (25, Figure 4), which is abundant in coffee, black tea and mate but is also present in apples, pears and berries [95]. This chlorogenic acid isolated from artichoke, displayed MIC values of 564 µM against B. subtilis, S. aureus, E. coli, S. typhimurium, P. aeruginosa and S. cerevisiae, and lower MIC values of 282 µM against Micrococcus luteus, Agrobacterium tumefaciens, Aspergillus niger, Penicillium oxalicum and Mucor mucedo [96]. It was even more active against the fungi Candida albicans, Candida lusitaniae, Saccharomyces carlsbergencis and Cladosporium cucumerinum with an MIC value of 141 µM [96]. MIC values for the natural cinnamic esters are summarized in Table 2. However the effect of (25), isolated from roasted coffee beans, against S. aureus and Streptococcus mutans was much less marked in the study of Daglia et al., with MIC values of 17.8 and 7.62 mM respectively [97]. The study of Xia et al., isolated the three position isomers of caffeoylquinic acid from the Prunus mume seeds, and evaluated their antimicrobial activity against a panel of microorganisms, finding for (25) MIC values of 282 µM against S. aureus, 423 µM against E. coli and C. albicans, 564 µM against S. enterica, S. cerevisiae and A. niger, and an MIC value of 705 µM against Vibrio parahaemolyticus [98]. This natural product isolated from Artemisia absinthium inhibited completely the growth of S. aureus at a concentration of 361 µM, being even more active against B. cereus, E. faecalis with an MIC value 181 µM [99]. In addition 5-O-caffeoylquinic acid demonstrated biofilm formation inhibition in S. aureus and E. faecalis. Controversial results have also been reported for 25, as the study of Alves et al. reported MIC values higher than 2.8 mM against all the evaluated microorganisms including E. coli [58]. The study of Lou et al., published in 2011, showed MIC values ranging from 54 µM against Streptococcus pneumoniae and Shigella disenteriae to 226 µM against E. coli [100]. The methyl ester of 25, namely methyl 5-O-caffeoylquinate isolated from the invasive plant Ageratina adenophora, inhibited the growth of S. aureus, Bacillus thuringensis, E. coli and S. enterica at a concentration of 89 µM [101], and was therefore comparatively more active than the parent chlorogenic acid. Transmission electron microscopy, membrane potential and nucleotide leakage studies on Shigella disenteriae led to the conclusion that the chlorogenic acid disrupted cell wall permeability and then depolarized the bacterial cell wall membrane causing cytoplasmic leakage [100].
Figure 4.
Chemical structures of cinnamic esters displaying antimicrobial activity.
Table 2.
Minimum inhibitory concentration values of natural and synthetic cinnamic esters 25–77.
| Compound | Microorganism Strain | MIC | Refs. |
|---|---|---|---|
| 5-O-caffeoylquinic acid (25) | Agrobacterium tumefaciens CGMCC 1.1415 | 282 µM | [96] |
| Aspergillus niger ATCC 10553 | 564 µM | [98] | |
| Aspergillus niger CGMCC 3.316 | 282 µM | [96] | |
| Bacillus subtilis CGMCC 1.1849 | 564 µM | [96] | |
| Bacillus subtilis 9372 | 108 µM | [100] | |
| Candida albicans ATCC 10231 | 141 µM | [96] | |
| Candida albicans ATCC 14053 | 423 µM | [98] | |
| Candida albicans DAY185 | 181 µM | [99] | |
| Candida lusitaniae ATCC 2201 | 141 µM | [96] | |
| Cladosporium cucumerinum ATCC 11279 | 141 µM | [96] | |
| Enterococcus faecalis OGRF1 | 181 µM | [99] | |
| Escherichia coli ATCC 25922 | 216 µM | [100] | |
| Escherichia coli ATCC 25922 | 423 µM | [98] | |
| Escherichia coli CGMCC 1.90 | 564 µM | [96] | |
| Micrococcus luteus CGMCC 1.880 | 282 µM | [96] | |
| Mucor mucedo CGMCC 3.15 | 282 µM | [96] | |
| Penicillium oxalicum CGMCC 3.4022 | 282 µM | [96] | |
| Pseudomonas aeruginosa CG-MCC 1.2031 | 564 µM | [96] | |
| Saccharomyces carlsbergensis ATCC 2166 | 141 µM | [96] | |
| Saccharomyces cerevisiae ATCC 36858 | 564 µM | [98] | |
| Saccharomyces cerevisiae IFFI 1611 | 564 µM | [96] | |
| Salmonella enterica ATCC 13076 | 564 µM | [98] | |
| Salmonella typhimurium CGMCC 1.1190 | 564 µM | [96] | |
| Salmonella typhimurium 50013 | 108 µM | [100] | |
| Shigella disenteriae 51302 | 54 µM | [100] | |
| Staphylococcus aureus | 17.8 mM | [97] | |
| Staphylococcus aureus 8325-4 | 361 µM | [99] | |
| Staphylococcus aureus 6538 | 108 µM | [100] | |
| Staphylococcus aureus ATCC 25923 | 282 µM | [98] | |
| Staphylococcus aureus ATCC 6358P | 564 µM | [96] | |
| Streptococcus mutans | 7.62 mM | [97] | |
| Streptococcus pneumoniae ATCC 49619 | 54 µM | [100] | |
| Vibrio parahaemolyticus ATCC 17802 | 705 µM | [98] | |
| 4-O-caffeoylquinic acid (26) | Aspergillus niger ATCC 10553 | 564 µM | [98] |
| Candida albicans ATCC 14053 | 564 µM | [98] | |
| Escherichia coli ATCC 25922 | 423 µM | [98] | |
| Saccharomyces cerevisiae ATCC 36858 | 705 µM | [98] | |
| Salmonella enterica ATCC 13076 | 705 µM | [98] | |
| Staphylococcus aureus ATCC 25923 | 423 µM | [98] | |
| 3-O-caffeoylquinic acid (27) | Aspergillus niger ATCC 10553 | 423 µM | [98] |
| Candida albicans ATCC 14053 | 423 µM | [98] | |
| Escherichia coli ATCC 25922 | 282 µM | [98] | |
| Saccharomyces cerevisiae ATCC 36858 | 564 µM | [98] | |
| Salmonella enterica ATCC 13076 | 564 µM | [98] | |
| Staphylococcus aureus ATCC 25923 | 282 µM | [98] | |
| Vibrio parahaemolyticus ATCC 17802 | 564 µM | [98] | |
| 1,3-di-O-caffeoylquinic acid (28) | Agrobacterium tumefaciens CGMCC 1.1415 | 194 µM | [96] |
| Aspergillus niger CGMCC 3.316 | 194 µM | [96] | |
| Bacillus subtilis CGMCC 1.1849 | 387 µM | [96] | |
| Candida albicans ATCC 10231 | 194 µM | [96] | |
| Candida lusitaniae ATCC 2201 | 194 µM | [96] | |
| Cladosporium cucumerinum ATCC 11279 | 194 µM | [96] | |
| Escherichia coli CGMCC 1.90 | 194 µM | [96] | |
| Micrococcus luteus CGMCC 1.880 | 194 µM | [96] | |
| Mucor mucedo CGMCC 3.15 | 194 µM | [96] | |
| Penicillium oxalicum CGMCC 3.4022 | 194 µM | [96] | |
| Pseudomonas aeruginosa CG-MCC 1.2031 | 194 µM | [96] | |
| Saccharomyces carlsbergensis ATCC 2166 | 194 µM | [96] | |
| Saccharomyces cerevisiae IFFI 1611 | 387 µM | [96] | |
| Salmonella typhimurium CGMCC 1.1190 | 387 µM | [96] | |
| Staphylococcus aureus ATCC 6358P | 387 µM | [96] | |
| 3,5-di-O-caffeoylquinic acid (29) | Agrobacterium tumefaciens CGMCC 1.1415 | 387 µM | [96] |
| Aspergillus niger CGMCC 3.316 | 194 µM | [96] | |
| Bacillus subtilis CGMCC 1.1849 | 387 µM | [96] | |
| Candida albicans ATCC 10231 | 387 µM | [96] | |
| Candida lusitaniae ATCC 2201 | 387 µM | [96] | |
| Cladosporium cucumerinum ATCC 11279 | 194 µM | [96] | |
| Escherichia coli CGMCC 1.90 | 387 µM | [96] | |
| Micrococcus luteus CGMCC 1.880 | 194 µM | [96] | |
| Mucor mucedo CGMCC 3.15 | 194 µM | [96] | |
| Penicillium oxalicum CGMCC 3.4022 | 194 µM | [96] | |
| Saccharomyces carlsbergensis ATCC 2166 | 387 µM | [96] | |
| Saccharomyces cerevisiae IFFI 1611 | 387 µM | [96] | |
| Salmonella typhimurium CGMCC 1.1190 | >387 µM | [96] | |
| Staphylococcus aureus ATCC 6358P | 387 µM | [96] | |
| 4,5-di-O-caffeoylquinic acid (30) | Agrobacterium tumefaciens CGMCC 1.1415 | 387 µM | [96] |
| Aspergillus niger CGMCC 3.316 | 387 µM | [96] | |
| Bacillus subtilis CGMCC 1.1849 | 387 µM | [96] | |
| Candida lusitaniae ATCC 2201 | 387 µM | [96] | |
| Cladosporium cucumerinum ATCC 11279 | 194 µM | [96] | |
| Escherichia coli CGMCC 1.90 | 194 µM | [96] | |
| Micrococcus luteus CGMCC 1.880 | 97 µM | [96] | |
| Mucor mucedo CGMCC 3.15 | 194 µM | [96] | |
| Penicillium oxalicum CGMCC 3.4022 | 194 µM | [96] | |
| Saccharomyces carlsbergensis ATCC 2166 | 387 µM | [96] | |
| Saccharomyces cerevisiae IFFI 1611 | 387 µM | [96] | |
| Staphylococcus aureus ATCC 6358P | 194 µM | [96] | |
| Acteoside (31) | Enterobacter cloacae P99 | 1.2 mM | [102] |
| Escherichia coli 507E | 4.8 mM | [102] | |
| Klebsiella oxytosa 1082E | 1.2 mM | [102] | |
| Klebsiella aerogenes 1522E | 2.4 mM | [102] | |
| Pseudomonas aeruginosa 9027 | 1.2 mM | [102] | |
| Staphylococcus aureus | 3.2 mM | [102] | |
| Staphylococcus aureus SG511 | 600 µM | [102] | |
| Streptococcus pyogenes A308 | 1.2 mM | [102] | |
| Campneoside I (32) | Enterobacter cloacae P99 | >917 µM | [102] |
| Escherichia coli 507E | >917 µM | [102] | |
| Klebsiella oxytosa 1082E | >917 µM | [102] | |
| Klebsiella aerogenes 1522E | >917 µM | [102] | |
| Pseudomonas aeruginosa 9027 | >917 µM | [102] | |
| Staphylococcus aureus | 200 µM | [102] | |
| Staphylococcus aureus SG511 | 229 µM | [102] | |
| Streptococcus pyogenes A308 | 229 µM | [102] | |
| Campneoside II (33) | Staphylococcus aureus | 2.0 mM | [102] |
| Caffeic acid ester (34) | Phomopsis longicolla | 19 µM | [103] |
| Rosmarinic acid (35) | Aspergillus niger ATCC 16404 | 2.8 mM | [106] |
| Bacillus cereus ATCC 10987 | 1.8 mM | [105] | |
| Bacillus subtilis ATCC 11060 | 1.8 mM | [105] | |
| Candida albicans ATCC 14053 | 694 µM | [106] | |
| Corynebacterium T25-17 | 6.9 mM | [104] | |
| Enterococcus faecalis C159-6 | 833 µM | [104] | |
| Escherichia coli ATCC 8739 | 1.4 mM | [106] | |
| Listeria monocytogenes ATCC 19115 | 1.8 mM | [105] | |
| Mycobacterium smegmatis 5003 | 3.3 mM | [104] | |
| Pseudomonas aeruginosa ATCC 27583 | 6.9 mM | [104] | |
| Pseudomonas aeruginosa ATCC 27853 | 1.8 mM | [105] | |
| Staphylococcus aureus ATCC 29213 | 888 µM | [105] | |
| Staphylococcus aureus ATCC 29737 | 1.4 mM | [106] | |
| Staphylococcus epidermis 5001 | 833 µM | [104] | |
| Staphylococcus lugdunensis T26A3 | 1.6 mM | [104] | |
| Staphylococcus warneri T12A12 | 3.3 mM | [104] | |
| Stenotrophomonas maltophilia | 833 µM | [104] | |
| Methyl rosmarinate (36) | Bacillus cereus ATCC 10987 | >3.4 mM | [105] |
| Bacillus subtilis ATCC 11060 | >3.4 mM | [105] | |
| Corynebacterium T25-17 | 3.2 mM | [104] | |
| Enterococcus faecalis C159-6 | 801 µM | [104] | |
| Listeria monocytogenes ATCC 19115 | >3.4 mM | [105] | |
| Mycobacterium smegmatis 5003 | 1.6 mM | [104] | |
| Pseudomonas aeruginosa ATCC 27583 | 3.2 mM | [104] | |
| Pseudomonas aeruginosa ATCC 27853 | >3.4 mM | [105] | |
| Staphylococcus aureus ATCC 29213 | >3.4 mM | [105] | |
| Staphylococcus epidermis 5001 | 801 µM | [104] | |
| Staphylococcus lugdunensis T26A3 | 1.6 mM | [104] | |
| Staphylococcus warneri T12A12 | 801 µM | [104] | |
| Stenotrophomonas maltophilia | 801 µM | [104] | |
| Caffeic acid phenethyl ester (37) | Escherichia coli ATCC 25922 | >800 µM | [108] |
| Staphylococcus aureus ATCC 6538P | 100 µM | [108] | |
| Enterococcus faecalis ATCC 29212 | 400 µM | [108] | |
| Listeria monocytogenes ATCC 7644 | 400 µM | [108] | |
| Methyl cinnamate (40) | Aspergillus flavus UBA 294 | >1.5 mM | [65] |
| Aspergillus niger | 61 µM | [44] | |
| Aspergillus niger ATCC 11394 | >1.5 mM | [65] | |
| Aspergillus terreus INM 031783 | >1.5 mM | [65] | |
| Bacillus subtilis | 301 µM | [44] | |
| Candida albicans | 50 µM | [44] | |
| Escherichia coli | 164 µM | [44] | |
| Staphylococcus aureus | 252 µM | [44] | |
| Ethyl cinnamate (41) | Aspergillus niger | 61 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 50 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 252 µM | [44] | |
| Propyl cinnamate (42) | Aspergillus niger | 43 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 59 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 301 µM | [44] | |
| Isopropyl cinnamate (43) | Aspergillus niger | 43 µM | [44] |
| Bacillus subtilis | 164 µM | [44] | |
| Candida albicans | 43 µM | [44] | |
| Escherichia coli | 139 µM | [44] | |
| Staphylococcus aureus | 139 µM | [44] | |
| Butyl cinnamate (44) | Aspergillus niger | 36 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 61 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 203 µM | [44] | |
| Isobutyl cinnamate (45) | Aspergillus niger | 12 µM | [44] |
| Bacillus subtilis | 43 µM | [44] | |
| Candida albicans | 14 µM | [44] | |
| Escherichia coli | 43 µM | [44] | |
| Staphylococcus aureus | 50 µM | [44] | |
| Octyl cinnamate (46) | Aspergillus niger | 43 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 43 µM | [44] | |
| Escherichia coli | 164 µM | [44] | |
| Staphylococcus aureus | 203 µM | [44] | |
| Phenyl cinnamate (47) | Aspergillus niger | 61 µM | [44] |
| Bacillus subtilis | 164 µM | [44] | |
| Candida albicans | 43 µM | [44] | |
| Escherichia coli | 252 µM | [44] | |
| Staphylococcus aureus | 203 µM | [44] | |
| Benzyl cinnamate (48) | Aspergillus niger | 50 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 43 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 252 µM | [44] | |
| 8-Hydroxyquinolyl cinnamate (49) | Aspergillus niger | 50 µM | [44] |
| Bacillus subtilis | 252 µM | [44] | |
| Candida albicans | 61 µM | [44] | |
| Escherichia coli | 164 µM | [44] | |
| Staphylococcus aureus | 164 µM | [44] | |
| Methyl 4-coumarate (50) | Aspergillus flavus UBA 294 | 702 µM | [65] |
| Aspergillus niger ATCC 11394 | 702 µM | [65] | |
| Aspergillus niger MTCC 8189 | 247 µM | [111] | |
| Aspergillus terreus INM 031783 | 702 µM | [65] | |
| Bacillus subtilis MTCC 2063 | 247 µM | [111] | |
| Candida albicans MTCC 227 | 247 µM | [111] | |
| Escherichia coli MTCC 1652 | 247 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 247 µM | [111] | |
| Ethyl 4-coumarate (51) | Aspergillus niger MTCC 8189 | 176 µM | [111] |
| Bacillus subtilis MTCC 2063 | 176 µM | [111] | |
| Candida albicans MTCC 227 | 176 µM | [111] | |
| Escherichia coli MTCC 1652 | 176 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 176 µM | [111] | |
| Propyl 4-coumarate (52) | Aspergillus niger MTCC 8189 | 2.3 mM | [111] |
| Bacillus subtilis MTCC 2063 | 137 µM | [111] | |
| Candida albicans MTCC 227 | 137 µM | [111] | |
| Escherichia coli MTCC 1652 | 137 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 137 µM | [111] | |
| Isopropyl 4-coumarate (53) | Aspergillus niger MTCC 8189 | 2.3 mM | [111] |
| Bacillus subtilis MTCC 2063 | 137 µM | [111] | |
| Candida albicans MTCC 227 | 137 µM | [111] | |
| Escherichia coli MTCC 1652 | 2.3 mM | [111] | |
| Staphylococcus aureus MTCC 2901 | 137 µM | [111] | |
| Butyl 4-coumarate (54) | Aspergillus niger MTCC 8189 | 1.9 mM | [111] |
| Bacillus subtilis MTCC 2063 | 107 µM | [111] | |
| Candida albicans MTCC 227 | 107 µM | [111] | |
| Escherichia coli MTCC 1652 | 1.9 mM | [111] | |
| Staphylococcus aureus MTCC 2901 | 107 µM | [111] | |
| Isopentyl 4-coumarate (55) | Aspergillus niger MTCC 8189 | 92 µM | [111] |
| Bacillus subtilis MTCC 2063 | 92 µM | [111] | |
| Candida albicans MTCC 227 | 92 µM | [111] | |
| Escherichia coli MTCC 1652 | 1.4 mM | [111] | |
| Staphylococcus aureus MTCC 2901 | 92 µM | [111] | |
| Cyclohexyl 4-coumarate (56) | Aspergillus niger MTCC 8189 | 78 µM | [111] |
| Bacillus subtilis MTCC 2063 | 9.1 µM | [111] | |
| Candida albicans MTCC 227 | 1.1 mM | [111] | |
| Escherichia coli MTCC 1652 | 41 mM | [111] | |
| Staphylococcus aureus MTCC 2901 | 78 µM | [111] | |
| Menthyl 4-coumarate (57) | Aspergillus niger MTCC 8189 | 1.9 mM | [111] |
| Bacillus subtilis MTCC 2063 | 107 µM | [111] | |
| Candida albicans MTCC 227 | 107 µM | [111] | |
| Escherichia coli MTCC 1652 | 1.9 mM | [111] | |
| Staphylococcus aureus MTCC 2901 | 107 µM | [111] | |
| Phenyl 4-coumarate (58) | Aspergillus niger MTCC 8189 | 1.2 mM | [111] |
| Bacillus subtilis MTCC 2063 | 85 µM | [111] | |
| Candida albicans MTCC 227 | 10 µM | [111] | |
| Escherichia coli MTCC 1652 | 47 mM | [111] | |
| Staphylococcus aureus MTCC 2901 | 85 µM | [111] | |
| 4-aminophenyl 4-coumarate (59) | Aspergillus niger MTCC 8189 | 67 µM | [111] |
| Bacillus subtilis MTCC 2063 | 8.5 µM | [111] | |
| Candida albicans MTCC 227 | 67 µM | [111] | |
| Escherichia coli MTCC 1652 | 905 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 67 µM | [111] | |
| 4-nitrophenyl 4-coumarate (60) | Aspergillus niger MTCC 8189 | 558 µM | [111] |
| Bacillus subtilis MTCC 2063 | 46 µM | [111] | |
| Candida albicans MTCC 227 | 46 µM | [111] | |
| Escherichia coli MTCC 1652 | 46 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 46 µM | [111] | |
| Benzyl 4-coumarate (61) | Aspergillus niger MTCC 8189 | 905 µM | [111] |
| Bacillus subtilis MTCC 2063 | 67 µM | [111] | |
| Candida albicans MTCC 227 | 67 µM | [111] | |
| Escherichia coli MTCC 1652 | 905 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 67 µM | [111] | |
| Methyl caffeate (62) | Aspergillus flavus UBA 294 | >1.3 mM | [65] |
| Aspergillus niger ATCC 11394 | >1.3 mM | [65] | |
| Aspergillus terreus INM 031783 | >1.3 mM | [65] | |
| Candida albicans ATCC 10231 | MIC50 = 659 µM | [78] | |
| Ethyl caffeate (63) | Candida albicans ATCC 10231 | MIC50 = 615 µM | [78] |
| Propyl caffeate (64) | Candida albicans ATCC 10231 | MIC50 = 576 µM | [78] |
| Isopropyl caffeate (65) | Candida albicans ATCC 10231 | MIC50 = 576 µM | [78] |
| 3-fluoro-4-methoxyphenyl caffeate (66) | Candida albicans ATCC 10231 | MIC50 = 421 µM | [78] |
| (5-nitrofuran-2-yl)methyl caffeate (67) | Candida albicans ATCC 10231 | MIC50 = 52 µM | [78] |
| Methyl ferulate (68) | Aspergillus flavus UBA 294 | >1.2 mM | [65] |
| Aspergillus niger | 4.0 mM | [80] | |
| Aspergillus niger ATCC 11394 | >1.2 mM | [65] | |
| Aspergillus terreus INM 031783 | >1.2 mM | [65] | |
| Bacillus subtilis | 6.0 mM | [80] | |
| Candida albicans | 4.0 mM | [80] | |
| Saccharomyces cerevisiae | 4.0 mM | [80] | |
| Staphylococcus aureus | 6.0 mM | [80] | |
| Ethyl ferulate (69) | Aspergillus niger | 4.0 mM | [80] |
| Bacillus subtilis | 2.0 mM | [80] | |
| Candida albicans | 4.0 mM | [80] | |
| Saccharomyces cerevisiae | 4.0 mM | [80] | |
| Staphylococcus aureus | 4.0 mM | [80] | |
| Butyl ferulate (70) | Aspergillus niger | >10 mM | [80] |
| Bacillus subtilis | 500 µM | [80] | |
| Candida albicans | 10 mM | [80] | |
| Saccharomyces cerevisiae | 500 µM | [80] | |
| Staphylococcus aureus | 500 µM | [80] | |
| Hexyl ferulate (71) | Aspergillus niger | >10 mM | [80] |
| Bacillus subtilis | 63 µM | [80] | |
| Candida albicans | >10 mM | [80] | |
| Saccharomyces cerevisiae | >10 mM | [80] | |
| Staphylococcus aureus | 125 µM | [80] | |
| 2-methyl-1-butyl ferulate (72) | Aspergillus niger | >10 mM | [80] |
| Bacillus subtilis | 125 µM | [80] | |
| Candida albicans | >10 mM | [80] | |
| Saccharomyces cerevisiae | 250 µM | [80] | |
| Staphylococcus aureus | 125 µM | [80] | |
| 4-t-butylphenyl ferulate (73) | Candida albicans ATCC 10231 | 391 µM | [81] |
| Candida krusei ATCC 6258 | 391 µM | [81] | |
| Enterococcus faecalis ATCC 29212 | 391 µM | [81] | |
| Escherichia coli ATCC 25922 | 782 µM | [81] | |
| Klebsiella pneumoniae RSKK 574 | 782 µM | [81] | |
| Staphylococcus aureus ATCC 29213 | 782 µM | [81] | |
| 4-isopropylphenyl ferulate (74) | Candida albicans ATCC 10231 | 204 µM | [81] |
| Candida krusei ATCC 6258 | 204 µM | [81] | |
| Enterococcus faecalis ATCC 29212 | 51 µM | [81] | |
| Escherichia coli ATCC 25922 | 818 µM | [81] | |
| Klebsiella pneumoniae RSKK 574 | 409 µM | [81] | |
| Staphylococcus aureus ATCC 29213 | 409 µM | [81] | |
| 4-chloro-3-methylphenyl ferulate (75) | Candida albicans ATCC 10231 | 201 µM | [81] |
| Candida krusei ATCC 6258 | 201 µM | [81] | |
| Enterococcus faecalis ATCC 29212 | 50 µM | [81] | |
| Escherichia coli ATCC 25922 | 812 µM | [81] | |
| Klebsiella pneumoniae RSKK 574 | 401 µM | [81] | |
| Staphylococcus aureus ATCC 29213 | <25 µM | [81] | |
| 4-methoxyphenyl ferulate (76) | Candida albicans ATCC 10231 | 425 µM | [81] |
| Candida krusei ATCC 6258 | 425 µM | [81] | |
| Enterococcus faecalis ATCC 29212 | 850 µM | [81] | |
| Escherichia coli ATCC 25922 | 850 µM | [81] | |
| Klebsiella pneumoniae RSKK 574 | 850 µM | [81] | |
| Staphylococcus aureus ATCC 29213 | 850 µM | [81] | |
| 4-acetamidophenyl ferulate (77) | Candida albicans ATCC 10231 | 390 µM | [81] |
| Candida krusei ATCC 6258 | 390 µM | [81] | |
| Enterococcus faecalis ATCC 29212 | 390 µM | [81] | |
| Escherichia coli ATCC 25922 | 780 µM | [81] | |
| Klebsiella pneumoniae RSKK 574 | 780 µM | [81] | |
| Staphylococcus aureus ATCC 29213 | 780 µM | [81] |
4-O-Caffeoylquinic acid (26) (Figure 4 and Table 2) was found to be slightly less active than the 5-O-isomer with MIC values of 423 µM against both S. aureus and E. coli, 564 µM against both fungi C. albicans and A. niger, and 705 µM against S. enterica and S. cerevisiae [98]. In contrast with 5-O-caffeoylquinic acid, 3-O-caffeoylquinic acid (27) displayed more pronounced growth inhibitory activity against bacteria than fungi (Table 2), achieving MIC values of 282 µM against E. coli and S. aureus [98]. The literature reports of the antimicrobial activity of the 3-O- and 4-O-isomers are scarce, in contrast with the 5-O-isomer. The fact that the esters 25, 26 and 27 have a different antimicrobial profile suggests that they have antimicrobial activity on their own, and the hydrolysis products caffeic and quinic acids, which could be released in the same amount, are not responsible for the antimicrobial activity of the three esters.
Among the chlorogenic acids formed by two caffeic acids residues, 1,3-di-O-caffeoylquinic acid (28), 3,5-di-O-caffeoylquinic acid (29) and 4,5-di-O-caffeoylquinic acid (30), the presence of caffeic residue in the position 3 of quinic acid increased the antifungal spectrum of activity, with 28 achieving MIC values below 200 µM against almost all fungi tested (Table 2), whereas the esterification in the position 5 of quinic acid increased the antibacterial potency of the acid, displaying MIC values of 194 µM against E. coli and S. aureus for 30 and even inhibiting completely the growth of Micrococcus luteus at 97 µM [96]. However discrepant MIC have also been reported. A report from 2011, published MIC values higher than 248 µM against S. aureus, B. cereus and higher than 496 µM against E. coli and C. albicans for 28, 29 and 30 [99]. Moreover the study found an impressive synergism (FICI < 0.002) between 30 and several fluoroquinolone antibiotics against S. aureus. Four caffeic acid glycosides isolated from Paulownia tomentosa displayed antibacterial activity [102]. Campneoside I (32) demonstrated the highest activity against Gram-positive bacteria achieving MIC values around 200 µM against different species of S. aureus, while both acteoside (31) and campneoside II (33) showed less antistaphylococcal activity [102].
The caffeic acid ester 34 (Figure 4) isolated from Zuccagnia punctata, showed antifungal activity against the phytopathogenic fungi Phomopsis longicolla with an MIC value of only 19 µM, being less active against other fungi [103]. Another caffeic acid ester known as rosmarinic acid (35), and its methyl ester 36 showed growth inhibition of several species of bacteria with MIC values ranging between 801 µM and 6.94 mM [104]. In the study, the methyl ester 36 was slightly more active than the free acid 35. However both rosmarinic acid (35) and its methyl ester 36 isolated from Rabdosia serra, were reported to have significantly higher MIC values against Gram-positive and Gram-negative bacteria [105], and surprisingly the methyl ester was found to be completely inactive with MIC values higher than 3.4 mM, whereas rosmarinic acid showed some activity [105] (Table 2). Moreover the study of Gohari et al., showed that Candida albicans was more susceptible to rosmarinic acid with an MIC value of 694 µM, in comparison with S. aureus, E. coli and Aspergillus niger [106]. Caffeic acid phenethyl ester (37) is a biologically active component of propolis showing interesting anticancer activity [107]. This ester demonstrated antibacterial activity against S. aureus, E. faecalis and L. monocytogenes with MIC values ranging between 100 and 400 µM, but did not show growth inhibition against P. aeruginosa and E. coli up to a concentration of 800 µM [108]. Trilepisiumic acid (38) is an ester of caffeic acid and protocatechuic acid, isolated from Trilepisium madagascariense, that shows antifungal activity against Candida albicans ATCC 9002 (MIC = 202 µM) while being less active against other fungi and bacteria [109]. Two cinnamic esters were isolated from the root of Ehretia longiflora, and identified as ehretiolide (39) and arachidyl ferulate [110]. Ehretiolide (39) showed growth inhibitory activity against M. tuberculosis H37Rv (MIC = 41 µM), which is impressive for an unmodified natural product.
All the reported esters of cinnamic acid 40–49, showed potent antifungal activity against A. niger and C. albicans, particularly isobutyl cinnamate (45) achieving MIC values of 12 and 14 µM respectively [44]. The MIC values of 40–49 against A. niger and C. albicans ranged between 12 and 61 µM, and between 14 and 61 µM respectively (Table 2). The MIC values of 40–49 were higher against Gram-positive and Gram-negative bacteria, ranging between 43 and 301 µM [44]. Isobutyl cinnamate (45), also showed the highest growth inhibitory activity against the bacterial species with MIC values between 43 and 50 µM. The esters of 4-coumaric acid 50–61 were comparatively less active compared to the esters of cinnamic acid, achieving MIC values between 67 µM and 2.3 mM against A. niger, and between 10 µM and 1.1 mM against C. albicans [111] (Table 2). The most active ester of 4-coumaric acid was 4-nitrophenyl 4-coumarate (60) showing MIC values of 46 µM against B. subtilis, C. albicans, E. coli and S. aureus [111]. The ester ethyl 4-methoxycinnamate, isolated from the plant Kaempferia galanga, showed anti-TB activity (MIC = 485 µM) against the virulent H37Rv and drug-resistant strains [112]. The esters of caffeic acid 62–67 were not assayed against different microorganisms, but only against some fungal species. Methyl caffeate (62) showed little antimicrobial activity against Aspergillus species with MIC values higher than 1.3 mM [65], however it was more active against C. albicans (MIC50 = 659 µM) [77,78].
The esters of caffeic acid showed moderate activity against C. albicans, and the most active was (5-nitrofuran-2-yl)methyl caffeate (67) with an MIC50 value of 52 µM against C. albicans [77,78]. The esters of ferulic acid 68–77 have been evaluated against different bacteria and fungi, and comparatively they have lower growth inhibitory activity compared to cinnamate and 4-coumarate esters. The MIC values of the ferulate esters 68–77 ranged between 201 µM and >10 mM against C. albicans, and between <25 µM and 6.0 mM against S. aureus [80,81]. The most active ferulate ester was identified to be 4-chloro-3-methylphenyl ferulate (75) achieving MIC values of 201 µM against C. albicans and C. krusei, of 50 µM against E. faecalis, 812 µM against E. coli, 401 µM against K. pneumoniae, and <25 µM against S. aureus [81].
3.2. Amides
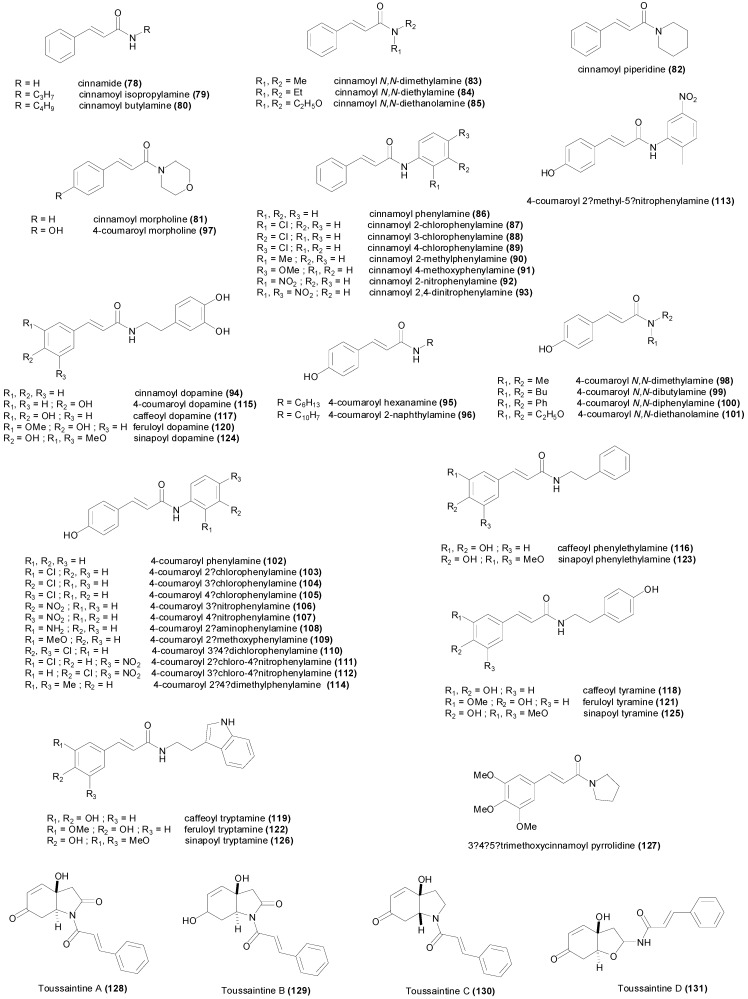
The simplest cinnamic amide is cinnamide (78) (Figure 5 and Table 3) which displayed growth inhibition against both fungi A. niger and C. albicans at a concentration of 60.8 µM, while being less active against bacteria, showing an MIC value of 252 µM against B. subtilis, E. coli and S. aureus [44]. All the cinnamoyl amides 79–94 were found to have a more potent effect against fungi compared to bacteria, except cinnamoyl dopamine (94) which showed little antimicrobial activity (MIC = 1.76 mM) [77]. Among the screened cinnamoyl amides, the compound with the highest antifungal activity was identified to be cinnamoyl N,N-diethylamide (84) achieving MIC values of 14.3 and 36.3 µM against A. niger and C. albicans respectively [44]. The most potent antibacterial cinnamoyl amide was cinnamoyl 2-methylphenylamine (90) with MIC values of 114, 139 and 139 µM against B. subtilis, E. coli and S. aureus respectively.
Figure 5.
Chemical structures of the cinnamic amides 78–131 with antimicrobial activity
Table 3.
Minimum inhibitory concentration values of cinnamic amides 78–131.
| Compound | Microorganism Strain | MIC | Refs. |
|---|---|---|---|
| Cinnamide (78) | Aspergillus niger | 60.8 µM | [44] |
| Bacillus subtilis | 252 µM | [44] | |
| Candida albicans | 60.8 µM | [44] | |
| Escherichia coli | 252 µM | [44] | |
| Staphylococcus aureus | 252 µM | [44] | |
| Cinnamoyl isopropylamine (79) | Aspergillus niger | 60.8 µM | [44] |
| Bacillus subtilis | 164 µM | [44] | |
| Candida albicans | 50.4 µM | [44] | |
| Escherichia coli | 164 µM | [44] | |
| Staphylococcus aureus | 252 µM | [44] | |
| Cinnamoyl butylamine (80) | Aspergillus niger | 43.5 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 43.5 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 164 µM | [44] | |
| Cinnamoyl morpholine (81) | Aspergillus niger | 60.8 µM | [44] |
| Bacillus subtilis | 164 µM | [44] | |
| Candida albicans | 60.8 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 203 µM | [44] | |
| Cinnamoyl piperidine (82) | Aspergillus niger | 43.5 µM | [44] |
| Bacillus subtilis | 164 µM | [44] | |
| Candida albicans | 50.4 µM | [44] | |
| Escherichia coli | 164 µM | [44] | |
| Staphylococcus aureus | 139 µM | [44] | |
| Cinnamoyl N,N-dimethylamine (83) | Aspergillus niger | 50.4 µM | [44] |
| Bacillus subtilis | 139 µM | [44] | |
| Candida albicans | 43.5 µM | [44] | |
| Escherichia coli | 139 µM | [44] | |
| Staphylococcus aureus | 139 µM | [44] | |
| Cinnamoyl N,N-diethylamine (84) | Aspergillus niger | 14.3 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 36.3 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 252 µM | [44] | |
| Cinnamoyl N,N-diethanolamine (85) | Aspergillus niger | 60.8 µM | [44] |
| Bacillus subtilis | 301 µM | [44] | |
| Candida albicans | 86 µM | [44] | |
| Escherichia coli | 301 µM | [44] | |
| Staphylococcus aureus | 301 µM | [44] | |
| Cinnamoyl phenylamine (86) | Aspergillus niger | 73.6 µM | [44] |
| Bacillus subtilis | 301 µM | [44] | |
| Candida albicans | 86 µM | [44] | |
| Escherichia coli | 301 µM | [44] | |
| Staphylococcus aureus | 203 µM | [44] | |
| Cinnamoyl 2-chlorophenylamine (87) | Aspergillus niger | 50.4 µM | [44] |
| Bacillus subtilis | 301 µM | [44] | |
| Candida albicans | 60.8 µM | [44] | |
| Escherichia coli | 301 µM | [44] | |
| Staphylococcus aureus | 301 µM | [44] | |
| Cinnamoyl 3-chlorophenylamine (88) | Aspergillus niger | 43.5 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 60.8 µM | [44] | |
| Escherichia coli | 164 µM | [44] | |
| Staphylococcus aureus | 203 µM | [44] | |
| Cinnamoyl 4-chlorophenylamine (89) | Aspergillus niger | 50.4 µM | [44] |
| Bacillus subtilis | 301 µM | [44] | |
| Candida albicans | 60.8 µM | [44] | |
| Escherichia coli | 301 µM | [44] | |
| Staphylococcus aureus | 301 µM | [44] | |
| Cinnamoyl 2-methylphenylamine (90) | Aspergillus niger | 73.6 µM | [44] |
| Bacillus subtilis | 114 µM | [44] | |
| Candida albicans | 86 µM | [44] | |
| Escherichia coli | 139 µM | [44] | |
| Staphylococcus aureus | 139 µM | [44] | |
| Cinnamoyl 4-methoxyphenylamine (91) | Aspergillus niger | 50.4 µM | [44] |
| Bacillus subtilis | 203 µM | [44] | |
| Candida albicans | 50.4 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 164 µM | [44] | |
| Cinnamoyl 2-nitrophenylamine (92) | Aspergillus niger | 86 µM | [44] |
| Bacillus subtilis | 164 µM | [44] | |
| Candida albicans | 73.6 µM | [44] | |
| Escherichia coli | 203 µM | [44] | |
| Staphylococcus aureus | 139 µM | [44] | |
| Cinnamoyl 2,4-dinitrophenylamine (93) | Aspergillus niger | 50.4 µM | [44] |
| Bacillus subtilis | 164 µM | [44] | |
| Candida albicans | 86 µM | [44] | |
| Escherichia coli | 164 µM | [44] | |
| Staphylococcus aureus | 164 µM | [44] | |
| Cinnamoyl dopamine (94) | Bacillus subtilis 1A95 | 1.76 mM | [77] |
| Candida albicans 62 | 1.76 mM | [77] | |
| Listeria monocytogenes C12 | 1.76 mM | [77] | |
| Staphylococcus aureus 209 | 1.76 mM | [77] | |
| Streptococcus pyogenes 10535 | 441 µM | [77] | |
| 4-coumaroyl hexanamine (95) | Aspergillus niger MTCC 8189 | 72.5 µM | [111] |
| Bacillus subtilis MTCC 2063 | 9.1 µM | [111] | |
| Candida albicans MTCC 227 | 72.5 µM | [111] | |
| Escherichia coli MTCC 1652 | 72.5 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 72.5 µM | [111] | |
| 4-coumaroyl 2-naphtylamine (96) | Aspergillus niger MTCC 8189 | 558 µM | [111] |
| Bacillus subtilis MTCC 2063 | 46.2 µM | [111] | |
| Candida albicans MTCC 227 | 15.5 mM | [111] | |
| Escherichia coli MTCC 1652 | 46.2 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 5.9 µM | [111] | |
| 4-coumaroyl morpholine (97) | Aspergillus niger MTCC 8189 | 1.4 mM | [111] |
| Bacillus subtilis MTCC 2063 | 91.6 µM | [111] | |
| Candida albicans MTCC 227 | 91.6 µM | [111] | |
| Escherichia coli MTCC 1652 | 91.6 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 1.4 mM | [111] | |
| 4-coumaroyl N,N-dimethylamine (98) | Aspergillus niger MTCC 8189 | 191 µM | [111] |
| Bacillus subtilis MTCC 2063 | 191 µM | [111] | |
| Candida albicans MTCC 227 | 191 µM | [111] | |
| Escherichia coli MTCC 1652 | 3.6 mM | [111] | |
| Staphylococcus aureus MTCC 2901 | 3.6 mM | [111] | |
| 4-coumaroyl N,N-dibutylamine (99) | Aspergillus niger MTCC 8189 | 676 µM | [111] |
| Bacillus subtilis MTCC 2063 | 53.6 µM | [111] | |
| Candida albicans MTCC 227 | 53.6 µM | [111] | |
| Escherichia coli MTCC 1652 | 676 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 53.6 µM | [111] | |
| 4-coumaroyl N,N-diphenylamine (100) | Aspergillus niger MTCC 8189 | 386 µM | [111] |
| Bacillus subtilis MTCC 2063 | 5.0 µM | [111] | |
| Candida albicans MTCC 227 | 34.6 µM | [111] | |
| Escherichia coli MTCC 1652 | 5.0 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 34.6 µM | [111] | |
| 4-coumaroyl N,N-diethanolamine (101) | Aspergillus niger MTCC 8189 | 72.5 µM | [111] |
| Bacillus subtilis MTCC 2063 | 9.1 µM | [111] | |
| Candida albicans MTCC 227 | 72.5 µM | [111] | |
| Escherichia coli MTCC 1652 | 72.5 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 72.5 µM | [111] | |
| 4-coumaroyl phenylamine (102) | Aspergillus niger MTCC 8189 | 1.2 mM | [111] |
| Bacillus subtilis MTCC 2063 | 84.7 µM | [111] | |
| Candida albicans MTCC 227 | 84.7 µM | [111] | |
| Escherichia coli MTCC 1652 | 84.7 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 10.3 µM | [111] | |
| 4-coumaroyl 2'-nitrophenylamine (103) | Aspergillus niger MTCC 8189 | 46.2 µM | [111] |
| Bacillus subtilis MTCC 2063 | 46.2 µM | [111] | |
| Candida albicans MTCC 227 | 6.3 µM | [111] | |
| Escherichia coli MTCC 1652 | 46.2 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 46.2 µM | [111] | |
| 4-coumaroyl 3'-chlorophenylamine (104) | Aspergillus niger MTCC 8189 | 676 µM | [111] |
| Bacillus subtilis MTCC 2063 | 7.1 µM | [111] | |
| Candida albicans MTCC 227 | 53.6 µM | [111] | |
| Escherichia coli MTCC 1652 | 53.6 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 7.1 µM | [111] | |
| 4-coumaroyl 4'-chlorophenylamine (105) | Aspergillus niger MTCC 8189 | 676 µM | [111] |
| Bacillus subtilis MTCC 2063 | 7.1 µM | [111] | |
| Candida albicans MTCC 227 | 53.6 µM | [111] | |
| Escherichia coli MTCC 1652 | 53.6 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 53.6 µM | [111] | |
| 4-coumaroyl 3'-nitrophenylamine (106) | Aspergillus niger MTCC 8189 | 46.2 µM | [111] |
| Bacillus subtilis MTCC 2063 | 46.2 µM | [111] | |
| Candida albicans MTCC 227 | 46.2 µM | [111] | |
| Escherichia coli MTCC 1652 | 558 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 46.2 µM | [111] | |
| 4-coumaroyl 4'-nitrophenylamine (107) | Aspergillus niger MTCC 8189 | 18 mM | [111] |
| Bacillus subtilis MTCC 2063 | 46.2 µM | [111] | |
| Candida albicans MTCC 227 | 46.2 µM | [111] | |
| Escherichia coli MTCC 1652 | 46.2 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 46.2 µM | [111] | |
| 4-coumaroyl 2'-aminophenylamine (108) | Aspergillus niger MTCC 8189 | 905 µM | [111] |
| Bacillus subtilis MTCC 2063 | 67.2 µM | [111] | |
| Candida albicans MTCC 227 | 67.2 µM | [111] | |
| Escherichia coli MTCC 1652 | 67.2 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 67.2 µM | [111] | |
| 4-coumaroyl 2'-methoxyphenylamine (109) | Aspergillus niger MTCC 8189 | 744 µM | [111] |
| Bacillus subtilis MTCC 2063 | 7.5 µM | [111] | |
| Candida albicans MTCC 227 | 57.7 µM | [111] | |
| Escherichia coli MTCC 1652 | 57.7 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 57.7 µM | [111] | |
| 4-coumaroyl 3',4'-dichlorophenylamine (110) | Aspergillus niger MTCC 8189 | 422 µM | [111] |
| Bacillus subtilis MTCC 2063 | 37.1 µM | [111] | |
| Candida albicans MTCC 227 | 37.1 µM | [111] | |
| Escherichia coli MTCC 1652 | 37.1 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 37.1 µM | [111] | |
| 4-coumaroyl 2'-chloro-4'-nitrophenylamine (111) | Aspergillus niger MTCC 8189 | 352 µM | [111] |
| Bacillus subtilis MTCC 2063 | 929 nM | [111] | |
| Candida albicans MTCC 227 | 32.2 µM | [111] | |
| Escherichia coli MTCC 1652 | 32.2 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 352 µM | [111] | |
| 4-coumaroyl 3'-chloro-4'-nitrophenylamine (112) | Aspergillus niger MTCC 8189 | 32.2 µM | [111] |
| Bacillus subtilis MTCC 2063 | 4.7 µM | [111] | |
| Candida albicans MTCC 227 | 32.2 µM | [111] | |
| Escherichia coli MTCC 1652 | 4.7 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 32.2 µM | [111] | |
| 4-coumaroyl 2'-methyl-5'-nitrophenylamine (113) | Aspergillus niger MTCC 8189 | 39.9 µM | [111] |
| Bacillus subtilis MTCC 2063 | 39.9 µM | [111] | |
| Candida albicans MTCC 227 | 39.9 µM | [111] | |
| Escherichia coli MTCC 1652 | 463 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 39.9 µM | [111] | |
| 4-coumaroyl 2',4'-dimethylphenylamine (114) | Aspergillus niger MTCC 8189 | 744 µM | [111] |
| Bacillus subtilis MTCC 2063 | 7.55 µM | [111] | |
| Candida albicans MTCC 227 | 57.7 µM | [111] | |
| Escherichia coli MTCC 1652 | 744 µM | [111] | |
| Staphylococcus aureus MTCC 2901 | 57.7 µM | [111] | |
| 4-coumaroyl dopamine (115) | Bacillus subtilis 1A95 | 1.67 mM | [77] |
| Candida albicans 62 | 1.67 mM | [77] | |
| Listeria monocytogenes C12 | 1.67 mM | [77] | |
| Staphylococcus aureus 209 | 418 µM | [77] | |
| Streptococcus pyogenes 10535 | 418 µM | [77] | |
| Caffeoyl phenylethylamine (116) | Bacillus subtilis 1A95 | 1.76 mM | [77] |
| Candida albicans 62 | 882 µM | [77] | |
| Listeria monocytogenes C12 | 441 µM | [77] | |
| Staphylococcus aureus 209 | 882 µM | [77] | |
| Streptococcus pyogenes 10535 | 882 µM | [77] | |
| Caffeoyl dopamine (117) | Bacillus subtilis 1A95 | 793 µM | [77] |
| Candida albicans 62 | 396 µM | [77] | |
| Listeria monocytogenes C12 | 793 µM | [77] | |
| Staphylococcus aureus 209 | 793 µM | [77] | |
| Streptococcus pyogenes 10535 | 793 µM | [77] | |
| Caffeoyl tyramine (118) | Bacillus subtilis 1A95 | 836 µM | [77] |
| Candida albicans 62 | 418 µM | [77] | |
| Listeria monocytogenes C12 | 836 µM | [77] | |
| Staphylococcus aureus 209 | 836 µM | [77] | |
| Streptococcus pyogenes 10535 | 836 µM | [77] | |
| Caffeoyl tryptamine (119) | Bacillus subtilis 1A95 | 1.55 mM | [77] |
| Candida albicans 62 | 1.55 mM | [77] | |
| Listeria monocytogenes C12 | 1.55 mM | [77] | |
| Staphylococcus aureus 209 | 388 µM | [77] | |
| Streptococcus pyogenes 10535 | 194 µM | [77] | |
| Feruloyl dopamine (120) | Bacillus subtilis 1A95 | 759 µM | [77] |
| Candida albicans 62 | 1.52 mM | [77] | |
| Listeria monocytogenes C12 | 1.52 mM | [77] | |
| Staphylococcus aureus 209 | 190 µM | [77] | |
| Streptococcus pyogenes 10535 | 380 µM | [77] | |
| Feruloyl tyramine (121) | Bacillus subtilis 1A95 | 798 µM | [77] |
| Candida albicans 62 | 1.59 mM | [77] | |
| Listeria monocytogenes C12 | 1.59 mM | [77] | |
| Staphylococcus aureus 209 | 199 µM | [77] | |
| Streptococcus pyogenes 10535 | 399 µM | [77] | |
| Feruloyl tryptamine (122) | Bacillus subtilis 1A95 | 743 µM | [77] |
| Candida albicans 62 | 1.49 mM | [77] | |
| Listeria monocytogenes C12 | 1.49 mM | [77] | |
| Staphylococcus aureus 209 | 372 µM | [77] | |
| Streptococcus pyogenes 10535 | 372 µM | [77] | |
| Sinapoyl phenylethylamine (123) | Bacillus subtilis 1A95 | 1.53 mM | [77] |
| Candida albicans 62 | 1.53 mM | [77] | |
| Listeria monocytogenes C12 | 1.53 mM | [77] | |
| Staphylococcus aureus 209 | 382 µM | [77] | |
| Streptococcus pyogenes 10535 | 382 µM | [77] | |
| Sinapoyl dopamine (124) | Bacillus subtilis 1A95 | 1.39 mM | [77] |
| Candida albicans 62 | 1.39 mM | [77] | |
| Listeria monocytogenes C12 | 1.39 mM | [77] | |
| Staphylococcus aureus 209 | 696 µM | [77] | |
| Streptococcus pyogenes 10535 | 696 µM | [77] | |
| Sinapoyl tyramine (125) | Bacillus subtilis 1A95 | 1.46 mM | [77] |
| Candida albicans 62 | 1.46 mM | [77] | |
| Listeria monocytogenes C12 | 1.46 mM | [77] | |
| Staphylococcus aureus 209 | 182 µM | [77] | |
| Streptococcus pyogenes 10535 | 182 µM | [77] | |
| Sinapoyl tryptamine (126) | Bacillus subtilis 1A95 | 1.36 mM | [77] |
| Candida albicans 62 | 1.36 mM | [77] | |
| Listeria monocytogenes C12 | 1.36 mM | [77] | |
| Staphylococcus aureus 209 | 682 µM | [77] | |
| Streptococcus pyogenes 10535 | 171 µM | [77] | |
| 3',4',5'-trimethoxycinnamoyl pyrrolidine (127) | Mycobacterium tuberculosis H37Ra | 600 µM | [118] |
| Toussaintine A (128) | Escherichia coli DSM 1103 | 34 µM | [119] |
| Staphylococcus aureus ATCC 25923 | >136 µM | [119] | |
| Toussaintine B (129) | Escherichia coli DSM 1103 | 67 µM | [119] |
| Staphylococcus aureus ATCC 25923 | 67 µM | [119] | |
| Toussaintine C (130) | Escherichia coli DSM 1103 | 34 µM | [119] |
| Staphylococcus aureus ATCC 25923 | >136 µM | [119] | |
| Toussaintine D (131) | Escherichia coli DSM 1103 | >136 µM | [119] |
| Staphylococcus aureus ATCC 25923 | 17 µM | [119] |
The 4-coumaroyl amides 95–115 were tested for antimicrobial activity, and the MIC results suggest that they are comparatively more active inhibitors of microbial growth than the cinnamoyl amides (Table 3). The 4-coumaroyl amides do not show a higher specificity for fungi compared to bacteria as was observed for the cinnamoyl amides. The MIC values of the 4-coumaroyl amides 95–115 ranged between 929 nM and 3.6 mM against bacteria, and between 6.3 µM and 18 mM against fungal species [77,111]. The 4-coumaroyl amide displaying the lowest MIC value against bacteria, was found to be 4-coumaroyl 2'-chloro-4'-nitrophenylamine (111) achieving an MIC value of 929 nM against B. subtilis MTCC 2063 [111], while the lowest MIC value against fungi was 6.3 µM attained by 4-coumaroyl 2'-nitrophenylamine (103) against C. albicans MTCC 227 [111]. Most of the 4-coumaroyl amides displayed higher activity against B. subtilis and S. aureus bacteria compared to E. coli (Table 3).
The caffeoyl amides 116–119 (Figure 5) showed moderate antimicrobial activity (Table 3) with MIC values ranging between 194 µM and 1.76 mM [77]. The lowest MIC value (194 µM) was reported for caffeoyl tryptamine (119) against S. pyogenes 10535, however the MIC values for the same amide against other bacteria were comparatively higher, being 1.55 mM against B. subtilis 1A95, C. albicans 62 and L. monocytogenes C12 [77]. The amides caffeoyl dopamine (117) and caffeoyl tyramine (118) showed growth inhibition against Gram-positive, Gram-negative and fungal species with MIC values between 396 and 793 µM, and 418 and 836 µM respectively, whereas the other caffeoyl amides displayed MIC values higher than 1.0 mM against at least one microorganism (Table 3). The three evaluated feruloyl amides 120–122 showed a very similar pattern of antimicrobial activity [77]. The feruloyl amides showed to be growth inhibitors of S. aureus 209 with MIC values between 190 and 372 µM. They were equally active against S. pyogenes but much less against B. subtilis (MIC around 743–798 µM), and C. albicans and L. monocytogenes. Interestingly all the sinapoyl amides 123–126 showed selective growth inhibition of Gram-positive cocci bacteria (MIC between 171 and 696 µM), while being much less effective inhibitors of B. subtilis, L. monocytogenes or C. albicans (MIC between 1.36 and 1.53 mM) [77].
The Piper amides, which are one of the major phytochemicals present in the medicinally important Piper species displayed a broad variety of biological activities [113]. The carboxylic acid residues of some Piper amides are in fact substituted cinnamic acids.
Piplartine (Figure 1), originally isolated from the roots of Piper tuberculatum [18], demonstrated antibacterial activity against K. pneumoniae, P. aeruginosa and S. aureus, however no MIC values were reported [114]. Among the amides isolated from the seeds of Piper tuberculatum, piplartine showed antifungal activity against Cladosporium sphaerospermun but no MIC value was determined [115,116]. The amide 3',4',5'-trimethoxycinnamoyl pyrrolidine (127), which is related to piplartine, was isolated from Piper sarmentosum, and showed MIC values higher than 600 µM against M. tuberculosis H37Ra [117,118]. The toussaintines are amides of cinnamic acid with indolidinones or benzofuranones, obtained from the African plant Toussaintia orientalis [119]. The toussaintine B (129) inhibited both S. aureus and E. coli with a low MIC value of 67 µM, while toussaintine A (128) inhibited E. coli at 34 µM but did not inhibit S. aureus, and toussaintine D (131) inhibited S. aureus at 17 µM but did not inhibit E. coli [119]. Reports of natural cinnamic amides displaying antimicrobial activity were very scarce in the literature.
4. Natural and Synthetic Cinnamic Aldehydes and Alcohols
Cinnamaldehyde (132) (Figure 6, Table 4) which is abundantly obtained from the essential oils of cinnamon [12,120], has demonstrated a significant antimicrobial activity particularly against Gram-negative bacteria. The aldehyde was found to be very active against Helicobacter pylori and E. coli with MIC values of 15 and 7.6 µM respectively [121]. Similar MIC values of 10.6 µM and < 1.5 µM have been reported against P. aeruginosa and E. coli respectively [122]. However controversial results have appeared for this compound, as other reports published higher MIC values against E. coli [12,123,124,125]. Against the Clostridium difficile bacteria, the aldehyde showed complete growth inhibition at a concentration of 605 µM [13]. A similar MIC value was observed against Legionella pneumophila, the causative bacteria of Legionnaire’s disease [126]. Cinnamaldehyde has also found to be more active against S. aureus than Gram-negative bacteria [124], and interestingly the MIC values against a susceptible S. aureus and a methicillin-resistant S. aureus (MRSA) were exactly the same. The antibiotic clindamycin actually was found to be synergistic with cinnamaldehyde, with a FICI value of 0.3125 [124]. In addition, cinnamaldehyde showed interaction with the FtsZ cell division protein using isothermal titration calorimetry, and inhibited the formation of FtsZ assembly in a dose-dependent manner, which phenotypically translated into elongated cells [124]. A report from 2003, showed that cinnamic aldehyde prevented Bacillus cereus daughter cells separation during division, forming filamentous cells [127], a result correlating with inhibition of FtsZ. Against S. aureus clinical isolates the reported MIC values were 5.0 and 10 mM, however cinnamaldehyde was found to inhibit biofilm formation at concentration five times higher than the MIC [128]. Cinnamaldehyde (25) showed an MIC value of 358 µM against Fusarium verticillioides, the phytopathogenic fungi infecting maize that can also produce mycotoxins during the storage of grains [129]. Treatment of F. verticillioides with the aldehyde produced morphological alterations of the hyphae and the cell wall, which resulted in cytoplasmic leakage. Cinnamaldehyde showed weak antifungal activity against C. albicans and Candida tropicalis with MIC values of 3.0 and 3.8 mM respectively [130], however lower MIC values have been reported [131,132]. Against wood decay fungi, Laetiporus betulina and Laetiporus sulphureus the potency was comparable, with MIC values around 750 µM [133,134].
Figure 6.
Chemical structures of the cinnamic aldehydes, alcohols and related natural products.
Table 4.
Minimum inhibitory concentration values of cinnamic aldehydes, alcohols and their derivatives (132–151).
| Compound | Microorganism Strain | MIC | Refs. |
|---|---|---|---|
| Cinnamaldehyde (132) | Aspergillus niger MTCC 404 | 1.89 mM | [139] |
| Aspergillus sydowii MTCC 4335 | 3.78 mM | [139] | |
| Aspergillus parasiticus MTCC 2797 | 3.78 mM | [139] | |
| Bacillus cereus ATCC 11778 | 3.2 mM | [127] | |
| Bacillus subtilis | 7.6 mM | [124] | |
| Bacillus subtilis MTCC 121 | 1.89 mM | [139] | |
| Burkholderia cepacea MTCC 438 | 1.89 mM | [139] | |
| Candida albicans ATCC 2091 | 1.1 mM | [131] | |
| Candida albicans ATCC 90028 | 459 µM | [132] | |
| Candida albicans MTCC 3017 | 470 µM | [139] | |
| Candida albicans STD-1141 | 3.0 mM | [130] | |
| Candida tropicalis STD-1118 | 3.8 mM | [130] | |
| Clostridium difficile PHTCC 107 | 605 µM | [13] | |
| Enterobacter cloacae MTCC 509 | 1.89 mM | [139] | |
| Enterococcus faecalis | 1.89 mM | [12] | |
| Escherichia coli | 3.78 mM | [124] | |
| Escherichia coli | 3.78 mM | [12] | |
| Escherichia coli ATCC 11105 | 3.78 mM | [125] | |
| Escherichia coli CGMCC 1.487 | 2.0 mM | [123] | |
| Escherichia coli MTCC 43 | 1.89 mM | [139] | |
| Escherichia coli NCIM-2089 | 7.6 µM | [121] | |
| Escherichia coli NTCT 8196 | < 1.5 µM | [122] | |
| Escherichia coli O157:H7 | 1.89 mM | [125] | |
| Fusarium verticillioides | 358 µM | [129] | |
| Gloeophyllum trabeum BCRC 31614 | 2.3 mM | [134] | |
| Helicobacter pylori ATCC 26695 | 15 µM | [121] | |
| Issatchenkia orientalis MTCC 231 | 470 µM | [139] | |
| Klebsiella pneumoniae MTCC 109 | 3.78 mM | [139] | |
| Laetiporus betulina | 750 µM | [133] | |
| Legionella pneumophila JCM 7571 | 620 µM | [126] | |
| Lenzites betulina BCRC 35296 | 757 µM | [134] | |
| Laetiporus sulphureus | 700 µM | [133] | |
| Laetiporus sulphurous BCRC 35305 | 757 µM | [134] | |
| Malassezia furfur IP305 | 757 µM | [131] | |
| Micrococcus luteus MTCC 2470 | 3.78 mM | [139] | |
| Pseudomonas aeruginosa | 7.57 mM | [12] | |
| Pseudomonas aeruginosa MTCC 424 | 1.89 mM | [139] | |
| Pseudomonas aeruginosa NCTC 9027 | 10.6 µM | [122] | |
| Sclerotinia sclerotiorum | 1.94 mM | [146] | |
| Staphylococcus aureus | 1.89 mM | [12] | |
| Staphylococcus aureus MTCC 121 | 1.89 mM | [139] | |
| Methicillin-resistant Staphylococcus aureus | 1.89 mM | [124] | |
| Tametes versicolor BCRC 35253 | 2.3 mM | [134] | |
| Trychophyton rubrum MTCC 296 | 470 µM | [139] | |
| Caffeic aldehyde (133) | Mycobacterium tuberculosis H37Rv | 154 µM | [135] |
| Coniferaldehyde (134) | Streptococcus mitis ATCC 49456T | 351 µM | [137] |
| Streptococcus mutans DMST 26095 | 1.4 mM | [137] | |
| Streptococcus pyogenes DMST 17020 | 351 µM | [137] | |
| Sinapaldehyde (135) | Streptococcus mitis ATCC 49456T | 601 µM | [137] |
| Streptococcus mutans DMST 26095 | 601 µM | [137] | |
| Streptococcus pyogenes DMST 17020 | 150 µM | [137] | |
| 2-Methoxycinnamaldehyde (136) | Aspergillus fumigatus Kuboyama | 617 µM | [138] |
| Aspergillus niger stA-2 | 1.2 mM | [138] | |
| Candida albicans stT-1 | 308 µM | [138] | |
| Cryptococcus neoformans stY-8 | 77 µM | [138] | |
| Escherichia coli E-2602 | >1.2 mM | [138] | |
| Microsporum canis stT-6 | 19 µM | [138] | |
| Salmonella typhimurium 75-276 | >1.2 mM | [138] | |
| Staphylococcus aureus 209P | 1.2 mM | [138] | |
| 4-Methoxycinnamaldehyde (137) | Aspergillus niger MTCC 404 | 3.08 mM | [139] |
| Aspergillus sydowii MTCC 4335 | 3.08 mM | [139] | |
| Aspergillus parasiticus MTCC 2797 | 3.08 mM | [139] | |
| Bacillus subtilis MTCC 121 | 6.16 mM | [139] | |
| Candida albicans MTCC 3017 | 770 µM | [139] | |
| Enterobacter cloacae MTCC 509 | 3.08 mM | [139] | |
| Escherichia coli MTCC 43 | 3.08 mM | [139] | |
| Issatchenkia orientalis MTCC 231 | 770 µM | [139] | |
| Klebsiella pneumoniae MTCC 109 | 3.08 mM | [139] | |
| Micrococcus luteus MTCC 2470 | 3.08 mM | [139] | |
| Pseudomonas aeruginosa MTCC 424 | 770 µM | [139] | |
| Staphylococcus aureus MTCC 121 | 3.08 mM | [139] | |
| Trychophyton rubrum MTCC 296 | 770 µM | [139] | |
| 3,4-dimethoxy-cinnamaldehyde (138) | Aspergillus niger MTCC 404 | 5.20 mM | [139] |
| Aspergillus sydowii MTCC 4335 | 5.20 mM | [139] | |
| Aspergillus parasiticus MTCC 2797 | 2.60 mM | [139] | |
| Bacillus subtilis MTCC 121 | 5.20 mM | [139] | |
| Burkholderia cepacea MTCC 438 | 5.20 mM | [139] | |
| Candida albicans MTCC 3017 | 2.60 mM | [139] | |
| Enterobacter cloacae MTCC 509 | 10.4 mM | [139] | |
| Escherichia coli MTCC 43 | 10.4 mM | [139] | |
| Issatchenkia orientalis MTCC 231 | 2.60 mM | [139] | |
| Klebsiella pneumoniae MTCC 109 | 10.4 mM | [139] | |
| Micrococcus luteus MTCC 2470 | 2.60 mM | [139] | |
| Pseudomonas aeruginosa MTCC 424 | 2.60 mM | [139] | |
| Staphylococcus aureus MTCC 121 | 5.20 mM | [139] | |
| Trychophyton rubrum MTCC 296 | 160 µM | [139] | |
| 3,4,6-trimethoxy-cinnamaldehyde (139) | Aspergillus sydowii MTCC 4335 | 4.50 mM | [139] |
| Candida albicans MTCC 3017 | 8.99 mM | [139] | |
| Issatchenkia orientalis MTCC 231 | 8.99 mM | [139] | |
| Micrococcus luteus MTCC 2470 | 2.25 mM | [139] | |
| Trychophyton rubrum MTCC 296 | 280 µM | [139] | |
| 3,4-methylenedioxy-cinnamaldehyde (140) | Aspergillus niger MTCC 404 | 1.42 mM | [139] |
| Aspergillus sydowii MTCC 4335 | 350 µM | [139] | |
| Aspergillus parasiticus MTCC 2797 | 710 µM | [139] | |
| Bacillus subtilis MTCC 121 | 2.84 mM | [139] | |
| Burkholderia cepacea MTCC 438 | 710 µM | [139] | |
| Candida albicans MTCC 3017 | 350 µM | [139] | |
| Enterobacter cloacae MTCC 509 | 1.42 mM | [139] | |
| Escherichia coli MTCC 43 | 2.84 mM | [139] | |
| Issatchenkia orientalis MTCC 231 | 350 µM | [139] | |
| Klebsiella pneumoniae MTCC 109 | 1.42 mM | [139] | |
| Micrococcus luteus MTCC 2470 | 2.84 mM | [139] | |
| Pseudomonas aeruginosa MTCC 424 | 350 µM | [139] | |
| Staphylococcus aureus MTCC 121 | 2.84 mM | [139] | |
| Trychophyton rubrum MTCC 296 | 40 µM | [139] | |
| 3-methoxy-4-ethoxy-cinnamaldehyde (141) | Aspergillus niger MTCC 404 | 4.85 mM | [139] |
| Aspergillus sydowii MTCC 4335 | 4.85 mM | [139] | |
| Aspergillus parasiticus MTCC 2797 | 2.42 mM | [139] | |
| Bacillus subtilis MTCC 121 | 4.85 mM | [139] | |
| Burkholderia cepacea MTCC 438 | 4.85 mM | [139] | |
| Candida albicans MTCC 3017 | 2.42 mM | [139] | |
| Enterobacter cloacae MTCC 509 | 9.70 mM | [139] | |
| Escherichia coli MTCC 43 | 9.70 mM | [139] | |
| Issatchenkia orientalis MTCC 231 | 4.85 mM | [139] | |
| Klebsiella pneumoniae MTCC 109 | 9.70 mM | [139] | |
| Micrococcus luteus MTCC 2470 | 4.85 mM | [139] | |
| Pseudomonas aeruginosa MTCC 424 | 4.85 mM | [139] | |
| Staphylococcus aureus MTCC 121 | 2.42 mM | [139] | |
| Trychophyton rubrum MTCC 296 | 600 µM | [139] | |
| 2-bromo-3,4-dimethoxy-cinnamaldehyde (142) | Trychophyton rubrum MTCC 296 | 460 µM | [139] |
| N,N-dimethyl-4-amino-cinnamaldehyde (143) | Enterobacter cloacae MTCC 509 | 11.4 mM | [139] |
| Micrococcus luteus MTCC 2470 | 11.4 mM | [139] | |
| Trychophyton rubrum MTCC 296 | 180 µM | [139] | |
| 2-nitro-4-methyl-cinnamaldehyde (144) | Bacillus subtilis MTCC 121 | 2.61 mM | [139] |
| Burkholderia cepacea MTCC 438 | 2.61 mM | [139] | |
| Enterobacter cloacae MTCC 509 | 10.5 mM | [139] | |
| Escherichia coli MTCC 43 | 2.61 mM | [139] | |
| Klebsiella pneumoniae MTCC 109 | 10.5 mM | [139] | |
| Micrococcus luteus MTCC 2470 | 2.61 mM | [139] | |
| Pseudomonas aeruginosa MTCC 424 | 2.61 mM | [139] | |
| Staphylococcus aureus MTCC 121 | 10.5 mM | [139] | |
| Trychophyton rubrum MTCC 296 | 160 µM | [139] | |
| Cinnamyl alcohol (145) | Aspergillus niger 101 | 7 mM | [143] |
| Aspergillus oryzae 102 | 8 mM | [143] | |
| Bacillus subtilis IFO 13721 | 10 mM | [143] | |
| Candida albicans IFO 0597 | 7 mM | [143] | |
| Epidermophyton floccosum IFO 9045 | 3 mM | [143] | |
| Escherichia coli | 7.5 mM | [12] | |
| Escherichia coli IFO 3545 | 9 mM | [143] | |
| Enterococcus faecalis | 7.5 mM | [12] | |
| Klebsiella pneumonia IFO 13541 | 10 mM | [143] | |
| Legionella pneumophila JCM 7571 | 7.6 mM | [126] | |
| Pseudomonas aeruginosa | 7.5 mM | [12] | |
| Rhizopus sp. 103 | 3 mM | [143] | |
| Saccharomyces cerevisiae Kyokai no. 8 | 8 mM | [143] | |
| Staphylococcus aureus | 3.7 mM | [12] | |
| Trichophyton rubrum IFO 9185 | 4 mM | [143] | |
| Trichophyton violaceum IFO 31064 | 4 mM | [143] | |
| 4-Coumaryl alcohol (146) | Bacillus subtilis 8649 | 8.0 mM | [68] |
| Escherichia coli 12210 | >8.0 mM | [68] | |
| Pseudomonas syringae 649 | >8.0 mM | [68] | |
| Saccharomyces cerevisiae 019391 | >8.0 mM | [68] | |
| Schizosaccharomyces pombe 039917 | >8.0 mM | [68] | |
| Sporobolomyces roseus 043529 | 8.0 mM | [68] | |
| Coniferyl alcohol (147) | Bacillus subtilis 8649 | >8.0 mM | [68] |
| Escherichia coli 12210 | >8.0 mM | [68] | |
| Pseudomonas syringae 649 | >8.0 mM | [68] | |
| Saccharomyces cerevisiae 019391 | >8.0 mM | [68] | |
| Schizosaccharomyces pombe 039917 | >8.0 mM | [68] | |
| Sporobolomyces roseus 043529 | >8.0 mM | [68] | |
| Sinapyl alcohol (148) | Bacillus subtilis 8649 | >8.0 mM | [68] |
| Escherichia coli 12210 | >8.0 mM | [68] | |
| Pseudomonas syringae 649 | >8.0 mM | [68] | |
| Saccharomyces cerevisiae 019391 | >8.0 mM | [68] | |
| Schizosaccharomyces pombe 039917 | >8.0 mM | [68] | |
| Sporobolomyces roseus 043529 | >8.0 mM | [68] | |
| Cinnamyl benzoate (149) | Aspergillus niger 101 | >10 mM | [143] |
| Aspergillus oryzae 102 | >10 mM | [143] | |
| Bacillus subtilis IFO 13721 | >10 mM | [143] | |
| Candida albicans IFO 0597 | >10 mM | [143] | |
| Epidermophyton floccosum IFO 9045 | 20 µM | [143] | |
| Escherichia coli IFO 3545 | >10 mM | [143] | |
| Klebsiella pneumoniae IFO 13541 | >10 mM | [143] | |
| Rhizopus sp. 103 | >10 mM | [143] | |
| Saccharomyces cerevisiae Kyokai no. 8 | >10 mM | [143] | |
| Trichophyton rubrum IFO 9185 | 20 µM | [143] | |
| Trichophyton violaceum IFO 31064 | 20 µM | [143] | |
| 4-coumaryl acetate (150) | Candida albicans ATCC 10231 | 5.2 mM | [144] |
| Microsporum canis ATCC 36299 | 10.4 mM | [144] | |
| Staphylococcus aureus ATCC 25923 | 6.5 mM | [144] | |
| Staphylococcus aureus VISA 24 | 203 µM | [144] | |
| Tricophyton rubrum ATCC 28188 | 10.4 mM | [144] | |
| 4-coumaryl diacetate (151) | Candida albicans ATCC 10231 | 2.7 mM | [144] |
| Microsporum canis ATCC 36299 | 2.7 mM | [144] | |
| Mycobacterium smegmatis mc2 155 | 215 µM | [145] | |
| Staphylococcus aureus ATCC 25923 | 2.7 mM | [144] | |
| Staphylococcus aureus VISA 24 | 672 µM | [144] | |
| Tricophyton rubrum ATCC 28188 | 2.7 mM | [144] |
From the medicinal plant Piper taiwanense, caffeic aldehyde (133) (Figure 6) was isolated and found to have significant antitubercular properties (MICH37Rv = 154 µM) [135]. The other natural brothers of cinnamaldehyde, coniferaldehyde (134) and sinapaldehyde (135) displayed moderate anticandidal activity [136]. More than 60 strains and clinical Candida isolates were evaluated for their susceptibility to the three aldehydes. Sinapaldehyde was found to exert the most potent effect with a MIC values between 480 and 960 µM, while the MIC value of coniferaldehyde and cinnamaldehyde varied between 914 µM and 1.83 mM, and between 1.14 and 3.78 mM respectively [136]. Phenotypic characterization of C. albicans treated with sub-inhibitory concentrations of the aldehydes showed alterations in cell morphology and the cell wall, as well as damage to the plasma membrane resulting in cell lysis and cytoplasmic leakage. The study suggested inhibition of membrane ATPases, by examination of H+ efflux rates in presence of the aldehydes and known ATPase inhibitors [136]. Both coniferaldehyde (134) and sinapaldehyde (135) showed growth inhibitory properties against three species of Streptococcus with MIC ranging from 150 µM to 1.4 mM, being 135 more active against S. pyogenes and S. mutans, while 134 was more active against S. mitis [137].
Interestingly the most active antifungal compound present in cinnamon was identified to be 2-methoxycinnamaldehyde (136), achieving MIC values as low as 19 µM against Microsporum canis [138], a fungi causing ringworms in pets and tinea capitis in humans. This aldehyde was also active against other fungi including C. albicans, with an MIC value of 308 µM, but was however less active against A. niger and different species of bacteria [138]. The positional isomer 4-methoxycinnamaldehyde (137) was less active with MIC values of 770 µM against C. albicans MTCC 3017, Issatchenkia orientalis MTCC 231, P. aeruginosa MTCC 424 and Trichophyton rubrum MTCC 296, and higher values (MIC > 3 mM) against other microorganisms [139]. Clearly, as observed for the cinnamic acids, the substitution on position 2 of the cinnamic skeleton, confer an increase in the antimicrobial effect. The presence of another methoxy groups in position 3, did not provide a substantial effect, as 3,4-dimethoxycinnamaldehyde (138) showed MIC values higher than 2.5 mM for all the evaluated microorganisms (Table 4), except against T. rubrum MTCC 296 (MIC = 160 µM) [139]. The aldehyde having three methoxyl groups, 3,4,6-trimethoxycinnamaldehyde (139), showed a similar profile of activity, being a weak growth inhibitor of species of Aspergillus, Candida, Issatchenkia and Micrococcus (MIC > 1.4 mM) but being active against the dermatophyte T. rubrum MTCC 296 with an MIC value of 280 µM [139]. The compound 3,4-methylenedioxycinnamaldehyde (140) showed to be a potent inhibitor of T. rubrum fungi (MIC = 40 µM), with moderate activity against A. sydowii, C. albicans, I. orientalis and P. aeruginosa with an MIC value of 350 µM [139]. The aldehyde was less active against other bacteria and fungi. The replacement of a methoxy group in position 4 of 138 for an ethoxy group, as in 3-methoxy-4-ethoxycinnamaldehyde (141), slightly decreased the MIC values for most of the compounds, however the specificity against T. rubrum and I. orientalis decreased substantially. Introduction of a bromide to 138, as in 2-bromo-3,4-dimethoxycinnamaldehyde (142) decreased the antifungal activity against T. rubrum, increasing the MIC value to 460 µM [139]. The compound N,N-dimethyl-4-aminocinnamaldehyde (143) was the only tested cinnamaldehyde having an amino group, and displayed an MIC of 180 µM against T. rubrum, but was inactive against the bacteria Enterobacter cloacae and Micrococcus luteus (MIC > 11 mM). The cinnamaldehyde having a nitro substituent 2,2-nitro-4-methylcinnamaldehyde (144) showed a similar antimicrobial profile as the other cinnamaldehydes, with a potent growth inhibitory effect against T. rubrum (MIC = 160 µM) but with low activity against several species of bacteria (MIC > 2.5 mM) [139].
Cinnamyl alcohol (145) is a component of several essential oils, particularly those of Cinnamomum species and other Lauraceae plants [140,141,142]. Cinnamyl alcohol showed little inhibitory effect against all the evaluated bacterial and fungal species (Table 4) [12,126,143]. The study of Barber et al. published in 2000, reported the antimicrobial activities of 4-coumaryl alcohol (146), coniferyl alcohol (147) and sinapyl alcohol (148) [68]. Comparatively the alcohols were found to be less active than their corresponding aldehydes, showing MIC values equal or higher than 8.0 mM against bacteria and yeasts. Cinnamyl benzoate (149) showed little activity against Gram-negative and Gram-positive bacteria (MIC > 10 mM), however it was higly active against dermatophyte fungi Trycophyton sp. and Epidermophyton floccosum IFO 9045 (MIC = 20 µM) [143]. Two acetate esters of coumaryl alcohol displayed growth inhibition of some fungi and S. aureus [144]. 4-Coumaryl acetate (150) displayed MIC values higher than 5 mM against C. albicans and other fungi, but was however more active against a vancomycin intermediate S. aureus (VISA) with an MIC value of 203 µM [144]. The diester 4-coumaryl diacetate (151) showed higher anti-fungal activity in comparison with 150, and additionally the diacetate demonstrated significant antimycobacterial activity with an MIC value of 215 µM against Mycobacterium smegmatis mc2-155 [145].
5. Synthetic Cinnamic Hybrids
The interest in making hybrids molecules result from the idea of combining the biological properties of two active molecules to yield a third molecule (chimera or mermaid) with a stronger effect [147]. The conjugation can be used to modulate selectivity, spectrum of activity, potency, and physicochemical parameters. Theoretically the hybrid molecule once administered to biological systems could be lysed to yield in situ the two active molecules with atomic economy, or the hybrid molecule could be made sterically flexible enough to act on the two domains of the biological targets as a single piece [148]. Several cinnamic hybrids have been prepared [149,150,151,152,153] but only a few of them have been evaluated for their antimicrobial properties.
The studied cephem hybrids 152–155 (Figure 6 and Table 5) are cephalosporin moieties linked covalently to a 2,5-dichlorocinnamic (Figure 4). These hybrids showed a potent growth inhibitory effect against Staphylococcus epidermidis A24548, Staphylococcus haemolyticus A21638 and S. pneumoniae A28272 with MIC values in the nanomolar range [154]. The cephem 155 having a cinnamic aldehyde and a 2-hydroxy-3-propylamine N-substitution on the pyridine ring, was the most active against S. epidermidis A24548 (MIC = 22 nM) and S. haemolyticus A21638 (MIC = 87 nM). The oxazolidinone hybrid (156) showed strong antibacterial activity against E. faecalis ATCC 29212 (MIC = 194 nM), Enterococcus faecium ATTC 700221 (MIC = 388 nM) and S. aureus ATCC 29213 (MIC = 194 nM) [155]. Against the methicillin-resistant S. aureus ATCC 33591, the cinnamic hybrid (156) displayed an MIC value of 1.6 µM. A peptide hybrid based on the human cysteine protease inhibitor cystatin C has been prepared and evaluated for its antimicrobial properties [156]. The peptide cystapep 1 (157), having a cinnamic acid residue, showed MIC values of 25.2 µM against S. aureus and Streptococcus pyogenes and slightly higher MIC values against other Streptococcus species. This is the only antimicrobial peptide cinnamic hybrid known so far. The rifamycin hybrid SV (T9) (158) (Figure 6) having a cinnamic acid residue linked through amide bond with the piperazine ring, is slightly more potent than rifampin (MICT9 = 106 nM vs. MICRIF = 243 nM) [157]. The molecule has demonstrated to be useful in clinical application [158]. The MIC values of the hybrid molecule were in the nanomolar range against the pathogenic mycobacteria Mycobacterium avium complex, Mycobacterium leprae and M. tuberculosis [157,159,160]. However the hybrid T9 is not active against rifampin-resistant M. tuberculosis strains.
Table 5.
Minimum inhibitory concentration values of cinnamic hybrids (152–184).
| Compound | Microorganism Strain | MIC | Refs. |
|---|---|---|---|
| cephem (152) | Streptococcus pneumoniae A28272 | 200 nM | [154] |
| Enterococcus faecalis A20688 | 1.6 µM | [154] | |
| Staphylococcus aureus MRSA A27223 | 3.2 µM | [154] | |
| Staphylococcus epidermidis A24548 | 95 nM | [154] | |
| Staphylococcus haemolyticus A21638 | 200 nM | [154] | |
| cephem (153) | Streptococcus pneumoniae A28272 | 760 nM | [154] |
| Enterococcus faecalis A20688 | 1.5 µM | [154] | |
| Staphylococcus aureus MRSA A27223 | 3.0 µM | [154] | |
| Staphylococcus epidermidis A24548 | 91 nM | [154] | |
| Staphylococcus haemolyticus A21638 | 190 nM | [154] | |
| cephem (154) | Streptococcus pneumoniae A28272 | 350 nM | [154] |
| Enterococcus faecalis A20688 | 1.4 mM | [154] | |
| Staphylococcus aureus MRSA A27223 | 2.8 mM | [154] | |
| Staphylococcus epidermidis A24548 | 42 nM | [154] | |
| Staphylococcus haemolyticus A21638 | 175 nM | [154] | |
| cephem (155) | Streptococcus pneumoniae A28272 | 727 nM | [154] |
| Enterococcus faecalis A20688 | 364 nM | [154] | |
| Staphylococcus aureus MRSA A27223 | 364 nM | [154] | |
| Staphylococcus epidermidis A24548 | 22 nM | [154] | |
| Staphylococcus haemolyticus A21638 | 87 nM | [154] | |
| oxazolidinone hybrid (156) | Enterococcus faecalis ATCC 29212 | 194 nM | [155] |
| Enterococcus faecium ATTC 700221 (VRE) | 388 nM | [155] | |
| Staphylococcus aureus ATCC 29213 | 194 nM | [155] | |
| Staphylococcus aureus ATCC 33591 (MRSA) | 1.6 µM | [155] | |
| cystapep 1 (157) | Staphylococcus aureus ATCC 29213 | 25.2 µM | [156] |
| Streptococcus agalactiae NTCC 8181 | 50.5 µM | [156] | |
| Streptococcus anginosus CCUG 27298 | 50.5 µM | [156] | |
| Streptococcus pneumoniae ATCC 49619 | 50.5 µM | [156] | |
| Streptococcus pyogenes type M1 | 25.2 µM | [156] | |
| rifamycin T9 (158) | Mycobacterium avium 101 | 15.9 nM | [159] |
| Mycobacterium avium N-260 | 831 nM | [157] | |
| Mycobacterium tuberculosis H37Rv | 31.9 nM | [159] | |
| Mycobacterium tuberculosis H37Rv | 106 nM | [157] | |
| Mycobacterium tuberculosis MTB9 (RIF-R) | 8.5 µM | [159] | |
| isoniazid hybrid (159) | Mycobacterium tuberculosis H37Rv | 300 nM | [161] |
| isoniazid hybrid (160) | Mycobacterium tuberculosis H37Rv | 1.1 µM | [161] |
| isoniazid hybrid (161) | Mycobacterium tuberculosis H37Rv | 1.3 µM | [161] |
| isoniazid hybrid (162) | Mycobacterium tuberculosis H37Rv | 2.2 µM | [161] |
| isoniazid hybrid (163) | Mycobacterium tuberculosis H37Rv | 2.3 µM | [161] |
| isoniazid hybrid (164) | Mycobacterium tuberculosis H37Rv | 1.9 µM | [161] |
| cycloserine hybrid (165) | Mycobacterium tuberculosis H37Rv | 950 µM | [161] |
| triazophtalazine hybrid (166) | Mycobacterium tuberculosis H37Rv | 1.4 µM | [161] |
| guanylhydrazone hybrid (167) | Mycobacterium tuberculosis H37Rv | 40.5 µM | [164] |
| guanylhydrazone hybrid (168) | Mycobacterium tuberculosis H37Rv | 8.9 µM | [164] |
| Fenchol hybrid (169) | Mycobacterium tuberculosis H37Rv | 6.7 µM | [165] |
| Fenchol hybrid (170) | Mycobacterium tuberculosis H37Rv | 6.7 µM | [165] |
| Fenchol hybrid (171) | Mycobacterium tuberculosis H37Rv | 2.4 µM | [165] |
| Fenchol hybrid (172) | Mycobacterium tuberculosis H37Rv | 540 nM | [165] |
| oleanolic acid hybrid (173) | Mycobacterium tuberculosis H37Rv | 85.2 µM | [166] |
| oleanolic acid hybrid (174) | Mycobacterium tuberculosis H37Rv | 10.4 µM | [166] |
| oleanolic acid hybrid (175) | Mycobacterium tuberculosis H37Rv | 323 µM | [166] |
| oleanolic acid hybrid (176) | Mycobacterium tuberculosis H37Rv | 19.8 µM | [166] |
| ursolic acid hybrid (177) | Mycobacterium tuberculosis H37Rv | >341 µM | [166] |
| ursolic acid hybrid (178) | Mycobacterium tuberculosis H37Rv | 10.4 µM | [166] |
| ursolic acid hybrid (179) | Mycobacterium tuberculosis H37Rv | 323 µM | [166] |
| ursolic acid hybrid (180) | Mycobacterium tuberculosis H37Rv | 4.95 µM | [166] |
| betulinic acid hybrid (181) | Mycobacterium tuberculosis H37Rv | >341 µM | [166] |
| betulinic acid hybrid (182) | Mycobacterium tuberculosis H37Rv | 10.4 µM | [166] |
| betulinic acid hybrid (183) | Mycobacterium tuberculosis H37Rv | 323 µM | [166] |
| betulinic acid hybrid (184) | Mycobacterium tuberculosis H37Rv | 316 µM | [166] |
Cinnamic acid hybrids have also been prepared with other antitubercular drugs including isoniazid 159–164 and cycloserine (165) (Figure 7).
Figure 7.
Chemical structures of the cinnamic hybrids 152–184.
The isoniazid 4-methoxycinnamoyl hybrid (159) was the most active (MIC = 300 nM) among the screened hybrids [161], but was however slightly less active than isoniazid itself (MIC = 182 nM) [162]. The presence of larger substituents on the 4-O-position, decreased anti-TB activity.
Among the prepared 4-coumaric hybrids from the same comprehensive study, the cycloserine hybrid 165 displayed an MIC value of 950 µM [161], while the literature MIC value of cycloserine is 245 µM [161,163]. Nonetheless the study identified the triazolophtalazine cinnamic derivative 166 with a significant MIC value of 1.4 µM against the H37Rv strain and a selectivity index around 320 in comparison to THP-1 cells [161]. Guanyl hydrazone hybrids have also been prepared and examined for antimycobacterial activity [164]. Hydroxy substitution of the benzaldehyde hydrazone increased the anti-TB activity, with the hybrid 167 having a hydroxyl in position 4 displaying an MIC value of 40.5 µM against the virulent H37Rv strain [164]. The presence of 3,4-dimethoxy substitution on the benzaldehyde hydrazone, as in the hybrid 168, increased growth inhibition (MIC = 8.9 µM).
Thirty-one fenchol hybrids were prepared with different substitutions on the cinnamic ring, and four hybrids 169–172 resulted with potent activity against M. tuberculosis H37Rv [165]. The fenchol hybrids 169 and 170 having respectively a 3,4-methylenedioxy and a 2-nitro substituents (170) on the aryl ring of the cinnamic moiety, displayed an MIC value of 6.7 µM (Table 5). The fenchol hybrid 171 with 3,4,5-trimethoxy substitution on the cinnamic acid showed higher potency (MIC = 2.4 µM), while the hybrid 172 having 4-dimethylamine substitution was the most active (MIC = 540 nM) against the H37Rv pathogenic strain [165].
In a study from 2008, three triterpenes, betulinic, oleanolic and ursolic acids, were esterified in the 3-hydroxyl position, with different cinnamic acids with the aim of generating novel molecules which could potentially inhibit the M. tuberculosis H37Rv bacteria [166]. The cinnamic acids employed for the preparation of the esters were cinnamic, 4-coumaric, caffeic and ferulic acids. The esters having the 4-coumaroyl moiety, 3-O-(4'-coumaroyl) oleanolic acid (174), 3-O-(4'-coumaroyl) ursolic acid (178) and 3-O-(4'-coumaroyl) betulinic acid (182) were among the esters with the highest anti-TB activity, achieving MIC values of 10.4 µM. The MIC values of the free triterpenoids were 109 µM for oleanolic and betulinic acids, and 27.4 µM for ursolic acid [166], and therefore conjugation with 4-coumaric acid increased the anti-TB potency. Moreover the ester with the highest activity, 3-O-feruloyl ursolic acid (180), was able to inhibit completely the growth of M. tuberculosis at a concentration of 4.95 µM. The cinnamate and caffeate esters showed moderate to little anti-TB activity, with MIC values higher than those obtained for the free triterpenoids.
6. Conclusions
This review summarizes the in vitro antimicrobial activity of several cinnamic-related molecules by collating the reported MICs in a comprehensive list. Because the MIC data included in this review was extracted from several studies (using different experimental methods), it is far from ideal to compare the MIC as absolute values, but rather the MICs should be used as relative numbers indicating the tendency of the compounds to inhibit certain microorganisms. This review primarily serves as a framework to quickly identify the cinnamic acids and related molecules that have been tested for their antimicrobial properties.
Among the cinnamic-related molecules with the highest antimicrobial activity, the hybrids between antibiotics and cinnamic acids, such as the rifamycin T9 (158) and the oxazolidinone 156 with MIC values in the nanomolar range, were the champions. However the activity of these hybrids is mostly due to the potent effect of the antibiotic component. It is unknown whether the conjugates actually hydrolyze in vivo to yield two active molecules with potential synergism, or it is the whole molecule responsible for the observed biological effect. Among the non-hybrid cinnamic-related molecules with the lowest MIC values 4-methoxycinnamic acid (9), 3-nitrocinnamic acid (11), all the caffeoyl quinic acids 25–30, most of the cinnamate and 4-coumarate esters 40–61, and most of the cinnamoyl and 4-coumaroyl amides 78–115 are worth mentioning. A remarkable antimicrobial activity was detected for isobutyl cinnamate (45) achieving a broad spectrum of activity against yeasts, Gram-positive and Gram-negative bacteria, with MIC values between 43 and 12 µM. Further studies need to confirm the potent antimicrobial activity observed, expanding the screening to other microorganisms and drug resistant-isolates, and finally evaluating its toxicity. This example illustrate how a simple substitution, for instance comparing isobutyl cinnamate (45) with butyl cinnamate (44), can have a significant impact on the biological properties of the molecules (4-fold MIC change against some microorganisms).
There is no doubt that the cinnamic acids, their derivatives and hybrid molecules display marked antimicrobial effects. Some microorganisms are more sensitive to a chemical class than others. For instance, fungal organisms are generally more susceptible to the cinnamic aldehydes, while the cinnamic acids, esters and amides tend to affect more importantly the bacteria. A noteworthy effect was observed for the cinnamic molecules against Mycobacterium tuberculosis. The growth of the TB-causing bacteria was repeatedly inhibited by micromolar concentrations of molecules containing the cinnamic acid moiety. However very little is known about the mechanism of action of cinnamic acid, and the essential structural features required for anti-TB activity. Detailed molecular studies of the biological targets of the cinnamic acids may help to design high-affinity cinnamic-based ligands which may be important for developing future therapeutic alternatives to the growing problem of drug-resistant microbial pathogens.
Acknowledgments
The author acknowledge financial research support from the Universidad del Norte (Internal Agenda 2014) and the Royal Society of Chemistry (Research Fund 2014).
Author Contributions
The author performed the literature searches, compiled the data, prepared the tables and figures and wrote the manuscript.
Conflicts of Interest
The author declares no conflict of interest.
References
- 1.Xu Z., Zhang D., Hu J., Zhou X., Ye X., Reichel K., Stewart N., Syrenne R., Yang X., Gao P., et al. Comparative genome analysis of lignin biosynthesis gene families across the plant kingdom. BMC Bioinform. 2009;10:S3. doi: 10.1186/1471-2105-10-S11-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lichtenthaler H.K., Schweiger J. Cell wall bound ferulic acid, the major substance of the blue-green fluorescence emission of plants. J. Plant Physiol. 1998;152:272–282. doi: 10.1016/S0176-1617(98)80142-9. [DOI] [Google Scholar]
- 3.Kroon P.A., Williamson G. Hydroxycinnamates in plants and food: current and future perspectives. J. Sci. Food Agric. 1999;79:355–361. doi: 10.1002/(SICI)1097-0010(19990301)79:3<355::AID-JSFA255>3.0.CO;2-G. [DOI] [Google Scholar]
- 4.Martin-Tanguy J., Cabanne F., Perdrizet E., Martin C. The distribution of hydroxycinnamic acid amides in flowering plants. Phytochemistry. 1978;17:1927–1928. doi: 10.1016/S0031-9422(00)88735-X. [DOI] [Google Scholar]
- 5.Vogt T. Phenylpropanoid biosynthesis. Mol. Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 6.Ververidis F., Trantas E., Douglas C., Vollmer G., Kretzschmar G., Panopoulos N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotech. J. 2007;2:1214–1234. doi: 10.1002/biot.200700084. [DOI] [PubMed] [Google Scholar]
- 7.Solecka D. Role of phenylpropanoid compounds in plant responses to different stress factors. Acta Physiol. Plant. 1997;19:257–268. doi: 10.1007/s11738-997-0001-1. [DOI] [Google Scholar]
- 8.Hyun M.W., Yun Y.H., Kim J.Y., Kim S.H. Fungal and plant phenylalanine ammonia-lyase. Mycobiology. 2011;39:257–65. doi: 10.5941/MYCO.2011.39.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pittman S. Cinnamon: It’s not just for making cinnamon rolls. Ethnobot. Leafl. 2011;2000:11. [Google Scholar]
- 10.Wang R., Wang R., Yang B. Extraction of essential oils from five cinnamon leaves and identification of their volatile compound compositions. Innov. Food Sci. Emerg. 2009;10:289–292. doi: 10.1016/j.ifset.2008.12.002. [DOI] [Google Scholar]
- 11.Ooi L.S.M., Li Y., Kam S.-L., Wang H., Wong E.Y.L., Ooi V.E.C. Antimicrobial Activities of cinnamon oil and cinnamaldehyde from the Chinese medicinal herb Cinnamomum cassia Blume. Am. J. Chin. Med. 2006;34:511–522. doi: 10.1142/S0192415X06004041. [DOI] [PubMed] [Google Scholar]
- 12.Chang S.-T., Chen P.-F., Chang S.-C. Antibacterial activity of leaf essential oils and their constituents from Cinnamomum osmophloeum. J. Ethnopharmacol. 2001;77:123–127. doi: 10.1016/S0378-8741(01)00273-2. [DOI] [PubMed] [Google Scholar]
- 13.Shahverdi A.R., Monsef-Esfahani H.R., Tavasoli F., Zaheri A., Mirjani R. Trans-cinnamaldehyde from Cinnamomum zeylanicum bark essential oil reduces the clindamycin resistance of Clostridium difficile in vitro. J. Food Sci. 2007;72:S055–S058. doi: 10.1111/j.1750-3841.2006.00204.x. [DOI] [PubMed] [Google Scholar]
- 14.Clifford M.N. Chlorogenic acids and other cinnamates—nature, occurrence and dietary burden. J. Sci. Food Agric. 1999;79:362–372. doi: 10.1002/(SICI)1097-0010(19990301)79:3<362::AID-JSFA256>3.0.CO;2-D. [DOI] [Google Scholar]
- 15.Zhang J., Yang J., Chang X., Zhang C., Zhou H., Liu M. Ozagrel for acute ischemic stroke: A meta-analysis of data from randomized controlled trials. Neurol. Res. 2012;34:346–353. doi: 10.1179/1743132812Y.0000000022. [DOI] [PubMed] [Google Scholar]
- 16.Wilensky A.J., Ojemann L.M., Friel P.N., Almes M.J., Levy R.H., Dodrill C.B. Cinromide in epilepsy: A pilot study. Epilepsia. 1983;24:401–409. doi: 10.1111/j.1528-1157.1983.tb04908.x. [DOI] [PubMed] [Google Scholar]
- 17.Bezerra D.P., Pessoa C., de Moraes M.O., Saker-Neto N., Silveira E.R., Costa-Lotufo L.V. Overview of the therapeutic potential of piplartine (piperlongumine) Eur. J. Pharm. Sci. 2012;48:453–463. doi: 10.1016/j.ejps.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Filho R.B., De Souza M.P., Mattos M.E.O. Piplartine-dimer A, a new alkaloid from Piper tuberculatum. Phytochemistry. 1981;20:345–346. doi: 10.1016/0031-9422(81)85125-4. [DOI] [Google Scholar]
- 19.Bezerra D.P., Pessoa C., de Moraes M.O., Silveira E.R., Lima M.A., Elmiro F.J., Costa-Lotufo L.V. Antiproliferative effects of two amides, piperine and piplartine, from Piper species. Z. Naturforsch. C Biol. Sci. 2005;60:539–543. doi: 10.1515/znc-2005-7-805. [DOI] [PubMed] [Google Scholar]
- 20.Bezerra D.P., Militao G.C., de Castro F.O., Pessoa C., de Moraes M.O., Silveira E.R., Lima M.A., Elmiro F.J., Costa-Lotufo L.V. Piplartine induces inhibition of leukemia cell proliferation triggering both apoptosis and necrosis pathways. Toxicol. In Vitro. 2007;21:1–8. doi: 10.1016/j.tiv.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 21.De P., Baltas M., Bedos-Belval F. Cinnamic acid derivatives as anticancer agents-a review. Curr. Med. Chem. 2011;18:1672–1703. doi: 10.2174/092986711795471347. [DOI] [PubMed] [Google Scholar]
- 22.De P., Veau D., Bedos-Belval F., Chassaing S., Baltas M. Cinnamic derivatives in tuberculosis. In: Cardona P.-J., editor. Understanding Tuberculosis—New Approaches to Fighting against Drug Resistance. InTech Publishing; Rijeka, Croatia: 2012. Chapter 15. [Google Scholar]
- 23.Wiesner J., Mitsch A., Wißner P., Jomaa H., Schlitzer M. Structure–activity relationships of novel anti-malarial agents. Part 2: Cinnamic acid derivatives. Bioorg. Med. Chem. Lett. 2001;11:423–424. doi: 10.1016/S0960-894X(00)00684-3. [DOI] [PubMed] [Google Scholar]
- 24.Tawata S., Taira S., Kobamoto N., Zhu J., Ishihara M., Toyama S. Synthesis and antifungal activity of cinnamic acid esters. Biosci. Biotechnol. Biochem. 1996;60:909–910. doi: 10.1271/bbb.60.909. [DOI] [PubMed] [Google Scholar]
- 25.Sova M. Antioxidant and antimicrobial activities of cinnamic acid derivatives. Mini Rev. Med. Chem. 2012;12:749–767. doi: 10.2174/138955712801264792. [DOI] [PubMed] [Google Scholar]
- 26.Lapeyre C., Delomenède M., Bedos-Belval F., Duran H., Nègre-Salvayre A., Baltas M. Design, synthesis, and evaluation of pharmacological properties of cinnamic derivatives as antiatherogenic agents. J. Med. Chem. 2005;48:8115–8124. doi: 10.1021/jm050454c. [DOI] [PubMed] [Google Scholar]
- 27.Simonyan A. Activity of cinnamic acid derivatives and new methods for their synthesis (review) Pharm. Chem. J. 1993;27:92–100. doi: 10.1007/BF00781068. [DOI] [Google Scholar]
- 28.Wall V.M., Eisenstadt A., Ager D.J., Laneman S.A. The Heck reaction and cinnamic acid synthesis by heterogeneous catalysis. Platin. Metals Rev. 1999;43:138–144. [Google Scholar]
- 29.Sharma P. Cinnamic acid derivatives: A new chapter of various pharmacological activities. J. Chem. Pharm. Res. 2011;3:403–423. [Google Scholar]
- 30.Bygbjerg I.C. Double burden of noncommunicable and infectious diseases in developing countries. Science. 2012;337:1499–1501. doi: 10.1126/science.1223466. [DOI] [PubMed] [Google Scholar]
- 31.Coker R.J., Hunter B.M., Rudge J.W., Liverani M., Hanvoravongchai P. Emerging infectious diseases in southeast Asia: Regional challenges to control. Lancet. 2011;377:599–609. doi: 10.1016/S0140-6736(10)62004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mabey D., Peeling R.W., Ustianowski A., Perkins M.D. Tropical infectious diseases: Diagnostics for the developing world. Nat. Rev. Microbiol. 2004;2:231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 33.Fauci A.S., Morens D.M. The perpetual challenge of infectious diseases. N. Eng. J. Med. 2012;366:454–461. doi: 10.1056/NEJMra1108296. [DOI] [PubMed] [Google Scholar]
- 34.WHO . The Global Burden of Disease: 2004 Update. World Health Organization; Geneva, Switzerland: 2008. pp. 7–22. [Google Scholar]
- 35.Powers J.H. Antimicrobial drug development—The past, the present, and the future. Clin. Microbiol. Infect. 2004;10:23–31. doi: 10.1111/j.1465-0691.2004.1007.x. [DOI] [PubMed] [Google Scholar]
- 36.Gold H.S., Moellering R.C. Antimicrobial-drug resistance. N. Eng. J. Med. 1996;335:1445–1453. doi: 10.1056/NEJM199609263351304. [DOI] [PubMed] [Google Scholar]
- 37.Cohen M.L. Epidemiology of drug resistance: Implications for a post—Antimicrobial era. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- 38.Sotgiu G., Centis R., D’Ambrosio L., Tadolini M., Castiglia P., Migliori G.B. Do we need a new Fleming époque: The nightmare of drug-resistant tuberculosis. Int. J. Mycobacteriol. 2013;2:123–125. doi: 10.1016/j.ijmyco.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 39.David M.Z., Daum R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacGowan A., Macnaughton E. Antibiotic resistance. Medicine. 2013;41:642–648. doi: 10.1016/j.mpmed.2013.08.002. [DOI] [Google Scholar]
- 41.Hancock R.E.W. Mechanisms of action of newer antibiotics for Gram-positive pathogens. Lancet Infect. Dis. 2005;5:209–218. doi: 10.1016/S1473-3099(05)70051-7. [DOI] [PubMed] [Google Scholar]
- 42.Korošec B., Sova M., Turk S., Kraševec N., Novak M., Lah L., Stojan J., Podobnik B., Berne S., Zupanec N., et al. Antifungal activity of cinnamic acid derivatives involves inhibition of benzoate 4-hydroxylase (CYP53) J. Appl. Microbiol. 2014;116:955–966. doi: 10.1111/jam.12417. [DOI] [PubMed] [Google Scholar]
- 43.Jitareanu A., Tataringa G., Zbancioc A.M., Tuchilus C., Stanescu U. Antimicrobial activity of some cinnamic acid derivatives. Rev. Med. Chir. Soc. Med. Nat. Iasi. 2011;115:965–971. [PubMed] [Google Scholar]
- 44.Narasimhan B., Belsare D., Pharande D., Mourya V., Dhake A. Esters, amides and substituted derivatives of cinnamic acid: Synthesis, antimicrobial activity and QSAR investigations. Eur. J. Med. Chem. 2004;39:827–834. doi: 10.1016/j.ejmech.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 45.Hemaiswarya S., Doble M. Synergistic interaction of phenylpropanoids with antibiotics against bacteria. J. Med. Microbiol. 2010;59:1469–1476. doi: 10.1099/jmm.0.022426-0. [DOI] [PubMed] [Google Scholar]
- 46.Wen A., Delaquis P., Stanich K., Toivonen P. Antilisterial activity of selected phenolic acids. Food Microbiol. 2003;20:305–311. doi: 10.1016/S0740-0020(02)00135-1. [DOI] [Google Scholar]
- 47.Kujumgiev A., Tsvetkova I., Serkedjieva Y., Bankova V., Christov R., Popov S. Antibacterial, antifungal and antiviral activity of propolis of different geographic origin. J. Ethnopharmacol. 1999;64:235–240. doi: 10.1016/S0378-8741(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 48.Simone-Finstrom M., Spivak M. Propolis and bee health: The natural history and significance of resin use by honey bees. Apidologie. 2010;41:295–311. doi: 10.1051/apido/2010016. [DOI] [Google Scholar]
- 49.Bankova V., Christov R., Kujumgiev A., Marcucci M.C., Popov S. Chemical composition and antibacterial activity of Brazilian propolis. Z. Naturforsch. C Bio. Sci. 1995;50:167–172. doi: 10.1515/znc-1995-3-402. [DOI] [PubMed] [Google Scholar]
- 50.Bankova V.S., Popov S.S., Marekov N.L. Isopentenyl cinnamates from poplar buds and propolis. Phytochemistry. 1989;28:871–873. doi: 10.1016/0031-9422(89)80133-5. [DOI] [Google Scholar]
- 51.Bankova V.S., de Castro S.L., Marcucci M.C. Propolis: Recent advances in chemistry and plant origin. Apidologie. 2000;31:3–16. doi: 10.1051/apido:2000102. [DOI] [Google Scholar]
- 52.Mohammadzadeh S., Shariatpanahi M., Hamedi M., Ahmadkhaniha R., Samadi N., Ostad S.N. Chemical composition, oral toxicity and antimicrobial activity of Iranian propolis. Food Chem. 2007;103:1097–1103. doi: 10.1016/j.foodchem.2006.10.006. [DOI] [Google Scholar]
- 53.Stepanović S., Antić N., Dakić I., Švabić-Vlahović M. In vitro antimicrobial activity of propolis and synergism between propolis and antimicrobial drugs. Microbiol. Res. 2003;158:353–357. doi: 10.1078/0944-5013-00215. [DOI] [PubMed] [Google Scholar]
- 54.Koo H., Gomes B.P.F.A., Rosalen P.L., Ambrosano G.M.B., Park Y.K., Cury J.A. In vitro antimicrobial activity of propolis and Arnica montana against oral pathogens. Arch. Oral Biol. 2000;45:141–148. doi: 10.1016/S0003-9969(99)00117-X. [DOI] [PubMed] [Google Scholar]
- 55.Hewitt W., Vincent S. Theory and Application of Microbiological Assay. Academic Press; San Diego, CA, USA: 1989. pp. 1–37. [Google Scholar]
- 56.Olasupo N.A., Fitzgerald D.J., Gasson M.J., Narbad A. Activity of natural antimicrobial compounds against Escherichia coli and Salmonella enterica serovar Typhimurium. Lett. Appl. Microbiol. 2003;37:448–451. doi: 10.1046/j.1472-765X.2003.01427.x. [DOI] [PubMed] [Google Scholar]
- 57.Rastogi N., Domadia P., Shetty S., Dasgupta D. Screening of natural phenolic compounds for potential to inhibit bacterial cell division protein FtsZ. Ind. J. Exp. Biol. 2008;46:783–787. [PubMed] [Google Scholar]
- 58.Alves M.J., Ferreira I.C., Froufe H.J., Abreu R.M., Martins A., Pintado M. Antimicrobial activity of phenolic compounds identified in wild mushrooms, SAR analysis and docking studies. J. Appl. Microbiol. 2013;115:346–357. doi: 10.1111/jam.12196. [DOI] [PubMed] [Google Scholar]
- 59.Prasad V.G.N.V., Krishna B.V., Swamy P.L., Rao T.S., Rao G.S. Antibacterial synergy between quercetin and polyphenolic acids against bacterial pathogens of fish. Asian Pac. J. Trop. Dis. 2014;4(Suppl. 1):S326–S329. doi: 10.1016/S2222-1808(14)60464-3. [DOI] [Google Scholar]
- 60.Guzman J.D., Mortazavi P.N., Munshi T., Evangelopoulos D., McHugh T.D., Gibbons S., Malkinson J., Bhakta S. 2-Hydroxy-substituted cinnamic acids and acetanilides are selective growth inhibitors of Mycobacterium tuberculosis. MedChemComm. 2014;5:47–50. doi: 10.1039/c3md00251a. [DOI] [Google Scholar]
- 61.Guzman J.D., Evangelopoulos D., Gupta A., Birchall K., Mwaigwisya S., Saxty B., McHugh T.D., Gibbons S., Malkinson J., Bhakta S. Antitubercular specific activity of ibuprofen and the other 2-arylpropanoic acids using the HT-SPOTi whole-cell phenotypic assay. BMJ Open. 2013;3:e002672–e002685. doi: 10.1136/bmjopen-2013-002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rastogi N., Goh K.S., Horgen L., Barrow W.W. Synergistic activities of antituberculous drugs with cerulenin and trans-cinnamic acid against Mycobacterium tuberculosis. FEMS Immunol. Med. Microbiol. 1998;21:149–157. doi: 10.1111/j.1574-695X.1998.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 63.Chen Y.L., Huang S.T., Sun F.M., Chiang Y.L., Chiang C.J., Tsai C.M., Weng C.J. Transformation of cinnamic acid from trans- to cis-form raises a notable bactericidal and synergistic activity against multiple-drug resistant Mycobacterium tuberculosis. Eur. J. Pharm. Sci. 2011;43:188–194. doi: 10.1016/j.ejps.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 64.Warbasse J.P. Cinnamic acid in the treatment of tuberculosis. Ann. Surg. 1894;19:102–111. doi: 10.1097/00000658-189401000-00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bisogno F., Mascoti L., Sanchez C., Garibotto F., Giannini F., Kurina-Sanz M., Enriz R. Structure–antifungal activity relationship of cinnamic acid derivatives. J. Agric. Food Chem. 2007;55:10635–10640. doi: 10.1021/jf0729098. [DOI] [PubMed] [Google Scholar]
- 66.Schmidt E., Bail S., Friedl S.M., Jirovetz L., Buchbauer G., Wanner J., Denkova Z., Slavchev A., Stoyanova A., Geissler M. Antimicrobial activities of single aroma compounds. Nat. Prod. Commun. 2010;5:1365–1368. [PubMed] [Google Scholar]
- 67.Lou Z., Wang H., Rao S., Sun J., Ma C., Li J. p-Coumaric acid kills bacteria through dual damage mechanisms. Food Control. 2012;25:550–554. doi: 10.1016/j.foodcont.2011.11.022. [DOI] [Google Scholar]
- 68.Barber M.S., McConnell V.S., DeCaux B.S. Antimicrobial intermediates of the general phenylpropanoid and lignin specific pathways. Phytochemistry. 2000;54:53–56. doi: 10.1016/S0031-9422(00)00038-8. [DOI] [PubMed] [Google Scholar]
- 69.Parkar S.G., Stevenson D.E., Skinner M.A. The potential influence of fruit polyphenols on colonic microflora and human gut health. Int. J. Food Microbiol. 2008;124:295–298. doi: 10.1016/j.ijfoodmicro.2008.03.017. [DOI] [PubMed] [Google Scholar]
- 70.Tunçel G., Nergiz C. Antimicrobial effect of some olive phenols in a laboratory medium. Lett. Appl. Microbiol. 1993;17:300–302. doi: 10.1111/j.1472-765X.1993.tb01472.x. [DOI] [Google Scholar]
- 71.García-Ruiz A., Bartolomé B., Cueva C., Martín-Álvarez P.J., Moreno-Arribas M.V. Inactivation of oenological lactic acid bacteria (Lactobacillus hilgardii and Pediococcus pentosaceus) by wine phenolic compounds. J. Appl. Microbiol. 2009;107:1042–1053. doi: 10.1111/j.1365-2672.2009.04287.x. [DOI] [PubMed] [Google Scholar]
- 72.Stead D. The effect of hydroxycinnamic acids on the growth of wine-spoilage lactic acid bacteria. J. Appl. Bacteriol. 1993;75:135–141. doi: 10.1111/j.1365-2672.1993.tb02758.x. [DOI] [Google Scholar]
- 73.Zabka M., Pavela R. Antifungal efficacy of some natural phenolic compounds against significant pathogenic and toxinogenic filamentous fungi. Chemosphere. 2013;93:1051–1056. doi: 10.1016/j.chemosphere.2013.05.076. [DOI] [PubMed] [Google Scholar]
- 74.Puupponen-Pimiä R., Nohynek L., Alakomi H.-L., Oksman-Caldentey K.-M. Bioactive berry compounds—Novel tools against human pathogens. Appl. Microbiol. Biotechnol. 2005;67:8–18. doi: 10.1007/s00253-004-1817-x. [DOI] [PubMed] [Google Scholar]
- 75.Gasparetto J.C., Guimarães de Francisco T.M., Campos F.R., Pontarolo R. Development and validation of two methods based on high-performance liquid chromatography-tandem mass spectrometry for determining 1,2-benzopyrone, dihydrocoumarin, o-coumaric acid, syringaldehyde and kaurenoic acid in guaco extracts and pharmaceutical preparations. J. Sep. Sci. 2011;34:740–748. doi: 10.1002/jssc.201000792. [DOI] [PubMed] [Google Scholar]
- 76.Herald P.J., Davidson P.M. Antibacterial activity of selected hydroxycinnamic acids. J. Food Sci. 1983;48:1378–1379. doi: 10.1111/j.1365-2621.1983.tb09243.x. [DOI] [Google Scholar]
- 77.Georgiev L., Chochkova M., Ivanova G., Najdenski H., Ninova M., Milkova T. Radical scavenging and antimicrobial activities of cinnamoyl amides of biogenic monoamines. Riv. Ital. Sost. Grasse. 2012;89:91–102. [Google Scholar]
- 78.De Vita D., Friggeri L., D’Auria F.D., Pandolfi F., Piccoli F., Panella S., Palamara A.T., Simonetti G., Scipione L., di Santo R., et al. Activity of caffeic acid derivatives against Candida albicans biofilm. Bioorg. Med. Chem. Lett. 2014;24:1502–1505. doi: 10.1016/j.bmcl.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 79.Borges A., Ferreira C., Saavedra M.J., Simoes M. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb. Drug Resist. 2013;19:256–265. doi: 10.1089/mdr.2012.0244. [DOI] [PubMed] [Google Scholar]
- 80.Nakauchi M., Ikemoto S., Yamanishi H., Ozaki Y., Tsuno T., Nomura E., Hosoda A., Taniguchi H. Antimicrobial activities of synthetic ferulic acid derivatives. Food Preserv. Sci. 2002;28:183–188. doi: 10.5891/jafps.28.183. [DOI] [Google Scholar]
- 81.Ergün B., Çoban T., Onurdag F., Banoglu E. Synthesis, antioxidant and antimicrobial evaluation of simple aromatic esters of ferulic acid. Arch. Pharm. Res. 2011;34:1251–1261. doi: 10.1007/s12272-011-0803-y. [DOI] [PubMed] [Google Scholar]
- 82.Klančnik A., Možina S.S., Zhang Q. Anti-Campylobacter activities and resistance mechanisms of natural phenolic compounds in Campylobacter. PLoS One. 2012;7:e51800. doi: 10.1371/journal.pone.0051800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Engels C., Schieber A., Gänzle M. Sinapic acid derivatives in defatted Oriental mustard (Brassica juncea L.) seed meal extracts using UHPLC-DAD-ESI-MS n and identification of compounds with antibacterial activity. Eur. Food Res. Technol. 2012;234:535–542. doi: 10.1007/s00217-012-1669-z. [DOI] [Google Scholar]
- 84.Nakazono Y., Watanabe Y., Hashinaga F., Tadera K. Studies on antimicrobial and antioxidative substance of Yuzu (Citrus junos hort. ex Tanaka) seed. J. Biol. Sci. 2006;6:135–139. doi: 10.3923/jbs.2006.135.139. [DOI] [Google Scholar]
- 85.Cheng M.-J., Wu M.-D., Yanai H., Su Y.-S., Chen I.-S., Yuan G.-F., Hsieh S.-Y., Chen J.-J. Secondary metabolites from the endophytic fungus Biscogniauxia formosana and their antimycobacterial activity. Phytochem. Lett. 2012;5:467–472. doi: 10.1016/j.phytol.2012.04.007. [DOI] [Google Scholar]
- 86.Tonari K., Mitsui K., Yonemoto K. Structure and antibacterial activity of cinnamic acid related compounds. J. Oleo Sci. 2002;51:271–273. doi: 10.5650/jos.51.271. [DOI] [Google Scholar]
- 87.Feasley C.F., Gwynn B.H., Degering F., Tetrault P.A. Bacteriostatic properties of phenylacetic acid. Effect of ortho-substituents on. J. Am. Pharm. Assoc. 1941;30:41–45. doi: 10.1002/jps.3030300203. [DOI] [Google Scholar]
- 88.Kanao S., Kashino S., Haisa M. Topochemical studies. XIII. Structures of 3-bromocinnamic acid and 3-chlorocinnamic acid. Acta Crystallograph. Sect. C Cryst. Struct. Commun. 1990;46:2436–2438. doi: 10.1107/S0108270190004334. [DOI] [Google Scholar]
- 89.Schaldach B., Grützmacher H.F. The fragmentations of substituted cinnamic acids after electron impact. Org. Mass Spectrom. 1980;15:175–181. doi: 10.1002/oms.1210150404. [DOI] [Google Scholar]
- 90.Newby B.M.Z., Cutright T., Barrios C., Xu Q. Zosteric acid—An effective antifoulant for reducing fresh water bacterial attachment on coatings. J. Coat. Technol. Res. 2006;3:69–76. doi: 10.1007/s11998-006-0007-4. [DOI] [Google Scholar]
- 91.Villa F., Pitts B., Stewart P., Giussani B., Roncoroni S., Albanese D., Giordano C., Tunesi M., Cappitelli F. Efficacy of zosteric acid sodium salt on the yeast biofilm model Candida albicans. Microb. Ecol. 2011;62:584–598. doi: 10.1007/s00248-011-9876-x. [DOI] [PubMed] [Google Scholar]
- 92.Feresin G.E., Tapia A., Gimenez A., Ravelo A.G., Zacchino S., Sortino M., Schmeda-Hirschmann G. Constituents of the Argentinian medicinal plant Baccharis grisebachii and their antimicrobial activity. J. Ethnopharmacol. 2003;89:73–80. doi: 10.1016/s0378-8741(03)00259-9. [DOI] [PubMed] [Google Scholar]
- 93.Hermans D., Martel A., van Deun K., van Immerseel F., Heyndrickx M., Haesebrouck F., Pasmans F. The cinnamon-oil ingredient trans-cinnamaldehyde fails to target Campylobacter jejuni strain KC 40 in the broiler chicken cecum despite marked in vitro activity. J. Food Prot. 2011;74:1729–1734. doi: 10.4315/0362-028X.JFP-10-487. [DOI] [PubMed] [Google Scholar]
- 94.Tesaki S., Tanabe S., Ono H., Fukushi E., Kawabata J., Watanabe M. 4-Hydroxy-3-nitrophenylacetic and sinapic acids as antibacterial compounds from mustard seeds. Biosci. Biotechnol. Biochem. 1998;62:998–1000. doi: 10.1271/bbb.62.998. [DOI] [PubMed] [Google Scholar]
- 95.Olthof M.R., Hollman P.C.H., Katan M.B. Chlorogenic acid and caffeic acid are absorbed in humans. J. Nutr. 2001;131:66–71. doi: 10.1093/jn/131.1.66. [DOI] [PubMed] [Google Scholar]
- 96.Zhu X., Zhang H., Lo R. Phenolic compounds from the leaf extract of artichoke (Cynara scolymus L.) and their antimicrobial activities. J. Agric. Food Chem. 2004;52:7272–7278. doi: 10.1021/jf0490192. [DOI] [PubMed] [Google Scholar]
- 97.Daglia M., Papetti A., Grisoli P., Aceti C., Spini V., Dacarro C., Gazzani G. Isolation, identification, and quantification of roasted coffee antibacterial compounds. J. Agric. Food Chem. 2007;55:10208–10213. doi: 10.1021/jf0722607. [DOI] [PubMed] [Google Scholar]
- 98.Xia D., Wu X., Shi J., Yang Q., Zhang Y. Phenolic compounds from the edible seeds extract of Chinese Mei (Prunus mume Sieb. et Zucc) and their antimicrobial activity. LWT—Food Sci. Technol. 2011;44:347–349. doi: 10.1016/j.lwt.2010.05.017. [DOI] [Google Scholar]
- 99.Fiamegos Y.C., Kastritis P.L., Exarchou V., Han H., Bonvin A.M.J.J., Vervoort J., Lewis K., Hamblin M.R., Tegos G.P. Antimicrobial and efflux pump inhibitory activity of caffeoylquinic acids from Artemisia absinthium against Gram-positive pathogenic bacteria. PLoS One. 2011;6:e18127. doi: 10.1371/journal.pone.0018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lou Z., Wang H., Zhu S., Ma C., Wang Z. Antibacterial activity and mechanism of action of chlorogenic acid. J. Food Sci. 2011;76:M398–M403. doi: 10.1111/j.1750-3841.2011.02213.x. [DOI] [PubMed] [Google Scholar]
- 101.Zhang M., Liu W.-X., Zheng M.-F., Xu Q.-L., Wan F.-H., Wang J., Lei T., Zhou Z.-Y., Tan J.-W. Bioactive quinic acid derivatives from Ageratina adenophora. Molecules. 2013;18:14096–14104. doi: 10.3390/molecules181114096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kang K., Jang S., Kim B.-K., Park M. Antibacterial phenylpropanoid glycosides from Paulownia tomentosa Steud. Arch. Pharm. Res. 1994;17:470–475. doi: 10.1007/BF02979128. [DOI] [PubMed] [Google Scholar]
- 103.Svetaz L., Tapia A., López S.N., Furlán R.L.E., Petenatti E., Pioli R., Schmeda-Hirschmann G., Zacchino S.A. Antifungal chalcones and new caffeic acid esters from Zuccagnia punctata acting against soybean infecting fungi. J. Agric. Food Chem. 2004;52:3297–3300. doi: 10.1021/jf035213x. [DOI] [PubMed] [Google Scholar]
- 104.Abedini A., Roumy V., Mahieux S., Biabiany M., Standaert-Vitse A., Riviere C., Sahpaz S., Bailleul F., Neut C., Hennebelle T. Rosmarinic acid and its methyl ester as antimicrobial components of the hydromethanolic extract of Hyptis atrorubens Poit. (Lamiaceae) Evid. Based Complement. Altern. Med. 2013;2013:604536:1–604536:11. doi: 10.1155/2013/604536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lin L., Zhu D., Zou L., Yang B., Zhao M. Antibacterial activity-guided purification and identification of a novel C-20 oxygenated ent-kaurane from Rabdosia serra (Maxim.) Hara. Food Chem. 2013;139:902–909. doi: 10.1016/j.foodchem.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 106.Gohari A.R., Saeidnia S., Shahverdi A.R., Yassa N., Malmir M., Mollazade K., Naghinejad A.R. Phytochemistry and antimicrobial compounds of Hymenocrater calycinus. EurAsian J. Biosci. 2009;3:64–68. doi: 10.5053/ejobios.2009.3.0.9. [DOI] [Google Scholar]
- 107.Grunberger D., Banerjee R., Eisinger K., Oltz E.M., Efros L., Caldwell M., Estevez V., Nakanishi K. Preferential cytotoxicity on tumor cells by caffeic acid phenethyl ester isolated from propolis. Experientia. 1988;44:230–232. doi: 10.1007/BF01941717. [DOI] [PubMed] [Google Scholar]
- 108.Velazquez C., Navarro M., Acosta A., Angulo A., Dominguez Z., Robles R., Robles-Zepeda R., Lugo E., Goycoolea F.M., Velazquez E.F., et al. Antibacterial and free-radical scavenging activities of Sonoran propolis. J. Appl. Microbiol. 2007;103:1747–1756. doi: 10.1111/j.1365-2672.2007.03409.x. [DOI] [PubMed] [Google Scholar]
- 109.Ango P.Y., Kapche D.W.F.G., Kuete V., Ngadjui B.T., Bezabih M., Abegaz B.M. Chemical constituents of Trilepisium madagascariense (Moraceae) and their antimicrobial activity. Phytochem. Lett. 2012;5:524–528. doi: 10.1016/j.phytol.2012.05.006. [DOI] [Google Scholar]
- 110.Chien Y.C., Lin C.H., Chiang M.Y., Chang H.S., Liao C.H., Chen I.S., Peng C.F., Tsai I.L. Secondary metabolites from the root of Ehretia longiflora and their biological activities. Phytochemistry. 2012;80:50–57. doi: 10.1016/j.phytochem.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 111.Khatkar A., Nanda A., Kumar P., Narasimhan B. Synthesis, antimicrobial evaluation and QSAR studies of p-coumaric acid derivatives. Arab. J. Chem. 2014 in press. [Google Scholar]
- 112.Lakshmanan D., Werngren J., Jose L., Suja K.P., Nair M.S., Varma R.L., Mundayoor S., Hoffner S., Kumar R.A. Ethyl p-methoxycinnamate isolated from a traditional anti-tuberculosis medicinal herb inhibits drug resistant strains of Mycobacterium tuberculosis in vitro. Fitoterapia. 2011;82:757–761. doi: 10.1016/j.fitote.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 113.Dyer L.A., Richards J., Dodson C.D. Isolation, synthesis, and evolutionary ecology of Piper amides. In: Dyer L.A., Palmer A.D.N., editors. Piper: A Model Genus for Studies of Phytochemistry, Ecology, and Evolution. Springer; New York, NY, USA: 2004. pp. 117–139. [Google Scholar]
- 114.Naika R., Prasanna K., Ganapathy P. Antibacterial activity of piperlongumine an alkaloid isolated from the methanolic root extract of Piper longum L. Pharmacophore. 2010;1:141–148. [Google Scholar]
- 115.Navickiene H.M.D., Alécio A.C., Kato M.J., Bolzani V.D.S., Young M.C.M., Cavalheiro A.J., Furlan M. Antifungal amides from Piper hispidum and Piper tuberculatum. Phytochemistry. 2000;55:621–626. doi: 10.1016/S0031-9422(00)00226-0. [DOI] [PubMed] [Google Scholar]
- 116.Vasques da Silva R., Debonsi Navickiene H.M., Kato M.J., Bolzani V.D.S., Méda C.I., Young M.C.M., Furlan M. Antifungal amides from Piper arboreum and Piper tuberculatum. Phytochemistry. 2002;59:521–527. doi: 10.1016/S0031-9422(01)00431-9. [DOI] [PubMed] [Google Scholar]
- 117.Rukachaisirikul T., Siriwattanakit P., Sukcharoenphol K., Wongvein C., Ruttanaweang P., Wongwattanavuch P., Suksamrarn A. Chemical constituents and bioactivity of Piper sarmentosum. J. Ethnopharmacol. 2004;93:173–176. doi: 10.1016/j.jep.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 118.Tuntiwachwuttikul P., Phansa P., Pootaeng-On Y., Taylor W.C. Chemical constituents of the roots of Piper sarmentosum. Chem. Pharm. Bull. 2006;54:149–151. doi: 10.1248/cpb.54.149. [DOI] [PubMed] [Google Scholar]
- 119.Samwel S., Odalo J.O., Nkunya M.H., Joseph C.C., Koorbanally N.A. Toussaintines A-E: Antimicrobial indolidinoids, a cinnamoylhydrobenzofuranoid and a cinnamoylcyclohexenoid from Toussaintia orientalis leaves. Phytochemistry. 2011;72:1826–1832. doi: 10.1016/j.phytochem.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 120.Jantan I.B., Karim Moharam B.A., Santhanam J., Jamal J.A. Correlation between chemical composition and antifungal activity of the essential oils of eight Cinnamomum species. Pharm. Biol. 2008;46:406–412. doi: 10.1080/13880200802055859. [DOI] [Google Scholar]
- 121.Ali S., Khan A., Ahmed I., Musaddiq M., Ahmed K., Polasa H., Rao L.V., Habibullah C., Sechi L., Ahmed N. Antimicrobial activities of eugenol and cinnamaldehyde against the human gastric pathogen Helicobacter pylori. Ann. Clin. Microbiol. Antimicrob. 2005;4:20–27. doi: 10.1186/1476-0711-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cox S.D., Markham J.L. Susceptibility and intrinsic tolerance of Pseudomonas aeruginosa to selected plant volatile compounds. J. Appl. Microbiol. 2007;103:930–936. doi: 10.1111/j.1365-2672.2007.03353.x. [DOI] [PubMed] [Google Scholar]
- 123.Pei R.-S., Zhou F., Ji B.-P., Xu J. Evaluation of combined antibacterial effects of eugenol, cinnamaldehyde, thymol, and carvacrol against E. coli with an improved method. J. Food Sci. 2009;74:M379–M383. doi: 10.1111/j.1750-3841.2009.01287.x. [DOI] [PubMed] [Google Scholar]
- 124.Domadia P., Swarup S., Bhunia A., Sivaraman J., Dasgupta D. Inhibition of bacterial cell division protein FtsZ by cinnamaldehyde. Biochem. Pharmacol. 2007;74:831–840. doi: 10.1016/j.bcp.2007.06.029. [DOI] [PubMed] [Google Scholar]
- 125.Kim H.O., Park S.W., Park H.D. Inactivation of Escherichia coli O157:H7 by cinnamic aldehyde purified from Cinnamomum cassia shoot. Food Microbiol. 2004;21:105–110. doi: 10.1016/S0740-0020(03)00010-8. [DOI] [Google Scholar]
- 126.Shimizu I., Isshiki Y., Nomura H., Sakuda K., Sakuma K., Kondo S. The antibacterial activity of fragrance ingredients against Legionella pneumophila. Biol. Pharm. Bull. 2009;32:1114–1117. doi: 10.1248/bpb.32.1114. [DOI] [PubMed] [Google Scholar]
- 127.Kwon J.A., Yu C.B., Park H.D. Bacteriocidal effects and inhibition of cell separation of cinnamic aldehyde on Bacillus cereus. Lett. Appl. Microbiol. 2003;37:61–65. doi: 10.1046/j.1472-765X.2003.01350.x. [DOI] [PubMed] [Google Scholar]
- 128.Jia P., Xue Y.J., Duan X.J., Shao S.H. Effect of cinnamaldehyde on biofilm formation and sarA expression by methicillin-resistant Staphylococcus aureus. Lett. Appl. Microbiol. 2011;53:409–416. doi: 10.1111/j.1472-765X.2011.03122.x. [DOI] [PubMed] [Google Scholar]
- 129.Xing F., Hua H., Selvaraj J.N., Zhao Y., Zhou L., Liu X., Liu Y. Growth inhibition and morphological alterations of Fusarium verticillioides by cinnamon oil and cinnamaldehyde. Food Control. 2014;46:343–350. doi: 10.1016/j.foodcont.2014.04.037. [DOI] [Google Scholar]
- 130.Shreaz S., Sheikh R.A., Rimple B., Hashmi A.A., Nikhat M., Khan L.A. Anticandidal activity of cinnamaldehyde, its ligand and Ni(II) complex: Effect of increase in ring and side chain. Microb. Pathog. 2010;49:75–82. doi: 10.1016/j.micpath.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 131.Ferhout H., Bohatier J., Guillot J., Chalchat J.C. Antifungal activity of selected essential oils, cinnamaldehyde and carvacrol against Malassezia furfur and Candida albicans. J. Essent. Oil Res. 1999;11:119–129. doi: 10.1080/10412905.1999.9701086. [DOI] [Google Scholar]
- 132.Rajput S.B., Karuppayil S.M. Small molecules inhibit growth, viability and ergosterol biosynthesis in Candida albicans. SpringerPlus. 2013;2:26–28. doi: 10.1186/2193-1801-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yen T.-B., Chang S.-T. Synergistic effects of cinnamaldehyde in combination with eugenol against wood decay fungi. Bioresour. Technol. 2008;99:232–236. doi: 10.1016/j.biortech.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 134.Hsu F.-L., Chang H.-T., Chang S.-T. Evaluation of antifungal properties of octyl gallate and its synergy with cinnamaldehyde. Bioresour. Technol. 2007;98:734–738. doi: 10.1016/j.biortech.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 135.Chen S., Huang H.-Y., Cheng M.-J., Wu C.-C., Ishikawa T., Peng C.-F., Chang H.-S., Wang C.-J., Wong S.-L., Chen I.-S. Neolignans and phenylpropanoids from the roots of Piper taiwanense and their antiplatelet and antitubercular activities. Phytochemistry. 2013;93:203–209. doi: 10.1016/j.phytochem.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 136.Shreaz S., Bhatia R., Khan N., Muralidhar S., Manzoor N., Khan L.A. Influences of cinnamic aldehydes on H+ extrusion activity and ultrastructure of Candida. J. Med. Microbiol. 2013;62:232–240. doi: 10.1099/jmm.0.036145-0. [DOI] [PubMed] [Google Scholar]
- 137.Meerungrueang W., Panichayupakaranant P. Antimicrobial activities of some Thai traditional medical longevity formulations from plants and antibacterial compounds from Ficus foveolata. Pharm. Biol. 2014;52:1104–1109. doi: 10.3109/13880209.2013.877493. [DOI] [PubMed] [Google Scholar]
- 138.Morozumi S. Isolation, purification, and antibiotic activity of o-methoxycinnamaldehyde from cinnamon. Appl. Environ. Microbiol. 1978;36:577–583. doi: 10.1128/aem.36.4.577-583.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sharma U., Sood S., Sharma N., Rahi P., Kumar R., Sinha A., Gulati A. Synthesis and SAR investigation of natural phenylpropene-derived methoxylated cinnamaldehydes and their novel Schiff bases as potent antimicrobial and antioxidant agents. Med. Chem. Res. 2013;22:5129–5140. doi: 10.1007/s00044-013-0484-9. [DOI] [Google Scholar]
- 140.Cheng S.-S., Liu J.-Y., Tsai K.-H., Chen W.-J., Chang S.-T. Chemical composition and mosquito larvicidal activity of essential oils from leaves of different Cinnamomum osmophloeum provenances. J. Agric. Food Chem. 2004;52:4395–4400. doi: 10.1021/jf0497152. [DOI] [PubMed] [Google Scholar]
- 141.Caredda A., Marongiu B., Porcedda S., Soro C. Supercritical carbon dioxide extraction and characterization of Laurus nobilis essential oil. J. Agric. Food Chem. 2002;50:1492–1496. doi: 10.1021/jf0108563. [DOI] [PubMed] [Google Scholar]
- 142.Simić A., Soković M., Ristić M., Grujić-Jovanović S., Vukojević J., Marin P. The chemical composition of some Lauraceae essential oils and their antifungal activities. Phytother. Res. 2004;18:713–717. doi: 10.1002/ptr.1516. [DOI] [PubMed] [Google Scholar]
- 143.Koga T., Matsuse I., Teramoto Y., Ueda S. Synthesis and antimycotic activity of cinnamyl benzoate. J. Ferment. Bioengineer. 1993;76:524–526. doi: 10.1016/0922-338X(93)90253-5. [DOI] [Google Scholar]
- 144.Aziz A.N., Ibrahim H., Rosmy Syamsir D., Mohtar M., Vejayan J., Awang K. Antimicrobial compounds from Alpinia conchigera. J. Ethnopharmacol. 2013;145:798–802. doi: 10.1016/j.jep.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 145.Roy S.K., Pahwa S., Nandanwar H., Jachak S.M. Phenylpropanoids of Alpinia galanga as efflux pump inhibitors in Mycobacterium smegmatis mc2 155. Fitoterapia. 2012;83:1248–1255. doi: 10.1016/j.fitote.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 146.Jiang Z., Jiang H., Xie P. Antifungal activities against Sclerotinia sclerotiorum by Cinnamomum cassia oil and its main components. J. Essent. Oil Res. 2013;25:444–451. [Google Scholar]
- 147.Mehta G., Singh V. Hybrid systems through natural product leads: An approach towards new molecular entities. Chem. Soc. Rev. 2002;31:324–334. doi: 10.1039/b204748a. [DOI] [PubMed] [Google Scholar]
- 148.Meunier B. Hybrid molecules with a dual mode of action: Dream or reality? Acc. Chem. Res. 2008;41:69–77. doi: 10.1021/ar7000843. [DOI] [PubMed] [Google Scholar]
- 149.Wang L.-J., Geng C.-A., Ma Y.-B., Luo J., Huang X.-Y., Chen H., Zhou N.-J., Zhang X.-M., Chen J.-J. Design, synthesis, and molecular hybrids of caudatin and cinnamic acids as novel anti-hepatitis B virus agents. Eur. J. Med. Chem. 2012;54:352–365. doi: 10.1016/j.ejmech.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Voisin-Chiret A.S., Bazin M.-A., Lancelot J.-C., Rault S. Synthesis of new L-ascorbic ferulic acid hybrids. Molecules. 2007;12:2533–2545. doi: 10.3390/12112533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Teixeira J., Silva T., Benfeito S., Gaspar A., Garrido E.M., Garrido J., Borges F. Exploring nature profits: Development of novel and potent lipophilic antioxidants based on galloyl–cinnamic hybrids. Eur. J. Med. Chem. 2013;62:289–296. doi: 10.1016/j.ejmech.2012.12.049. [DOI] [PubMed] [Google Scholar]
- 152.Pérez B., Teixeira C., Gut J., Rosenthal P.J., Gomes J.R.B., Gomes P. Cinnamic acid/chloroquinoline conjugates as potent agents against chloroquine-resistant Plasmodium falciparum. ChemMedChem. 2012;7:1537–1540. doi: 10.1002/cmdc.201200257. [DOI] [PubMed] [Google Scholar]
- 153.Pérez B.C., Teixeira C., Figueiras M., Gut J., Rosenthal P.J., Gomes J.R.B., Gomes P. Novel cinnamic acid/4-aminoquinoline conjugates bearing non-proteinogenic amino acids: Towards the development of potential dual action antimalarials. Eur. J. Med. Chem. 2012;54:887–899. doi: 10.1016/j.ejmech.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 154.Springer D.M., Luh B.-Y., Goodrich J., Bronson J.J. Anti-MRSA cephems. Part 2: C-7 cinnamic acid derivatives. Bioorg. Med. Chem. 2003;11:265–279. doi: 10.1016/S0968-0896(02)00336-X. [DOI] [PubMed] [Google Scholar]
- 155.Michalska K., Karpiuk I., Król M., Tyski S. Recent development of potent analogues of oxazolidinone antibacterial agents. Bioorg. Med. Chem. 2013;21:577–591. doi: 10.1016/j.bmc.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 156.Jasir A., Kasprzykowski F., Kasprzykowska R., Lindström V., Schalén C., Grubb A. New antimicrobial cystatin C-based peptide active against gram-positive bacterial pathogens, including methicillin-resistant Staphylococcus aureus and multiresistant coagulase-negative staphylococci. Acta Pathol. Microbiol. Immunol. Scand. 2003;111:1004–1010. doi: 10.1111/j.1600-0463.2003.t01-1-apm1111110.x. [DOI] [PubMed] [Google Scholar]
- 157.Sato K., Shimizu T., Dimova V., Tomioka H. Antimicrobial activities of cinnamyl rifamycin derivatives, T-9 and T-11, against Mycobacterium tuberculosis and Mycobacterium avium complex (MAC) with special reference to the activities against intracellular MAC. Microbiol. Immunol. 2006;50:621–623. doi: 10.1111/j.1348-0421.2006.tb03836.x. [DOI] [PubMed] [Google Scholar]
- 158.Velichka D., Ivana A., Haruaki T., Katsumasa S., Venkata R., Nadadhur G., Donna D., Todor K., Arvind D., Yurii F., et al. Experimental and clinical studies on Rifacinna—The new effective antituberculous drug (review) Recent Pat. Antiinfect. Drug Discover. 2010;5:76–90. doi: 10.2174/157489110790112572. [DOI] [PubMed] [Google Scholar]
- 159.Reddy V.M., Nadadhur G., Daneluzzi D., Dimova V., Gangadharam P.R. Antimycobacterial activity of a new rifamycin derivative, 3-(4-cinnamylpiperazinyl iminomethyl) rifamycin SV (T9) Antimicrob. Agents Chemother. 1995;39:2320–2324. doi: 10.1128/AAC.39.10.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Dhople A.M. Comparative in vitro activities of rifamycin analogues against Mycobacterium leprae. Indian J. Lepr. 1997;69:377–384. [PubMed] [Google Scholar]
- 161.De P., Koumba Yoya G., Constant P., Bedos-Belval F., Duran H., Saffon N., Daffé M., Baltas M. Design, synthesis, and biological evaluation of new cinnamic derivatives as antituberculosis agents. J. Med. Chem. 2011;54:1449–1461. doi: 10.1021/jm101510d. [DOI] [PubMed] [Google Scholar]
- 162.TBAlliance Isoniazid. Tuberculosis. 2008;88:112–116. doi: 10.1016/S1472-9792(08)70011-8. [DOI] [PubMed] [Google Scholar]
- 163.TBAlliance Cycloserine. Tuberculosis. 2008;88:100–101. doi: 10.1016/S1472-9792(08)70007-6. [DOI] [PubMed] [Google Scholar]
- 164.Bairwa R., Kakwani M., Tawari N.R., Lalchandani J., Ray M.K., Rajan M.G. R., Degani M.S. Novel molecular hybrids of cinnamic acids and guanylhydrazones as potential antitubercular agents. Bioorg. Med. Chem. Lett. 2010;20:1623–1625. doi: 10.1016/j.bmcl.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 165.Slavchev I., Dobrikov G.M., Valcheva V., Ugrinova I., Pasheva E., Dimitrov V. Antimycobacterial activity generated by the amide coupling of (−)-fenchone derived aminoalcohol with cinnamic acids and analogues. Bioorg. Med. Chem. Lett. 2014 doi: 10.1016/j.bmcl.2014.09.021. in press. [DOI] [PubMed] [Google Scholar]
- 166.Tanachatchairatana T., Bremner J.B., Chokchaisiri R., Suksamrarn A. Antimycobacterial activity of cinnamate-based esters of the triterpenes betulinic, oleanolic and ursolic acids. Chem. Pharm. Bull. 2008;56:194–198. doi: 10.1248/cpb.56.194. [DOI] [PubMed] [Google Scholar]