Abstract
Although liquid‐based cytology (LBC) has increased the sensitivity of cytological diagnosis of endometrial cancer (EC) compared with conventional smear cytology, the sensitivity of LBC for the detection of EC is between 70% and 96% and remains unsatisfactory. In the present study, we compared the efficacy of LBC with liquid‐based genetic diagnosis (LBGDx) by amplicon sequencing of five genes including PTEN,PIK3CA,CTNNB1,KRAS, and TP53 in 48 LBC subjects who underwent endometrial screening. Consequently, LBC classified 15 samples as “positive or suspicious for malignancy” and the 15 were later confirmed as EC. However, LBC failed to identify five cases who were diagnosed as EC by additional transvaginal ultrasound and endometrial curettage, indicating that the sensitivity of cytology alone was 75% (15/20). LBGDx identified 11 pathogenic PTEN variants in 10 subjects, six PIK3CA variants in nine, three CTNNB1 variants in five, two KRAS variants in four, and three TP53 variants in three. Collectively, at least one pathogenic variant was identified in 19 subjects, which included 17 EC (15 endometrioid carcinoma and 2 endometrial carcinosarcomas), and one cervical adenocarcinoma. However, LBGDx did not identify any pathogenic mutations in three of the 20 EC, indicating that the sensitivity of LBGDx alone was 85% (17/20). Although five EC were negative for malignancy by LBC and three were negative for pathogenic mutations by LBGDx, the combination of LBC and LBGDx would successfully diagnose all 20 EC. These data suggested that LBGDx is a useful strategy to improve the sensitivity of screening of EC by LBC.
Keywords: amplicon sequencing, endometrial cancer, endometrial cytology, genetic diagnosis, liquid‐based cytology
1. INTRODUCTION
Endometrial cancer (EC) is the most common gynecological malignancy and its incidence is increasing.1 Although a great majority of patients with uterine‐limited EC (stage I, II) generally show a favorable prognosis, those with stage III and IV disease show poor prognosis with five‐year survival of approximately 82.9% and 26.7%, respectively.1 Factors affecting the prognosis include type 2 histological subtypes, increased depth of myometrial invasion, and lymph node metastasis.2 To improve the prognosis, early detection and diagnosis of cancerous and precancerous lesions are of great importance. Transvaginal ultrasound is a non‐invasive method to screen tumors in the uterine, fallopian tube, and ovary, but it is difficult to detect tumors at a very early stage and discriminate malignant tumors from non‐malignant changes. Although endometrial hysteroscopy and/or curettage may be used for the diagnosis, this intervention is difficult to apply for screening because it is invasive, time‐consuming, and expensive. Endometrial cytology by brush sampling has been widely applied for the screening of endometrial neoplasms.3 This method is a simple, painless, inexpensive and relatively accurate strategy to obtain materials for cancer detection, showing a sensitivity of 79% and a specificity of 99%.4 However, there are some factors that interfere with conventional smear quality, and the presence of excess blood is one of the main factors.5 Therefore, standardization of cytological diagnostic criteria and appropriate handling of samples are problems to be improved for the augmentation of its sensitivity.6, 7, 8
Liquid‐based cytology (LBC) has increased the sensitivity up to 100% for postmenopausal subjects, but the sensitivity remains at approximately 53% for premenopausal women.9 LBC has a great advantage in cell preservation and clearance of background, both of which are crucial for accurate diagnoses.5 Although LBC is useful for precise observation of cell components in the endometrial samples, evaluation of glandular architecture relies indirectly on the morphology of cell clumps. Therefore, LBC alone is insufficient for the detection of early EC, and the development of new diagnostic approaches is a matter of concern.
Next generation sequencing (NGS) has increased the sensitivity of detection of somatic mutations. NGS has also enabled the detection of somatic mutations at low frequencies in cancer tissues, and the identification of tumor DNA derived from cancer tissues (ctDNA) in circulating peripheral blood. Genomic profiles of ctDNA have been shown to closely associate with those of the malignant tumor, suggesting that ctDNA could contribute to the diagnosis and personalized treatment of cancer.10 In addition to peripheral blood, other body fluids, such as ascites, urine and pleural effusion, may contain tumor‐derived genetic information. Recent studies showed that genetic analysis of endometrial material obtained by the Papanicolaou (Pap) test could be used for the diagnosis of endometrial cancer.11, 12
To investigate the efficacy of liquid‐based genetic diagnosis (LBGDx) of EC, we selected PTEN, PIK3CA, CTNNB1, KRAS, and TP53, because these five genes are highly mutated in EC and their distribution is relatively narrow. Using 48 endometrial LBC samples, we analyzed mutations in the five genes by amplicon sequencing.
2. MATERIALS AND METHODS
2.1. Patients and their clinical information
This project was approved by the institutional review boards of Sapporo Medical University (SMU‐IRB, 292‐77) and Institute of Medical Science, the University of Tokyo (IMSUT‐IRB, 29‐52‐A1123). All clinical investigation was conducted according to the principles expressed in the Declaration of Helsinki. A total of 184 individuals were enrolled in this study at their endometrial screening in Sapporo Medical University Hospital from October 2017 to March 2018. Written informed consent was obtained from all participants. Endometrial LBC samples were collected. When the cytological result was positive or suspicious for malignancy, or when there were other clinical data raising suspicion for endometrial neoplasms, the patients underwent transvaginal ultrasound and/or endometrial curettage. For the analysis of LBGDx, we selected 48 women from whom more than 5 μg DNA was recovered from their LBC samples. The 48 subjects included patients with endometrioid carcinomas (n = 18), endometrial carcinosarcoma (n = 2), cervical cancer (n = 2), ovarian cancer (n = 2), benign gynecological diseases (n = 21), and women without involvement of gynecological organs (n = 3) (Table 1). Benign gynecological diseases comprised myoma uteri (n = 12), adenomyosis (n = 2), CIN cervical intraepithelial neoplasia (n = 2), and lobular endocervical glandular hyperplasia (n = 2), endometrial hyperplasia (n = 2), and benign ovarian tumor (n = 1).
Table 1.
Diagnosis, histology, and stage of the 48 subjects
| Diagnosis | Histology | Stageb | No. of cases |
|---|---|---|---|
| Endometrial cancer (n = 20) | Endometrioid carcinoma (grade 1‐2a) | Ia | 12 |
| Ib | 1 | ||
| IIIa | 2 | ||
| IIIc | 1 | ||
| IVb | 1 | ||
| Endometrioid carcinoma (grade 3a) | Ib | 1 | |
| Carcinosarcoma | Ib | 1 | |
| IIIb | 1 | ||
| Cervical cancer | Adenocarcinoma | Ia | 2 |
| Ovarian cancer | 2 | ||
| Benign gynecological disease | 21 | ||
| No gynecological disease | 3 | ||
| Total | 48 | ||
WHO (World Health Organization) classification 2014.
FIGO (International Federation of Gynecology and Obstetrics) 2008.
2.2. Sampling and cytological examination
For endometrial LBC, Scree brush (Soft Medical Co., Ltd, Tokyo, Japan) was gently inserted to the level of the uterine fundus. The outer sheath was then pulled back, and the brush was rotated in the uterine cavity. Then the outer sheath was pushed in again, and the brush was removed. The sample was placed into 10 mL of LBC Prep liquid (Muto Pure Chemicals Co., Ltd, Tokyo, Japan), and 8 mL of the liquid was used for cytological diagnosis and the remaining 2 mL was used for genetic analysis. Cytological diagnosis was carried out and evaluated according to the guidelines provided by The Japanese Society of Clinical Cytology. At the Institute of Medical Science, the University of Tokyo, genetic analysis was carried out independently in a blinded method without prior cytological, histological, or clinical information.
2.3. Extraction of DNA and PCR
DNA was extracted from the 2 mL of the LBC sample using a QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations, and the amount of DNA was subsequently quantified using NanoDrop (Thermo Fisher Scientific K.K., Tokyo, Japan). Regions including mutation hot spots in PTEN, PIK3CA, CTNNB1, KRAS, and TP53 were amplified using gene‐specific primer sets (Table S1). DNA (25 ng) was mixed with 10 pmol of forward and reverse primers in a reaction mixture of 25 μL containing five units of KOD plus NEO Taq polymerase (TOYOBO, Osaka, Japan). All reactions involved initial denaturation at 95°C for 2 minutes followed by 35 cycles at 98°C for 30 seconds, 60°C for 30 s, and 68°C for 30 seconds, and final extension at 68°C for 5 minutes on a Veriti Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA). The PCR products were quantified using LabChip GX Touch (PerkinElmer, Waltham, MA, USA).
2.4. Sequencing and analysis of data
Amplicon sequencing was carried out using MiSeq platforms with paired‐end reads of 101 bp according to the manufacturer's instructions (Illumina, San Diego, CA, USA). Sequence libraries were prepared using 1 μg of a mixture of PCR products from the same sample and barcodes. For data processing, fastq files were aligned to human reference sequence (hg19) and bam files were created using MiSeq Reporter (Illumina). Variants were determined by comparison of the bam files with the reference sequence using MiSeq Reporter.
3. RESULTS
3.1. Cytological diagnosis
Cytological assessment of the 48 LBC showed that 16 samples were “positive or suspicious for malignancy” and the remaining 32 were “negative for malignancy” (Figure 1). However, LBC in combination with subsequent transvaginal ultrasound and curettage/biopsy analysis finally diagnosed 22 uterine neoplasms including 20 EC (18 endometrioid carcinomas and two endometrial carcinosarcomas), and two cervical adenocarcinomas. Clinicopathological features of the 20 EC are shown in Table S2. Thirteen of the 18 endometrioid carcinomas were at early stages (Ia and Ib) of grade 1‐2. In the remaining 26 cases, no uterine neoplasms were identified by cytology or transvaginal ultrasound. Among the 20 EC, 15 were “positive or suspicious for malignancy” (Table S2) indicating that the sensitivity of cytology alone was 75%. The remaining five cases were all stage I cases that were diagnosed by transvaginal ultrasound and curettage/biopsy analysis, although they were negative for malignancy by LBC. The five false‐negative cases included three of seven premenopausal EC patients and two of 13 postmenopausal patients (Table S2). This data may imply a tendency for decreased sensitivity for cytological EC diagnosis in premenopausal patients than postmenopausal patients, although a significant difference was not observed. The two carcinosarcomas were correctly diagnosed as “positive or suspicious for malignancy” by LBC. Only one of the two cervical adenocarcinomas was identified as positive by the cytology. These data suggest that there is a limit for the early detection of EC by LBC alone.
Figure 1.
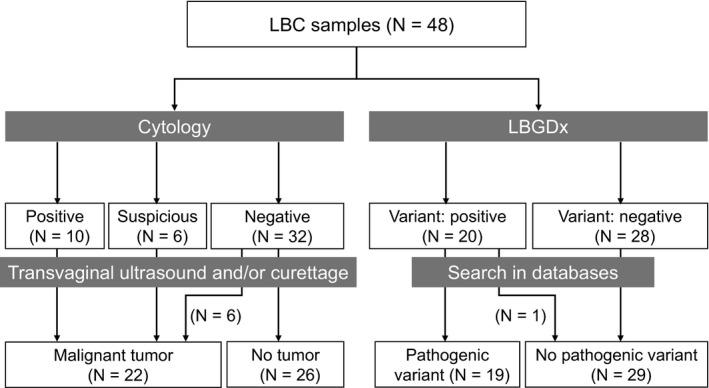
Number of subjects classified by cytology in combination with transvaginal ultrasound and/or endometrial curettage, and that by liquid‐based genetic diagnosis (LBGDx). LBC, liquid‐based cytology
3.2. Detection of variants in the five genes
First, we tested the extraction of DNA from 2 mL of LBC samples and successfully obtained more than 400 ng of DNA in 153 (83.1%) of the 184 samples, and more than 200 ng of DNA in 175 (95.1%) of the 184 samples (Table S3). The amount ranged from 0 to 63.4 μg and the average was 4.7 μg. In the present study, we randomly selected 48 samples with an amount of DNA more than 5 μg and screened variants in the five genes including PTEN, PIK3CA, CTNNB1, KRAS, and TP53. As shown in Table S4, the average of read‐depth in the 13 regions ranged from 4703 to 59 501. Although we could not obtain enough sequence reads to evaluate exon 3 of KRAS in two subjects (LB‐130 and LB‐51), we had enough reads across the 13 regions in the 48 samples yielding an assessment rate of 99.7% (622/624). Consequently, we identified a total of 11 PTEN variants in 10 women, six PIK3CA variants in nine, three CTNNB1 variants in five, two KRAS variants in four, and three TP53 variants in three (Table 2).
Table 2.
List of variants in five genes that are highly mutated in endometrial carcinoma
| Gene | Variant | Type | Significancea | No. of subjects | Sample ID |
|---|---|---|---|---|---|
| PTEN | c.275A>G (p.D92G) | Nonsynonymous | Pathogenic | 1 | LB‐65 |
| c.388C>G (p.R130G) | Nonsynonymous | Pathogenic | 1 | LB‐78 | |
| c.388C>T (p.R130*) | Nonsense | Pathogenic | 1 | LB‐184 | |
| c.389G>C (p.R130P) | Nonsynonymous | Pathogenic | 1 | LB‐176 | |
| c.469G>T (p.E157*) | Nonsense | Pathogenic | 1 | LB‐105 | |
| c.517C>T (p.R173C) | Nonsynonymous | Pathogenic | 1 | LB‐176 | |
| c.518G>A (p.R173H) | Nonsynonymous | Pathogenic | 1 | LB‐184 | |
| c.569delC (p.P190 fs*9) | Deletion | Pathogenic | 1 | LB‐51 | |
| c.697C>T (p.R233*) | Nonsense | Pathogenic | 3 | LB‐1, LB‐177, LB‐184 | |
| c.800delA (p.K267 fs*9) | Deletion | Pathogenic | 1 | LB‐140 | |
| c. 850G>T (p.E284*) | Nonsense | Pathogenic | 1 | LB‐3 | |
| PIK3CA | c.1624G>A (p.E542K) | Nonsynonymous | Pathogenic | 2 | LB‐105, LB‐154 |
| c.1633G>A (p.E545K) | Nonsynonymous | Pathogenic | 1 | LB‐41 | |
| c.1638G>T (p.Q546H) | Nonsynonymous | Pathogenic | 1 | LB‐64 | |
| c.3075C>T (p.T1025T) | Synonymous | Neutral | 1 | LB‐103 | |
| c.3140A>G (p.H1047R) | Nonsynonymous | Pathogenic | 3 | LB‐2, LB‐53, LB‐173 | |
| c.3145G>C (p.G1049R) | Nonsynonymous | Pathogenic | 1 | LB‐184 | |
| CTNNB1 | c.97T>G (p.S33A) | Nonsynonymous | Pathogenic | 2 | LB‐78, LB‐173 |
| c.98C>G (p.S33C) | Nonsynonymous | Pathogenic | 2 | LB‐64, LB‐154 | |
| c.110C>T (p.S37F) | Nonsynonymous | Pathogenic | 1 | LB‐53 | |
| KRAS | c.35G>A (p.G12D) | Nonsynonymous | Pathogenic | 3 | LB‐41, LB‐171, LB‐184 |
| c.175G>A (p.A59T) | Nonsynonymous | Pathogenic | 1 | LB‐176 | |
| TP53 | c.473G>T (p.R158L) | Nonsynonymous | Pathogenic | 1 | LB‐178 |
| c.584T>C (p.I195T) | Nonsynonymous | Pathogenic | 1 | LB‐59 | |
| c.818G>A (p.R273H) | Nonsynonymous | Pathogenic | 1 | LB‐3 |
FATHMM prediction in COSMIC database (https://cancer.sanger.ac.uk/cosmic).
Regarding the PTEN mutations in the LBC samples, the allele frequencies of each mutation ranged from 8% to 56% with an average of 31.1%, suggesting a high proportion of tumor cells in the LBC samples. Although the average frequencies of pathogenic mutations in PIK3CA, CTNNB1, and TP53, were 32.1%, 25.8%, and 44.7%, respectively, the frequency of KRAS mutation ranged from 8% to 9% with an average of 8.5%. Notably, in LB‐176, the frequency of KRAS mutation was 8% but the frequencies of two PTEN mutations were 24% and 26%. These data suggest that the difference in mutation frequencies may reflect intratumor heterogeneity of EC, and that KRAS mutation(s) may be acquired after the accumulation of trunk mutation(s) in the PI3K‐AKT and/or other pathways.
3.3. Analysis of variations
The 11 PTEN variants comprised five nonsynonymous variants (p.D92G, p.R130G, p.R130P, p.R173C, and p.R173H), four nonsense variants (p.R130*, p.E157*, p.R233*, and p.E284*), and two deletions (c.568delC and c.800delA). The 11 variants were categorized as pathogenic mutations according to the COSMIC database in the Sanger Institute. Because the 10 individuals did not show clinical features associated with Cowden disease, a hereditary disease caused by germline mutations in PTEN, the 11 variants were considered somatic mutations. Although eight of the 10 cases had one PTEN mutation, a case (LB‐176) carried two (p.R130P and p.R173C) and the remaining case (LB‐184) had three PTEN mutations (p.R130*, p.R173H, and p.R233*), suggesting that PTEN is a typical tumor suppressor gene that is inactivated by genetic alterations in both alleles. Regarding PIK3CA, we identified a total of six variants in nine women. Five of the six were nonsynonymous variants and were categorized as pathogenic mutations, but the remaining one was a synonymous single nucleotide polymorphism (rs17849079) and was regarded as a non‐pathogenic variant in the COSMIC database. Mutations of CTNNB1, KRAS, and TP53 were found in five, four, and three cases, respectively, and all were regarded as pathogenic mutations. Taken together, 19 of the 48 subjects carried at least one pathogenic variant in the five genes (Figure 1).
3.4. Association of mutations with clinical information
Among the 19 subjects with at least one pathogenic mutation, 15 were diagnosed as endometrioid carcinoma, two as endometrial carcinosarcoma, and one as cervical adenocarcinoma. Although the remaining case carried a KRAS mutation, no cytological or ultrasound abnormalities were identified in the patient who suffered from familial adenomatous (FAP) polyposis of the colon. In the 15 endometrioid carcinoma cases, 11 patients were at stage I (Figure 2, Table S2), implying that LBGDx can detect endometrioid cancer at early stages. It is of note that three of the 20 EC had no pathogenic mutation in the five genes, suggesting that screening of the five genes or hot spot regions may not be sufficient to identify all EC cases.
Figure 2.
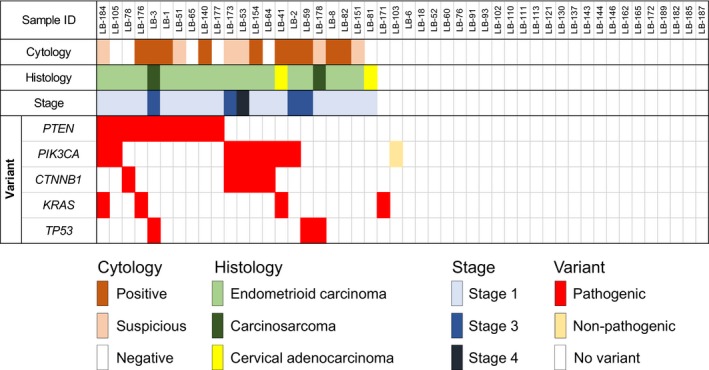
Variations in the five genes and their association with cytological, histological, and WHO stage classifications
3.5. Comparison between LBC with LBGDx
Liquid‐based cytology correctly diagnosed 15 of the 20 EC (nine malignant and six suspicious cases) but failed to identify five early endometrial cancers (75% sensitivity) (Figure 2, Table S2). All 16 cytologically positive or suspected cases turned out to be endometrial or cervical tumors (15 EC and one cervical cancer) suggesting 100% specificity of LBC. Meanwhile, LBGDx successfully diagnosed 17 of the 20 EC (85% sensitivity). Among the 19 mutation‐positive cases, 18 turned out to be endometrial or cervical disease, suggesting 96% specificity of LBGDx. It is worthwhile to note that all 20 EC could be correctly diagnosed when LBC was combined with LBGDx.
4. DISCUSSION
In the present study, we identified frequent mutations in PTEN (10/20), PIK3CA (7/20), CTNNB1 (5/20), KRAS (2/20), and TP53 (3/20) in EC by amplicon sequencing of the five genes and showed that LBGDx contributes to enhanced sensitivity of cytological screening for endometrial cancer. Although the number of samples analyzed in this study is limited, the frequencies of mutations in the five genes are in good agreement with previous reports.13, 14 It is of note that frequent mutations of TP53 have been found in aggressive endometrial carcinomas including high‐grade serous types and carcinosarcomas.15 In addition, TP53 (75.5%) and PIK3CA (34.0%) mutations are most frequently observed in uterine carcinosarcoma.16 Consistent with these reports, we identified TP53 mutations in the two patients with carcinosarcomas and in a patient with endometrioid carcinoma at stage IIIc2. Additionally, PIK3CA and KRAS mutations were identified in one of the two cases with cervical adenocarcinoma.
Kinde et al reported that DNA extracted from liquid uterine specimen is useful for the diagnosis of EC and ovarian cancer.11 This group searched for somatic mutations in 12 genes by sequencing DNA from Pap brush smear specimen, which they called “PapGene” test. Their sampling method of liquid specimen collection was designed for the detection of cervical cancer using a brush to collect cells from the ectocervix. In contrast, the sampling method in the present study used a brush for the screening of endometrial cancer. Therefore, we expected to recover a greater number of endometrial cells shedding from the endometrioid layer of the uterus in LBC samples compared with Pap brush sampling. Consistent with this view, frequencies of mutation alleles in our analysis were much higher than those in the analysis by Kinde et al (an average of 31.1% vs 0.04% for PTEN, 32.1% vs 6.8% for PIK3CA, 25.8% vs 4.3% for CTNNB1, and 44.7% vs 23% for TP53).11 Although the frequencies of mutant alleles were less than ours, they identified at least one somatic mutation in all 12 EC patients. As they analyzed a greater number of genes than in our study (12 genes vs 5 genes), the increase of the target genes and/or regions for amplicon sequencing could have augmented the sensitivity.
The same group recently published the results of a larger study including 382 patients with EC, 245 with ovarian cancer, and 714 control women without cancer.12 They applied Pap brush samples for panel sequencing of 18 genes. They detected somatic mutations in 81% of patients with EC, and the median of mutated allele frequency was 0.54%. Furthermore, they carried out the same panel sequencing with samples using a Tao brush for endometrial cytology from 123 patients with EC, 51 with ovarian cancers, and 125 control women without cancer. Consequently, they identified at least one driver gene mutation in 93% of samples from EC patients, and the median of mutant allele frequency was 24.7%, which was much higher than the Pap brush samples. Our data are comparable with theirs, as we identified pathogenic mutations in 85% of EC in LBC samples by amplicon sequencing of five genes, and the median of mutant allele frequencies in the five genes was 26%.
Maritschnegg et al also determined tumor‐specific mutations using cells that were collected through a lavage of the uterine cavity.17 They analyzed a total of 65 patients, including 30 with ovarian cancer, five with EC at stage Ia, three with other malignancies, and 27 with benign lesions involving gynecological organs. As a result, they identified mutations in all five EC patients. Nair et al applied cellular and cell‐free DNA in uterine lavage fluid recovered from 107 women undergoing hysteroscopy for ultra‐deep amplicon sequencing of 56 and 12 gene sets.18 They identified cancer‐associated somatic mutations in all seven endometrial cancer subjects including six endometrioid cancer at stage Ia and one grade 3 carcinosarcoma at stage IIIa.
We identified a driver mutation (KRAS G12D) with a mutant allele frequency of 12% in one of the three women without any gynecological disease. No pathogenic mutations were found in the remaining two women without gynecological disease or in the 21 women with benign disease. Wang Y. and colleagues reported that somatic mutations were detected in nine Pap brush samples from 714 control women without cancer.12 However, Maritschnegg et al documented that they determined somatic mutations in eight of 27 patients with benign disease, and that six of the eight mutations were detected in KRAS.17 Surprisingly, Nair et al. disclosed that uterine lavage fluid from 51 of 95 women without histopathological evidence of cancer carried mutations in cancer‐associated genes.18 The mutations included KRAS G12S (10 cases), KRAS G12C (eight cases), and PIK3CA H1047R (eight cases), and their allele fractions were relatively high ranging from 1.0% to 30.4% (average: 3.0%). They identified driver and/or potential driver mutations in 31 of 34 women with endometrial polyp(s), premalignant lesions of endometrial cancer.18 A recent study showed that 10 of 39 infiltrating endometriosis had somatic mutations in genes associated with endometrial cancers such as ARID1A, PIK3CA, KRAS, and PPP2R1A, suggesting that somatic mutations in these genes are involved in endometriosis, a benign disease with clonal proliferation of endometrial cells.19 Recently, Suda et al sequenced ovarian endometriotic and normal uterine endometrial epithelium samples obtained from subjects with benign gynecological disease.20 They also identified somatic mutations in cancer‐associated genes such as KRAS and PIK3CA in histologically normal endometrial glands. Therefore, we should carefully assess the subjects with mutations in these genes. These data may imply that the KRAS mutation found in the FAP patient may be derived from non‐pathogenic or premalignant lesion(s). In addition, we may need to consider growth of non‐neoplastic endometrial epithelial cell(s) carrying a few driver mutations, similar to clonal hematopoiesis observed in bone marrow.21 Nevertheless, we should not exclude the possibility of occult cancer in the uterus or ovary of this patient. Therefore, we recommended her to undergo careful checkup for gynecological tumors.
Germline mutations in PTEN are responsible for Cowden syndrome. In the present study, we found 11 PTEN variants at a frequency ranging from 8% to 56%, and five cases showed more than 40%. Therefore, it is difficult to discriminate somatic mutations from germline variants in the five cases. However, Cowden syndrome is a rare autosomal dominant disorder and the frequency of this disease is considered to be approximately one in 200 000 individuals.22 In addition, no abnormality was found in 240 patients with endometrial cancer in the screening of PTEN mutations.23 Therefore, we assumed that the PTEN variants were somatic mutations.
We have shown herein that LBGDx could diagnose EC at a sensitivity of 85%. Among 17 EC cases diagnosed by LBGDx, five cases were negative for cytological diagnosis, corroborating that it is difficult to diagnose EC by cytology alone.6, 7, 8 Kilicci et al reported that the sensitivity is lower in premenopausal cases than in postmenopausal cases,9 and our study showed a similar result. If the cytological examination was combined with LBGDx, all endometrial neoplasms could be correctly diagnosed and additional histological examination was not necessary for five of the six suspicious cases by cytological examination alone. Presumably, increase in the number of genes will increase the sensitivity of genetic diagnosis. However, without the analysis of the corresponding non‐cancerous tissue, it may become difficult to judge variations detected in LBC samples alone.
In this report, we have shown that LBGDx should be a useful strategy to improve the diagnostic sensitivity and specificity of endometrial screening and that LBC coupled with LBGDx will decrease invasive examinations for screening of endometrial neoplasm. In addition, LBGDx may help to detect occult lesions and/or premalignant lesions, and thus contribute to the identification of malignant diseases at early stages. Furthermore, identification of pathogenic mutations at endometrial screening may lead to the selection of anticancer drugs for neoadjuvant chemotherapy prior to surgical intervention. Although assessment of pathogenic mutations in women without evidence of malignancy needs to be studied in future projects, implementation of genetic analysis for screening of endometrial cancer will reduce the mortality and increase the quality of life of patients with EC by early detection of neoplastic lesions.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.
Supporting information
ACKNOWLEDGMENTS
We thank Seira Hatakeyama (The University of Tokyo) for her technical assistance. This study was supported in part by a Grant for Joint Research Project of the Institute of Medical Science, the University of Tokyo.
Matsuura M, Yamaguchi K, Tamate M, et al. Efficacy of liquid‐based genetic diagnosis of endometrial cancer. Cancer Sci. 2018;109:4025–4032. 10.1111/cas.13819
Funding information
Grant for Joint Research Project of the Institute of Medical Science, the University of Tokyo, (Grant/Award Number: ‘2018‐2110’).
REFERENCES
- 1. Matsuura M, Takahashi A, Nomura H, et al. Analysis of a single para‐aortic lymph node metastasis in endometrial cancer. J Cancer Sci Ther. 2018;10(2):19‐21. [Google Scholar]
- 2. Matsuura M, Suzuki T, Morishita M, Tanaka R, Ito E, Saito T. Chemotherapy (CT) with radiotherapy versus CT alone for FIGO stage3c endometrial cancer. Eur J Gynaec Oncol. 2009;30(1):40‐44. [PubMed] [Google Scholar]
- 3. Fulciniti F, Yanoh K, Karakitsos P, et al. The Yokohama system for reporting directly sampled endometrial cytology: the quest to develop a standardized terminology. Diagn Cytopathol. 2018;46(5):400‐412. [DOI] [PubMed] [Google Scholar]
- 4. Yanoh K, Hirai Y, Sakamoto A, et al. New terminology for intrauterine endometrial samples: a group study by the Japanese society of clinical cytology. Acta Cytol. 2012;56(3):233‐241. [DOI] [PubMed] [Google Scholar]
- 5. Lv S, Wang R, Wang Q, et al. A novel solution configuration on liquid‐based endometrial cytology. PLoS ONE. 2018;13(2):e0190851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Norimatsu Y, Yanoh K, Kobayashi TK. The role of liquid‐based preparation in the evaluation of endometrial cytology. Acta Cytol. 2013;57(5):423‐435. [DOI] [PubMed] [Google Scholar]
- 7. Byrne AJ. Endocyte endometrial smears in the cytodiagnosis of endometrial carcinoma. Acta Cytol. 1990;34(3):373‐381. [PubMed] [Google Scholar]
- 8. Fujiwara H, Takahashi Y, Takano M, et al. Evaluation of endometrial cytology: cytohistological correlations in 1441 cancer patients. Oncology. 2015;88(2):86‐94. [DOI] [PubMed] [Google Scholar]
- 9. Kilicci C, Cogendez E, Kumru P, Bostanci EE, Koc N, Abide YC. Cytological assessment of endometrial washings obtained with Karman cannula using a liquid‐based preparation method for the detection of endometrial pathologies. Arch Gynecol Obstet. 2018;298(1):171‐177. [DOI] [PubMed] [Google Scholar]
- 10. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531‐548. [DOI] [PubMed] [Google Scholar]
- 11. Kinde I, Bettegowda C, Wang Y, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013; 5(167): 167ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y, Li L, Douville C, et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci Transl Med. 2018; 10(433): eaap8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cancer Genome Atlas Research Network , Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013; 497(7447): 67‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Forbes SA, Beare D, Boutselakis H, et al. COSMIC: somatic cancer genetics at high‐resolution. Nucleic Acids Res. 2017;45(D1):D777‐D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matias‐guiu X, Catasus L, Bussaglia E, et al. Molecular pathology of endometrial hyperplasia and carcinoma. Hum Pathol. 2001;32(6):569‐577. [DOI] [PubMed] [Google Scholar]
- 16. Le Gallo M, Rudd ML, Urick ME, et al. The FOXA2 transcription factor is frequently somatically mutated in uterine carcinosarcoma and carcinomas. Cancer. 2018;124(1):65‐73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maritschnegg E, Wang Y, Pecha N, et al. Lavage of the uterine cavity for molecular detection of Mullerian duct carcinomas: A proof‐of‐concept study. J Clin Oncol. 2015;33(36):4293‐4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nair N, Camacho‐Vanegas O, Rykunov D, et al. Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: a prospective cross‐sectional study. PLoS Med. 2016;13(12):e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anglesio MS, Papadopoulos N, Ayhan A, et al. Cancer‐associated mutations in endometriosis without cancer. N Engl J Med. 2017;376:1835‐1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suda K, Nakaoka H, Yoshihara K, et al. Clonal expansion and diversification of cancer associated mutations in endometriosis and normal endometrium. Cell Rep. 2018;24(7):1777‐1789. [DOI] [PubMed] [Google Scholar]
- 21. Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science. 2015;349(6255):1483‐1489. [DOI] [PubMed] [Google Scholar]
- 22. Pilarski R. Cowden syndrome: a critical review of the clinical literature. J Genet Couns. 2009;18(1):13‐27. [DOI] [PubMed] [Google Scholar]
- 23. Black D, Bogomolniy F, Robson ME, Offit K, Barakat RR, Boyd J. Evaluation of germline PTEN mtations in endometrial cancer patients. Gynecol Oncol. 2005;96(1):21‐24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


