Abstract
Long non‐coding RNA MIF‐AS1 (lncMIF‐AS1) has been found to be upregulated in the tumor tissues of gastric cancer; however, its importance for the progression of gastric cancer remains unknown. Thus, the present study was designed to determine the role of the lncMIF‐AS1‐based signal transduction pathway in mediating the proliferation and apoptosis of gastric cancer cells. Differentially expressed lncRNAs and mRNAs were screened out using microarray analysis, based on the published data (GSE63288), and validated using quantitative RT‐PCR. Target relationships between lncRNA‐micro RNA (miRNA) and miRNA‐mRNA were predicted by bioinformatics analysis and verified by dual‐luciferase reporter assay. Protein expression of NDUFA4, COX6C and COX5B was detected by western blot. Cell proliferation, cell cycle and apoptosis were determined using colony formation assay and flow cytometry analysis. Oxidative phosphorylation in gastric cancer cells was assessed by levels of oxygen consumption and ATP synthase activity. Expression of lncMIF‐AS1 and NDUFA4 were upregulated in gastric cancer tissues and cells as compared with non‐cancerous gastric tissues and cells (P < .05). MiR‐212‐5p was identified as the most important miRNA linker between lncMIF‐AS1 and NDUFA4, which was negatively regulated by lncMIF‐AS1 and its depletion is the main cause of NDUFA4 overexpression (P < .01). The upregulated expression of NDUFA4 then greatly promoted the proliferation and decreased the apoptosis of gastric cancer cells through activation of the oxidative phosphorylation pathway. Taken together, the present study implies that inhibition of lncMIF‐AS1/miR‐212‐5p/NDUFA4 signal transduction may provide a promising therapeutic target for the treatment of gastric cancer.
Keywords: cell proliferation, gastric cancer, lncMIF‐AS1, miR‐212‐5p, NDUFA4
1. INTRODUCTION
Gastric cancer is the fifth most common cancer1 and the third leading cause of cancer‐related death worldwide.2 The incidence of gastric cancer varies considerably by geographical region; the incidence has decreased globally, but in the Chinese population it is increasing.3 To date, surgical resection is the most effective therapy for early gastric cancer without distant or locoregional metastasis.4 However, for patients with advanced gastric cancer, the 5‐year recurrence‐free survival rate is only approximately 25% even when the whole tumor is completely resected and adjuvant chemoradiotherapy is given.5 Endoscopic screening is the most reliable method for gastric cancer diagnosis, but its cost and invasive nature have largely limited its use.6 Therefore, there is an urgent need for developing less‐invasive but more efficient biomarkers to detect gastric cancer at an early stage.
In the past few years, increasing evidence indicates that long non‐coding (lnc)RNAs, a class of non‐coding RNAs that are greater than 200 nucleotides in size, can mediate the progression of tumorigenesis and metastasis in a variety of cancers through inducing the dysregulation of gene products at the transcriptional, post‐transcriptional or epigenetic levels.7, 8, 9 Aberrant expression profiles of lncRNAs in gastric cancer have been uncovered by several previous studies.10, 11, 12 For example, upregulation of oncogenic ZFAS1 significantly promoted the proliferation of gastric cancer cells by repressing the expression of KLF2 and NKD2.10 However, our understanding of the functional characteristics of lncRNAs in gastric cancer is still insufficient, and many members of the dysexpressed lncRNAs remain uncharacterized.
In the present study, based on the analysis of publicly available microarray data from Gene Expression Omnibus (GEO) datasets (https://www.ncbi.nlm.nih.gov/gds), upregulated lncMIF‐AS1 and NDUFA4 mRNA in gastric cancer largely attracted our attention. To the best of our knowledge, the role of lncMIF‐AS1 in the initiation and progression of cancers remains unknown. The present study showed that lncMIF‐AS1 can positively regulate the expression of NDUFA4. NDUFA4 encodes a protein belonging to the respiratory chain of mitochondria,13, 14 which has been implicated in the development of various cancers including renal cell carcinoma and lung cancer.15, 16, 17 Herein, our study demonstrates that the expression of NDUFA4 is positively regulated by lncMIF‐AS1 in a micro RNA (miR)‐212‐5p‐targeting competitive endogenous way. The overexpressed NDUFA4 further promotes the proliferation and inhibits the apoptosis of gastric cancer cells through activation of the oxidative phosphorylation pathway.
2. MATERIALS AND METHODS
2.1. Clinical samples
Twenty tumor tissues and their adjacent non‐cancerous tissues were collected from patients with primary gastric cancer undergoing radical gastrostomy at the Department of General Surgery of The First People's Hospital of Yunnan Province, The Affiliated Hospital of Kunming University of Science and Technology. No patients received any preoperative treatment including radiotherapy, chemotherapy or other adjuvant anticancer therapies. All samples were immediately frozen in liquid nitrogen and stored at −80°C until subsequent analysis. Sample collection, usage and all procedures carried out in studies involving human participants were approved by the ethics committee of Cancer Hospital of the First People's Hospital of Yunnan Province, The Affiliated Hospital of Kunming University of Science and Technology (IRB approval number: 2017KY047), and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all patients.
2.2. Cell lines and culture
Human normal gastric mucosa cell line (GES‐1), gastric adenocarcinoma cell lines (AGS, SGC‐7901) and gastric cancer cell lines (MKN‐28, MKN‐45) were purchased from BeNa Culture Collection (Beijing, China). All cells were grown in RPMI‐1640 medium (Sigma‐Aldrich, St Louis, MO, USA) supplemented with 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA), in a humidified atmosphere of 5% CO2 at 37°C.
2.3. Microarray analysis
MicroRNA and mRNA expression profiles were established by microarray analysis, based on the published data of Chang et al,18 which were generated from the tumor tissues of patients with early gastric cancer by high‐throughput RNA sequencing. These data were downloaded from GEO datasets under accession number GSE63288 (https://www.ncbi.nlm.nih.gov/gds). Here, three tumor samples and their matched non‐cancerous adjacent samples were randomly selected and then analyzed on the GPL13393 platform. All lncRNAs and mRNAs with changed expression were shown by heat map.
2.4. Dual‐luciferase reporter assay
Firefly luciferase‐carrying pmirGLO and Renilla luciferase‐carrying pRL‐TK vectors were commercially provided by Promega (Madison, WI, USA). Complete fragments of lncMIF‐AS1 and the 3′‐UTR of NDUFA4 were prepared by PCR amplification and then subcloned into the pmirGLO vector downstream of firefly luciferase, named p‐MIF‐AS1‐WT and p‐NDUFA4‐WT. Mimics of miR‐212‐5p, miR‐29a‐3p, miR‐339a‐5p, and miR‐199a‐5p were purchased from Ribobio (Guangzhou, China). HEK293T cells were preplated in 24‐well plates at a density of 1 × 105 cells per well and transfected 24 hours later. Transfection was carried out using Lipofectamine 3000 reagents (Invitrogen, Carlsbad, CA, USA). Each reaction contained 10 ng p‐MIF‐AS1‐WT or p‐NDUFA4‐WT vectors, 20 ng Renilla vector (pRL‐TK, Promega) and 50 nmol/L miRNA mimics. Forty‐eight hours post‐transfection, cells were harvested and activities of Firefly and Renilla luciferase were measured using the dual‐luciferase reporter assay system (Promega) and a luminometer (PerkinElmer Life Sciences, Boston, MA, USA), according to the manufacturer's protocol.
2.5. RNA extraction and quantitative RT‐PCR
Total RNA was extracted from the tissues and cells using TRIZOL™ (Invitrogen). cDNA was synthesized by using PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Tokyo, Japan). Quantitative PCR was carried out by using QuantiTect SYBR Green RT‐PCR Kit (QIAGEN, Düsseldorf, Germany). Primer sequences are shown in Table 1. Relative expression was normalized to U6 (for miRNA) or GAPDH (for lncRNA and mRNA) by method.
Table 1.
Primer sequences for qRT‐PCR
| Gene | Sequence (5′‐3′) |
|---|---|
| MIF‐AS1 | |
| Forward primer | ACATCGGCATGATGGCAGAA |
| Reverse primer | TCACAAAAGGCGGGACCAC |
| miR‐212‐5p | |
| Forward primer | ACCTTGGCTCTAGACTGCTT |
| Reverse primer | GTATCCAGTGCGAATACCTC |
| NDUFA4 | |
| Forward primer | AAGCATCCGAGCTTGATCCC |
| Reverse primer | TGGGACCCAGTTTGTTCCAG |
| GAPDH | |
| Forward primer | TCGGAGTCAACGGATTTGGT |
| Reverse primer | TTCCCGTTCTCAGCCTTGAC |
| U6 | |
| Forward primer | CACAGCACACCAGAATCA |
| Reverse primer | GCAGTCCTTGAATCCTTGT |
2.6. Western blot
Tissues and cells were lysed by RIPA lysate buffer (Sigma‐Aldrich). Protein in the lysates was quantified using a BCA protein assay kit (Pierce, Rockford, IL, USA), and 100 μg total protein was separated using 10% SDS‐PAGE electrophoresis and transferred onto PVDF membrane. Membranes were blocked in 5% milk under room temperature for 1 hour, followed by the incubation with the following primary antibodies at recommended concentration in TBST overnight at 4°C: anti‐NDUFA4 (1:500, PA5‐35296; Invitrogen), anti‐NDUFA8 (1:500, PA5‐59642; Invitrogen), anti‐COX6C (1:250, PA5‐49921; Invitrogen), anti‐COX5B (1:250, PA5‐56963; Invitrogen) and anti‐GAPDH (1:250, A21994; Invitrogen). After washing with PBST three times, membranes were incubated with fluorescent‐tagged secondary antibody (1:200, A32727; Invitrogen) and detected using a chemiluminescence plus system (Life Technologies Corporation, Gaithersburg, MD, USA). GAPDH was used as an endogenous control.
2.7. Cell transfection
MicroRNA‐212‐5p mimics and control mimics were purchased from GenePharma (Shanghai, China). Full‐length sequences of lncMIF‐AS1 (2192 bp; NR_038911.1) and NDUFA4 (CDS: 246 bp; NM_002489.3) were cloned into pcDNA3.1 (Invitrogen), named pcDNA3.1‐MIR‐AS1 and pcDNA3.1‐NDUFA4. AGS cells were divided into six different groups as follows: (i) negative control (NC) group; (ii) MIF‐AS1 group transfected with p‐MIF‐AS1; (iii) miR‐212‐5p group transfected with miR‐212‐5p mimics; (iv) NDUFA4 group transfected with pcDNA3.1‐NDUFA4; (v) mix 1 group, cotransfected with miR‐212‐5p mimics and pcDNA3.1‐MIF‐AS1; and (vi) mix 2 group cotransfected with pcDNA3.1‐NDUFA4 and miR‐212‐5p mimics. Moreover, for detecting interactions among lncMIF‐AS1, miR‐212‐5p and NDUFA4, specific lncMIF‐AS1‐targeting shRNA or miR‐212‐5p inhibitor were individually transfected into the AGS cells. Transfection was carried out after the cells reached 70% confluence using Lipofectamine 3000 reagents (Invitrogen).
2.8. Colony formation assay
Cell proliferation was assessed using colony formation assay. Briefly, the transfected cells were cultivated for 48 hours, harvested, trypsinized, and resuspended into fresh RPMI‐1640 medium containing 10% FBS at a density of 500 cells per milliliter. Then, 2 mL cell suspension was plated into six‐well cell culture plates followed by 2‐week incubation under optimum conditions of 37°C and 5% CO2. Cells were fixed by 4% paraformaldehyde and visualized by 0.5% crystal violet (in methanol) for 5 minutes. Colonies with at least 50 cells were counted manually.
2.9. Cell cycle experiments
Cells were harvested by trypsinization after transfection for 72 hours, washed twice with ice‐cold PBS and centrifuged at 110 g for 5 minutes. Cell precipitates were fixed with 75% ethanol at 4°C for 4 hours, washed three times with ice‐cold PBS and stained with 40 μg propidium iodide (PI) and 1 mL 100 μg RNase staining solution (BD Biosciences, San Jose, CA, USA) in a light‐resistance condition at room temperature for 15 minutes. After staining, cell cycles were detected using a FACS Calibur (BD Biosciences) and statistical analyses were carried out using FACS Diva (BD Biosciences).
2.10. Oxygen consumption test
Cells were grown in 24‐well cell culture plates (3 × 104 cells/well). The assay plate was hydrated on the first day and incubated overnight at 37°C. Cells should show a normal state under microscopic observation and basically cover the bottom on test day. The original medium was replaced with test medium (Seahorse Bioscience, North Billerica, MA, USA) and then incubated for 1 hour at 37°C. The test plate final dosing is as follows: oligomycin 1 μmol/L, FCCP (cyanogen 4‐trifluoromethoxyphenylhydrazone) 1 μmol/L, antimycin A and rotenone 1 μmol/L each. Base level of rate of oxygen consumption in each group was calculated according to the final results.
2.11. ATP synthase activity test
For detecting the oxidative phosphorylation activity of transfected cells, the ATP synthase activity test was carried out. The mitochondrial extract was cultured at a recommended concentration in the steel plate preliminarily coated with an appropriate immunized antibody to allow its respective complexes to be immobilized. The mitochondrial extract was mixed with the base solution containing base and compound. The kinetic activity of the complex was recorded by spectroscopic M4 microplate spectrophotometer (Bio‐Rad, Hercules, CA, USA).
2.12. Statistical analysis
All the experiments were repeated more than three times to ensure accuracy, data are shown as mean ± standard deviation (SD). Statistical analyses were carried out using SPSS 16.0 (IBM, Armonk, NY, USA) and graphs were drawn by GraphPad Prism 6.0 (GraphPad Prism, La Jolla, CA, USA) or Cytoscape (National Resource for Network Biology, USA). Significance of difference was analyzed using Student's t‐test between two groups and one‐way ANOVA among more than two groups. A two‐tailed P‐value <0.05 was considered statistically significant. Particularly, the differentially expressed lncRNA and mRNA were identified at the threshold of P‐value <0.05 and log2 (fold change) >2.
3. RESULTS
3.1. Long non‐coding RNA MIF‐AS1 and messenger RNA NDUFA4 are upregulated and associated with oxidative phosphorylation pathway in gastric cancer
To investigate the expression profile of lncRNAs in gastric cancer tissues, microarray analyses were carried out, and the dysregulated lncRNAs are shown in Figure 1A. Of these lncRNAs, lncMIF‐AS1 showed the most significant increase in gastric cancer as compared with adjacent tissues. Moreover, the expression profile of mRNA was also surveyed using microarray analysis. Twenty mRNAs with the most significant alternations are shown, of which NDUFA4 was significantly upregulated in the cancerous gastric tissues compared to the normal gastric tissues (Figure 1B). Further correlation analysis indicated that the expression of NDUFA4 was positively associated with lncMIF‐AS1 (Figure 1C). In order to determine the NDUFA4‐associated biological pathway in the process of gastric cancer, the genes coexpression network was constructed using DigSee and STRING web servers. As shown in Figure 1D, essential members of the oxidative phosphorylation pathway were closely correlated with NDUFA4, including ATP6V0B and MT‐ATP8. Thus, oxygen consumption and ATP synthase in the fresh‐frozen tissues were measured and the results showed that both of the two indicators were significantly increased in gastric cancer tissues compared to para‐carcinoma tissues (Figure 1E,F).
Figure 1.

Long non‐coding RNA MIF‐AS1 and messenger RNA NDUFA4 are upregulated and associated with oxidative phosphorylation pathway in gastric cancer tissues. A,B, Heat map showing differentially expressed (A) lnc RNA and (B) mRNA levels in gastric cancer tissues (fold change value >2 and P < .01). C, LncMIF‐AS1 expression was positively correlated with NDUFA4 by analysis using R language and Cytoscape. D, NDUFA4 is predicted to be associated with oxidative phosphorylation pathways. (E) Oxygen consumption levels and (F) ATP synthase in 20 cases of gastric cancer and para‐cancerous tissues. *P < .05, **P < .01
Expression of lncMIF‐AS1 and NDUFA4 in human gastric cancer cell lines, including AGS, SGC‐7901, MKN‐28 and MKN‐45, were significantly higher than in the normal gastric epithelium cell line (GES‐1) (P < 0.05, Figure 2A,B). Notably, the AGS cell line showed the highest expression levels of lncMIF‐AS1 and NDUFA4 compared to other gastric cancer cell lines. Moreover, the overall expression of lncMIF‐AS1 and NDUFA4 were also higher in gastric cancer tissues as compared to the adjacent non‐cancerous tissues (P < 0.01, Figure 2C,D). Western blot results showed that the protein expression of NDUFA4 in gastric cancer tissues was greatly increased (P < 0.001, Figure 2E,F).
Figure 2.

Long non‐coding (lnc) RNA MIF‐AS1 and NDUFA4 expression levels were significantly upregulated in gastric cancer. A,B, qRT‐PCR results showing the mRNA levels of (A) lncMIF‐AS1 and (B) NDUFA4 in normal human gastric epithelium cell line (GES‐1) and four human gastric cancer cell lines (AGS, SGC‐7901, MKN‐28, MKN‐45). *P < .05, **P < .01. C,D, qRT‐PCR was used to detect the expression level of (C) lncMIF‐AS1 and (D) NDUFA4 in gastric cancer and adjacent non‐cancerous tissues. **P < .01. E,F, Western blotting assay shows the protein levels of NDUFA4 in gastric cancer and adjacent non‐cancerous tissues. GAPDH was used as a loading control. ***P < .001
3.2. MIF‐AS1 positivity regulates the expression of NDUFA4 by targeting miR‐212‐5p
To determine whether or not lncMIF‐AS1 acts as a competing endogenous RNA (ceRNA) to regulate NDUFA4 expression in gastric cancer (i.e. whether lncMIF‐AS1 regulates the expression of NDUFA4 in a miRNA‐dependent way), the lncRNA‐miRNA‐mRNA interaction network was predicted by an online bioinformatics analysis. As shown in Figure 3A, four promising miRNAs, including miR‐212‐5p, miR‐29a‐3p, miR‐339‐5p and miR‐199a‐5p, showed a potential binding site with both lncMIF‐AS1 and NDUFA4. Further dual‐luciferase reporter assay showed that the transfection of miR‐212‐5p and miR‐29a‐3p mimics significantly decreased the luciferase activity of firefly reporter vector carrying lncMIF‐AS1 and 3′‐UTR of NDUFA4, but the efficiency of miR‐212‐5p was higher than miR‐29a‐3p (Figure 3B). Therefore, expression levels of miR‐212‐5p and miR‐29a‐3p in gastric cancer cells were measured using qRT‐PCR. Compared to normal gastric mucosal cell line GES‐1, expression of miR‐212‐5p was significantly decreased in AGS (P < 0.01), SGC‐7901 (P < 0.01), MKN‐28 (P < 0.01) and MKN‐45 (P < 0.05) cell lines (Figure 3C), whereas miR‐29a‐3p was only decreased in AGS (P < 0.05) and SGC‐7901 (P < 0.05) cell lines (Figure 3D).
Figure 3.

Micro RNA (miR)‐212‐5p is the direct target of long non‐coding (lnc) MIF‐AS1 and reduces the expression of NDUFA4 in gastric cancer. A, Four potential miRNA linkers between potential lncMIF‐AS1 and NDUFA4 were predicted by bioinformatics methods. B, Dual‐luciferase reporter assay shows that the predicted miRNAs affected lncMIF‐AS1 and NDUFA4 activity, especially miR‐212‐5p. *P < .05, **P < .01. C,D, Relative expression levels of (C) miR‐212‐5p and (D) miR‐29a‐3p were measured in non‐cancerous gastric cells or gastric cancer cells by qRT‐PCR. *P < .05, **P < .01
Compared to the para‐carcinoma normal tissues, expression of miR‐212‐5p was significantly lower in gastric cancer tissues (Figure 4A). After transfection with specific lncMIF‐AS1‐targeting shRNA (sh‐MIF‐AS1), AGS cells showed upregulated expression of miR‐212‐5p and downregulated expression of NDUFA4 (Figure 4B). Conversely, we observed lower expression of miR‐212‐5p and higher expression of NDUFA4 if AGS cells were transfected with miR‐212‐5p inhibitors (Figure 4C).
Figure 4.
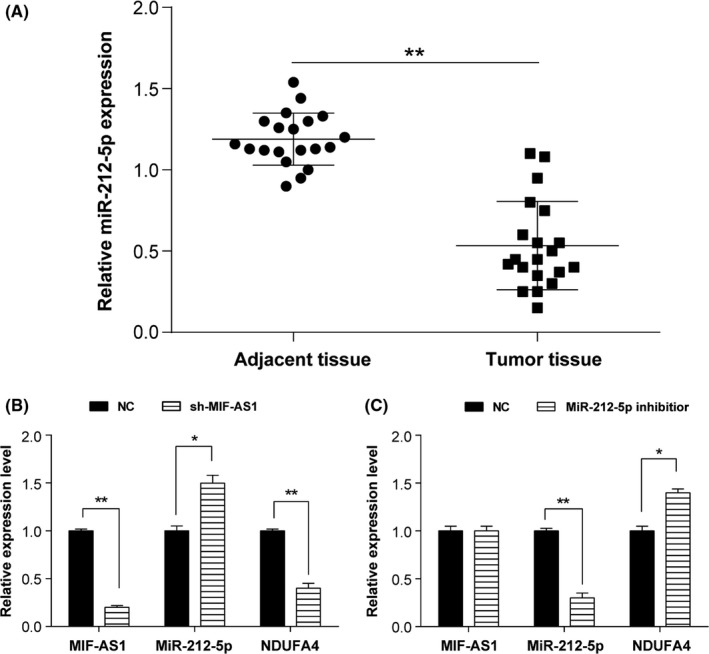
Micro RNA (miR)‐212‐5p repressed the expression of NDUFA4 in gastric cancer. A, miR‐212‐5p was expressed at a low level in gastric cancer tissues by qRT‐PCR. **P < .01. B,C, Relative expression of long non‐coding (lnc) MIF‐AS1, miR‐212‐5p, and NDUFA4 was measured after knockdown of (B) lncMIF‐AS1 or (C) miR‐212‐5p by qRT‐PCR. *P < .05, **P < .01
3.3. NDUFA4 promotes proliferation and reduces apoptosis of AGS cells
Protein expression of NDUFA4 in the transfected AGS cells was detected using western blot. As shown in Figure 5A,B, transfection of p‐MIF‐AS1 and p‐NDUFA4 largely increased the expression of NDUFA4, but transfection of miR‐212‐5p mimics caused the expression of NDUFA4 to be significantly decreased. Cotransfection of p‐MIR‐AS1 or p‐NDUFA4 with miR‐212‐5p mimics failed to change the expression level of NDUFA4 in AGS cells.
Figure 5.
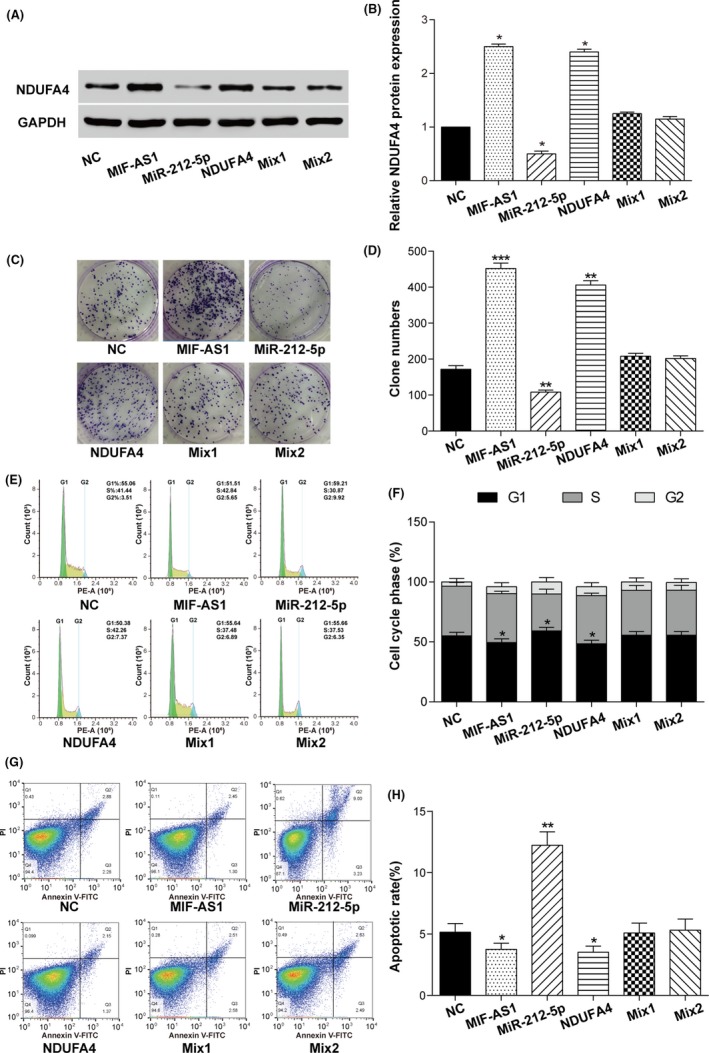
Long non‐coding (lnc) MIF‐AS1 promotes cell proliferation and reduces apoptosis of gastric cancer by regulating micro RNA (miR)‐212‐5p and NDUFA4. A,B, Relative protein expression of NDUFA4 was analyzed in gastric cancer cells with lncMIF‐AS1, miR‐212‐5p or NDUFA4 transfection. *P < .05. C,D, Proliferation ability of different overexpressed groups was verified by colony formation experiment. **P < .01, ***P < .001. E,F, The cell cycle was detected by flow cytometry in gastric cancer cells with lncMIF‐AS1, miR‐212‐5p or NDUFA4 transfection. *P < .05. G,H, Apoptotic rate of gastric cancer cells with lncMIF‐AS1, miR‐212‐5p or NDUFA4 transfection. *P < .05, **P < .01
Proliferation ability of transfected cells was analyzed using colony formation assay. As shown in Figure 5C,D, overexpression of lncMIF‐AS1 and NDUFA4 enhanced the proliferation ability of AGS cells, whereas overexpression of miR‐212‐5p significantly impaired the proliferation ability. Cotransfection of lncMIF‐AS1 or NDUFA4 with miR‐212‐5p failed to promote or suppress colony formation ability of AGS cells. The cell cycle was then analyzed using flow cytometry and the results showed that the proportion in G1 phase was obviously reduced in AGS cells with overexpression of lncMIF‐AS1 and NDUFA4, whereas it was significantly increased in cells with miR‐212‐5p overexpression compared to cells transfected with empty pcDNA3.1 vector and miR‐control (Figure 5E,F). Furthermore, flow cytometry was also used to detect cell apoptosis as shown in Figure 5G,H. The apoptosis rate of AGS cells was significantly decreased when lncMIF‐AS1 or NDUFA4 was overexpressed and increased when miR‐212‐5p was overexpressed. Cotransfection of lncMIF‐AS1 or NDUFA4 with miR‐212‐5p did not influence the proliferation and apoptosis of AGS cells.
3.4. NDUFA4 regulates the oxidative phosphorylation pathway in gastric cancer cells
As mentioned above, activation of the oxidative phosphorylation pathway was predicted to be associated with the expression of NDUFA4 (Figure 1C); this speculation was then verified in vitro. Results showed that both oxygen consumption and the ATP synthase activity of AGS cells were increased when lncMIF‐AS1 or NDUFA4 was overexpressed and decreased when miR‐212‐5p was overexpressed (Figure 6A,B). Moreover, expression levels of COX6C, COX5B and NDUFA8, the critical biomarkers in the oxidative phosphorylation pathway, were significantly increased in AGS cells with the overexpression of lncMIF‐AS1 or NDUFA4 and decreased in miR‐212‐5p‐overexpressed AGS cells (Figure 6C,D). Cotransfection of p‐MIR‐AS1 or p‐NDUFA4 with miR‐212‐5p mimics did not change the expression of COX6C, COX5B and NDUFA8, thus indicating failure to promote or suppress oxygen consumption and the activity of ATP synthase in AGS cells.
Figure 6.
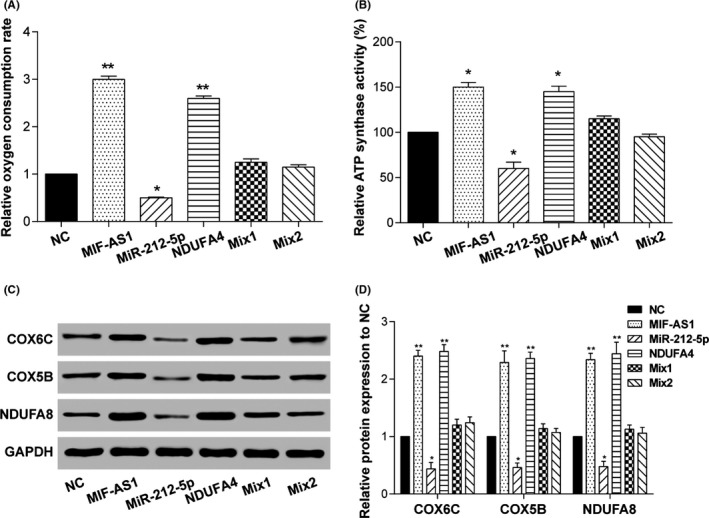
Long non‐coding (lnc) MIF‐AS1 affected cell proliferation through the oxidative phosphorylation pathway. (A) Oxygen consumption and (B) ATP synthase activities of gastric cancer cells transfected with lncMIF‐AS1, micro RNA (miR)‐212‐5p or NDUFA4. *P < .05, **P < .01. C,D, Expression of oxidative phosphorylation pathway‐related proteins in gastric cancer cells transfected with lncMIF‐AS1, miR‐212‐5p or NDUFA4. GAPDH was used as a loading control. *P < .05, **P < .01
4. DISCUSSION
Gastric cancer is one of the most common types of human cancer worldwide, and has a high incidence and mortality in Asia, especially in the Chinese population.19 Lack of available, efficient and less‐invasive diagnosis is the main limitation for early detection of gastric cancer.20 At present, the prognosis of patients with advanced gastric cancer remains poor, as the 5‐year survival rate is only about 20%‐30%.21, 22 Therefore, the molecular mechanism of gastric cancer progression needs to be explored in‐depth to provide available therapeutic targets.
In the present study, we analyzed the expression profile of lncRNAs in human gastric cancer, based on the previously published data by Chang et al18 showing that several lncRNAs significantly changed their expression in tumor tissues. Upregulated lncMIF‐AS1 was found to significantly promote the proliferation and inhibit the apoptosis of gastric cancer cells. To the best of our knowledge, this is the first study to show the role of lncMIF‐AS1 in cancer progression. LncRNAs are an extensive class of evolutionarily conserved non‐coding RNAs that are not translated into proteins; this “junk RNA” accounts for the major component of human transcripts.23 In the past few years, growing evidence suggests that lncRNAs play an important role in regulating many cell processes including proliferation, differentiation and apoptosis through regulation of the target gene at transcriptional, post‐transcriptional and epigenetic levels.24
To date, a large number of reports have shown that dysregulation of lncRNAs significantly contributes to the initiation and progression of gastric cancer through different molecular mechanisms.23, 25 For example, lncRNA OR3A4 has been found to promote tumorigenicity and metastasis of gastric cancer through regulating the activation of PDLIM2, MACC1, NTN4 and GNB2L1.26 ZFAS1 was upregulated in gastric cancer and its overexpression contributed to poor prognosis and shorter survival of patients through repression of KLF2 and NKD2.10 Moreover, LINC00668,27 BC005927,28 and GAS529 have also been implicated in the progression of gastric cancer by regulating the protein expression of target genes at the epigenetic level. In addition to direct epigenetic regulation, lncRNA‐induced dysregulation of mRNA in gastric cancer may be dependent on miRNA‐mediated indirect regulation.30 Shao et al31 found that lncRNA RMRP (RNA component of Mitochondrial RNA Processing endoribonuclease) can promote carcinogenesis of gastric cancer by acting as a miR‐206 sponge. Liu et al32 showed the tumor‐promoting role of HOTAIR in gastric cancer progression through positively regulating HER2 expression by sponging miR‐331‐3p. In addition, expression of several miRNAs, including miR‐675,33 miR‐145‐5p,34 miR‐152,35 miR‐145,36 miR‐23b‐3p,37 and miR‐335‐5p,38 are significantly dysregulated by lncRNAs and then change the expression of target mRNAs in gastric cancer.
Herein, we found that the tumor‐promoting role of lncMIF‐AS1 in gastric cancer was mainly dependent on positive regulation of NDUFA4. In particular, lncMIF‐AS1 may function as an endogenous “sponge” for miR‐212‐5p and thus attenuate the inhibition of miR‐212‐5p on NDUFA4 expression. miR‐212‐5p can bind to lncMIF‐AS1 but failed to cause degradation or decreased levels of lncMIF‐AS1. Based on bioinformatics analysis, four miRNAs (including miR‐212‐5p, miR‐29a‐3p, miR‐339‐5p and miR‐199a‐5p) showed the highest potential to target to lncMIF‐AS1 and NDUFA4, but only miR‐212‐5p and miR‐29a‐3p were confirmed. Further studies showed that enrichment of miR‐212‐5p in gastric cancer cells was negatively regulated by lncMIF‐AS1 and the decreased miR‐212‐5p largely promoted the expression of NDUFA4 in gastric cancer cells. During fungal infection, expression of several immune‐related target genes (such as KLF4, FKBP1B, and SPN) in dendritic cells were fine‐tuned by miR‐212‐5p, which, in turn, positively enhanced the anti‐fungal ability of dendritic cells.39 miR‐212‐5p was downregulated in prostate cancer, which promoted starvation‐induced autophagy in prostate cancer cells by targeting sirtuin 1.40 Guo et al41 found that overexpression of miR‐212‐5p decreased the protein level of stearoyl‐CoA desaturase‐1 (SCD1). The downregulated SCD1 has been found to significantly reduce the formation and metastasis of melanoma in vitro and in vivo.42 However, the elevated expression level of miR‐212‐5p has been reported in head and neck squamous cell carcinomas43 and in melanoma.44 Herein, we showed the tumor‐suppressing role of miR‐212‐5p in gastric cancer in that miR‐212‐5p overexpression obviously attenuated the proliferation ability and increased the apoptotic rate of gastric cancer cells in vitro. Whether the anti‐cancer property of miR‐212‐5p also exists in other cancer types will be determined in our subsequent studies.
NDUFA4 gene is located on chromosome 7p21.3, which encodes a subunit of the electron transport chain complex belonging to the respiratory chain of mitochondria to produce ATP.16, 45 Lei et al17 found that downregulation of NDUFA4 could contribute to the growth and metastasis of human lung cancer cells through altering the transduction of the Akt and Erk pathways. Downregulated NDUFA4 has also been detected in clear cell renal cell carcinoma,16 lung cancer,46 and esophageal squamous cell carcinoma.47 However, Liu et al48 observed that NDUFA4 was overexpressed in clear cell renal cell carcinoma and predicted the poor prognosis of patients. In addition, overexpression of NDUFA4 has been found to inhibit the apoptosis of neurons.16 In the present study, NDUFA4 significantly increased its expression in gastric cancer and positively promoted the proliferation and inhibited the apoptosis of gastric cancer cells. Based on the overexpression of NDUFA4, the oxidative phosphorylation pathway in gastric cancer cells was activated, represented by increased oxygen consumption and ATP synthase activity.
Our research has some limitations. First, several lncRNAs and mRNAs changed their expression in microarray analysis, but their roles in gastric cancer were not explored in depth in the present study. Next, miR‐29a‐3p showed the potential to act as a linker between lncMIF‐AS1 and NDUFA4, although the significance was slightly less than for miR‐212‐5p, but its role in gastric cancer was also not explored in depth herein.
In summary, our findings showed the role of the lncMIF‐AS1/NDUFA4/miR‐212‐5p axis in the tumorigenesis and progression of gastric cancer. LncMIF‐AS1 positively regulates NDUFA4 expression in gastric cancer cells through completely binding the miR‐212‐5p to attenuate miR‐212‐5p‐induced degradation or expression inhibition on NDUFA4 mRNA. The upregulated NDUFA4 significantly enhanced the proliferation ability and inhibited the apoptosis rate of gastric cancer cells in vitro through the activation of the oxidative phosphorylation pathway. Thus, inhibition of the lncMIF‐AS1/miR‐212‐5p/NDUFA4 pathway may be one of the potential therapeutic targets for controlling and treating gastric cancer.
CONFLICTS OF INTEREST
Authors declare no conflicts of interest for this article.
Li L, Li Y, Huang Y, et al. Long non‐coding RNA MIF‐AS1 promotes gastric cancer cell proliferation and reduces apoptosis to upregulate NDUFA4. Cancer Sci. 2018;109:3714–3725. 10.1111/cas.13801
L. Li and Y. Li equally contributed to this study.
Contributor Information
Ying Yuan, Email: km_ostrich@163.com.
Kunmei Gong, Email: kunhuagongkunmei@163.com.
REFERENCES
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7‐30. [DOI] [PubMed] [Google Scholar]
- 2. Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu X, Mao Y, Huang T, et al. High mitochondrial DNA copy number was associated with an increased gastric cancer risk in a Chinese population. Mol Carcinog. 2017;56:2593‐2600. [DOI] [PubMed] [Google Scholar]
- 4. Macdonald JS, Smalley SR, Benedetti J, et al. Chemoradiotherapy after surgery compared with surgery alone for adenocarcinoma of the stomach or gastroesophageal junction. N Engl J Med. 2001;345:725‐730. [DOI] [PubMed] [Google Scholar]
- 5. Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): a phase 3, open‐label, randomised controlled trial. Lancet. 2010;376:687‐697. [DOI] [PubMed] [Google Scholar]
- 6. Huang YK, Yu JC. Circulating microRNAs and long non‐coding RNAs in gastric cancer diagnosis: an update and review. World J Gastroenterol. 2015;21:9863‐9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L, Sun L, Dong L, et al. The role of long noncoding RNA‐LET in cell proliferation and invasion of nasopharyngeal carcinoma and its mechanism. OncoTargets Ther. 2017;10:2769‐2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schmitt AM, Chang HY. Long noncoding RNAs: at the intersection of cancer and chromatin biology. Cold Spring Harb Perspect Med. 2017;7:a026492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adams BD, Kasinski AL, Slack FJ. Aberrant regulation and function of microRNAs in cancer. Curr Biol. 2014;24:R762‐R776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nie F, Yu X, Huang M, et al. Long noncoding RNA ZFAS1 promotes gastric cancer cells proliferation by epigenetically repressing KLF2 and NKD2 expression. Oncotarget. 2017;8:38227‐38238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasionin gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571‐1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao JH, Sun JX, Song YX, et al. A novel long noncoding RNA‐LOWEG is low expressed in gastric cancer and acts as a tumor suppressor by inhibiting cell invasion. J Cancer Res Clin Oncol. 2016;142:601‐609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balsa E, Marco R, Perales‐Clemente E, et al. NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab. 2012;16:378‐386. [DOI] [PubMed] [Google Scholar]
- 14. Garbian Y, Ovadia O, Dadon S, Mishmar D. Gene expression patterns of oxidative phosphorylation complex I subunits are organized in clusters. PLoS ONE. 2010;5:e9985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gogvadze V, Orrenius S, Zhivotovsky B. Mitochondria in cancer cells: what is so special about them? Trends Cell Biol. 2008;18:165‐173. [DOI] [PubMed] [Google Scholar]
- 16. Muller FE, Braun M, Syring I, et al. NDUFA4 expression in clear cell renal cell carcinoma is predictive for cancer‐specific survival. Am J Cancer Res. 2015;5:2816‐2822. [PMC free article] [PubMed] [Google Scholar]
- 17. Lei L, Chen C, Zhao J, et al. Targeted expression of miR‐7 Operated by TTF‐1 promoter inhibited the growth of human lung cancer through the NDUFA4 pathway. Mol Ther Nucleic Acids. 2017;6:183‐197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang HR, Nam S, Kook MC, et al. HNF4alpha is a therapeutic target that links AMPK to WNT signalling in early‐stage gastric cancer. Gut. 2016;65:19‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang Q, Zhang RW, Sui PC, He HT, Ding L. Dysregulation of non‐coding RNAs in gastric cancer. World J Gastroenterol. 2015;21:10956‐10981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. da Silva Oliveira KC, Thomaz Araujo TM, Albuquerque CI, et al. Role of miRNAs and their potential to be useful as diagnostic and prognostic biomarkers in gastric cancer. World J Gastroenterol. 2016;22:7951‐7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tsai MM, Wang CS, Tsai CY, et al. Potential diagnostic, prognostic and therapeutic targets of MicroRNAs in human gastric cancer. Int J Mol Sci. 2016;17:945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. GASTRIC Group , Oba K, Paoletti X, et al. Role of chemotherapy for advanced/recurrent gastric cancer: an individual‐patient‐data meta‐analysis. Eur J Cancer. 2013;49:1565‐1577. [DOI] [PubMed] [Google Scholar]
- 23. Dykes IM, Emanueli C. Transcriptional and post‐transcriptional gene regulation by long non‐coding RNA. Genomics Proteomics Bioinformatics. 2017;15:177‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Della Ragione F, Gagliardi M, D'Esposito M, Matarazzo MR. Non‐coding RNAs in chromatin disease involving neurological defects. Front Cell Neurosci. 2014;8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun W, Yang Y, Xu C, Xie Y, Guo J. Roles of long noncoding RNAs in gastric cancer and their clinical applications. J Cancer Res Clin Oncol. 2016; 142: 2231‐2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo X, Yang Z, Zhi Q, et al. Long noncoding RNA OR3A4 promotes metastasis and tumorigenicity in gastric cancer. Oncotarget. 2016;7:30276‐30294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang E, Yin D, Han L, et al. E2F1‐induced upregulation of long noncoding RNA LINC00668 predicts a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically silencing of CKIs. Oncotarget. 2016;7:23212‐23226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu X, Wang Y, Sun L, et al. Long noncoding RNABC005927 upregulates EPHB4 and promotes gastric cancer metastasis under hypoxia. Cancer Sci. 2018;109:988‐1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sun M, Jin FY, Xia R, et al. Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer. 2014;14:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang G, Pian C, Chen Z, et al. Identification of cancer‐related miRNA‐lncRNA biomarkers using a basic miRNA‐lncRNA network. PLoS ONE. 2018;13:e0196681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shao Y, Ye M, Li Q, et al. LncRNA‐RMRP promotes carcinogenesis by acting as a miR‐206 sponge and is used as a novel biomarker for gastric cancer. Oncotarget. 2016;7:37812‐37824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR‐331‐3p in gastric cancer. Mol Cancer. 2014;13:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yan J, Zhang Y, She Q, et al. Long noncoding RNA H19/miR‐675 axis promotes gastric cancer via FADD/Caspase 8/Caspase 3 signaling pathway. Cell Physiol Biochem. 2017;42:2364‐2376. [DOI] [PubMed] [Google Scholar]
- 34. Ren K, Li Z, Li Y, Zhang W, Han X. Long noncoding RNA taurine‐upregulated gene 1 promotes cell proliferation and invasion in gastric cancer via negatively modulating miRNA‐145‐5p. Oncol Res. 2017;25:789‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li T, Meng XL, Yang WQ. Long noncoding RNA PVT1 acts as a “Sponge” to inhibit microRNA‐152 in gastric cancer cells. Dig Dis Sci. 2017;62:3021‐3028. [DOI] [PubMed] [Google Scholar]
- 36. Hu CE, Du PZ, Zhang HD, Huang GJ. Long noncoding RNA CRNDE promotes proliferation of gastric cancer cells by targeting miR‐145. Cell Physiol Biochem. 2017;42:13‐21. [DOI] [PubMed] [Google Scholar]
- 37. YiRen H, YingCong Y, Sunwu Y, et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR‐23b‐3p sequestration in gastric cancer. Mol Cancer. 2017;16:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang LL, Zhang LF, Guo XH, Zhang DZ, Yang F, Fan YY. Downregulation of miR‐335‐5p by long noncoding RNA ZEB1‐AS1 in gastric cancer promotes tumor proliferation and invasion. DNA Cell Biol. 2018;37:46‐52. [DOI] [PubMed] [Google Scholar]
- 39. Dix A, Czakai K, Leonhardt I, et al. Specific and novel microRNAs are regulated as response to fungal infection in human dendritic cells. Front Microbiol. 2017;8:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ramalinga M, Roy A, Srivastava A, et al. MicroRNA‐212 negatively regulates starvation induced autophagy in prostate cancer cells by inhibiting SIRT1 and is a modulator of angiogenesis and cellular senescence. Oncotarget. 2015;6:34446‐34457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guo Y, Yu J, Wang C, et al. miR‐212‐5p suppresses lipid accumulation by targeting FAS and SCD1. J Mol Endocrinol. 2017;59:205‐217. [DOI] [PubMed] [Google Scholar]
- 42. Liu G, Feng S, Jia L, Wang C, Fu Y, Luo Y. Lung fibroblasts promote metastatic colonization through upregulation of stearoyl‐CoA desaturase 1 in tumor cells. Oncogene. 2018;37:1519‐1533. [DOI] [PubMed] [Google Scholar]
- 43. Allen B, Schneider A, Victoria B, et al. Blood Serum from head and neck squamous cell carcinoma patients induces altered microRNA and target gene expression profile in treated cells. Front Oncol. 2018;8:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chai L, Kang XJ, Sun ZZ, et al. MiR‐497‐5p, miR‐195‐5p and miR‐455‐3p function as tumor suppressors by targeting hTERT in melanoma A375 cells. Cancer Manage Res. 2018;10:989‐1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fu F, Li Y, Li R, et al. NDUFA4 enhances neuron growth by triggering growth factors and inhibiting neuron apoptosis through Bcl‐2 and cytochrome C mediated signaling pathway. Am J Transl Res. 2018;10:164‐174. [PMC free article] [PubMed] [Google Scholar]
- 46. Puissegur MP, Mazure NM, Bertero T, et al. miR‐210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF‐1 activity. Cell Death Differ. 2011;18:465‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang Y, Li Z, Shi ZX. [Mechanisms of the suppression of proliferation and invasion ability mediated by microRNA‐147b in esophageal squamous cell carcinoma]. Zhonghua Yi Xue Za Zhi. 2018;98:2092‐2098. [DOI] [PubMed] [Google Scholar]
- 48. Liu L, Lan G, Peng L, et al. NDUFA4L2 expression predicts poor prognosis in clear cell renal cell carcinoma patients. Ren Fail. 2016;38:1199‐1205. [DOI] [PubMed] [Google Scholar]


