Abstract
Indoleamine 2,3‐dioxygenase 1 (IDO1) is a tryptophan‐metabolizing enzyme that is widely distributed in normal or malignant tissues and contributes to immunologic tolerance and immune escape. However, in hepatocellular carcinoma (HCC), the characteristics and mechanism of IDO1 expression have not been well defined. In this study, IDO1 expression in tumor cells (T‐IDO1) was frequently detected (109/112) by immunohistochemistry in formalin‐fixed paraffin‐embedded specimens from HCC patients, and the expression patterns were mostly focal (102/109). Expression of T‐IDO1 was significantly associated with the infiltration of CD8+ T cells (P = .043), as well as younger age (<50 years old, P = .02). It was also found that IDO1 had diffuse expression in inflammatory cells in all specimens, which were defined as antigen‐presenting cells. Significant correlations among IDO1,IFNG, and CD8A transcriptional levels were observed in freshly resected HCC specimens; moreover, no constitutive IDO1 expression was detected in HCC cell lines until stimulated by interferon‐γ through the JAK2‐STAT1 signaling pathway, but not type I interferon. Survival analyses showed that increased T‐IDO1 and CD8+ T cell infiltration were significantly associated with superior overall survival (OS) (T‐IDO1, P = .003; CD8+ T cells, P = .004), and T‐IDO1 expression is an independent prognosis factor in both OS and disease‐free survival (OS, P = .007; disease‐free survival, P = .044). These findings indicated that T‐IDO1 expression in HCC is common and is dominantly driven by the host antitumor immune response, which is a favorable prognostic factor in HCC.
Keywords: CD8, HCC, IDO1, IFN‐γ, inducible expression
Abbreviations
- APC
antigen‐presenting cell
- DC
dendritic cell
- DFS
disease‐free survival
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- IDO1
indoleamine 2,3‐dioxygenase 1
- IFN
interferon
- LAMP3
lysosomal associated membrane protein‐3
- NK
natural killer
- OS
overall survival
- ROI
region of interest
- TCGA
The Cancer Genome Atlas
1. INTRODUCTION
Hepatocellular carcinoma is a typical malignancy that slowly unfolds on a background of chronic inflammation mainly triggered by exposure to infectious agents (hepatotropic viruses) or toxic compounds (ethanol).1 In the tumorigenic process, the accumulation of genomic alterations creates various neoantigens that can elicit host antitumor immune responses.2 Indeed, intense lymphocytic infiltration within the tumor, especially CD8+ T cells, as a favorable predictor in HCC, is a major effector against HCC.3, 4 Unexpectedly, tumors appear to establish an immunosuppressive microenvironment to brake host immune killing. Recently, immune checkpoint inhibitors blocking cytotoxic T‐lymphocyte‐associated antigen‐4 and programmed cell death‐1/programmed cell death‐ligand 1 have been widely applied in HCC practice, but they have been less effective than anticipated, suggesting that additional immunosuppressive fences might need to be overcome to maximize therapeutic efficacy in HCC.5, 6 A recent study showed that IDO1 plays a key role in adaptive resistance to immune checkpoint inhibitors in HCC.7
Indoleamine 2,3‐dioxygenase 1 is a rate‐limiting enzyme in the catabolism of the essential amino acid tryptophan along the kynurenine pathway, and it is widely distributed in various types of tumors.8, 9 Broad evidence has indicated that local tryptophan deprivation and the accumulation of kynurenines produced by IDO1 elevation crucially facilitate the escape of malignant tumors from host immune killing with multiple effects, including the suppression of T and NK cells, the generation and activation of regulatory T cells and myeloid‐derived suppressor cells, and the promotion of tumor angiogenesis.10, 11, 12, 13, 14, 15
As an immune escape mechanism, high constitutive IDO1 expression, driven by intrinsic cellular changes such as gene mutations associated with carcinogenesis, represents an adverse prognostic factor in colorectal cancer, endometrial cancer, ovarian carcinoma, and a number of other cancers.16, 17, 18, 19, 20 In contrast, in breast carcinoma, early stage cervical cancer, and melanoma, IDO1 expression is accompany the host antitumor immune response, and it tends to positively correlate with survival.21, 22, 23, 24 However, in another study, the high inductive IDO1 expression in primary melanoma negatively affected progression‐free survival.25 Under the heterogeneity of tumor gene mutations and the tumor microenvironment, both constitutive and inducible IDO1 expression can co‐exist regulated by tumor intrinsic and extrinsic mechanisms, reflecting either the absence or presence of antitumor immunity in specific tumor microenvironments. Thus, higher IDO1 expression could correlate with either worse or improved prognosis in different tumors.26
Based on previous studies, the features and mechanisms of IDO1 expression in HCC tumor cells remain controversial, and its prognostic value must also be clarified.27, 28, 29 In this study, we characterize IDO1 expression in both tumor cells (T‐IDO1) and inflammatory cells (I‐IDO1) in a series of 112 surgically resected HCCs. In vitro and ex vivo experiments were carried out to explore the mechanism controlling T‐IDO1 expression, and survival analyses were undertaken to explore the significance of T‐IDO1 and I‐IDO1 expression in prognosis.
2. MATERIAL AND METHODS
2.1. Cell lines and culture
Two cell lines derived from human HCC (Hep3B and Huh7; ATCC, Manassas, VA, USA) were authenticated by short tandem repeat profiling‐based PCR and were analyzed to determine IDO1 protein expression. All cells were cultured at 37°C, 5% CO2 in DMEM supplemented with 10% FBS and 1% penicillin‐streptomycin. These cell lines have been tested for mycoplasma contamination by PCR.
2.2. Patients and samples
This research project was approved by the Ethics Committee of Sun Yat‐sen University Cancer Center (Guangzhou, China), and all HCC patients enrolled in the research had signed informed written consent before the start of the research. To reduce interference with survival analysis from complex follow‐up treatment, a total of 112 patients who underwent resection of HCC as an initial treatment between December 2007 and December 2010 were randomly selected and analyzed. Liver function was evaluated by the Child‐Pugh scoring system and albumin‐bilirubin grade.30 Clinical data were assessed according to the 7th edition of the AJCC/UICC TNM classification system and Barcelona Clinic Liver Cancer staging system. Surgically resected specimens were fixed in formalin and embedded in paraffin for routine histological diagnosis, and the histopathological findings were classified by the Edmondson grading system. Further prognostic analysis was carried out for patients receiving curative hepatic resection (complete removal of the tumor).
2.3. Western blot analysis
Cells were pretreated with or without a JAK2 inhibitor (BMS‐911543, 10 μmol/L; #S7144, Selleck Chemicals, Houston, TX, USA), a JAK1/3 inhibitor (ZM 39923 HCl, 10 μmol/L; #S8004, Selleck Chemicals), or a STAT1 inhibitor (fludarabine, 10 μmol/L; #S1491, Selleck Chemicals) in DMEM complete medium for 2 hours. The cells were then cultured in the presence or absence of IFN‐γ (20 ng/mL) for 24 hours. The other cells were cultured in the presence of IFN‐α (1000 IU/mL and 20 ng/mL), IFN‐β (1000 IU/mL and 20 ng/mL), or IFN‐γ (20 ng/mL) for 48 hours. The cells were washed twice with ice‐cold PBS, lysed in RIPA containing protease and phosphatase inhibitors, and then centrifuged for 5 minutes at 12 000 g at 4°C. The protein concentration in the supernatant was quantified utilizing the BCA protein assay kit (#P0012S; Beyotime, Shanghai, China). Protein extracts from each sample were separated on SDS‐PAGE and transferred onto a PVDF membrane. The membranes were blocked with 5% dry milk or 5% BSA (for phospho‐STAT1) in 1× TBST for 1 hour at room temperature and probed with mouse anti‐IDO1 mAb (4.16H1 Ab, 1:1000; a kind gift from Benoit J. Van den Eynde, Ludwig Institute for Cancer Research, de Duve Institute [Universite catholique de Louvain], Brussels, Belgium), rabbit anti‐phospho‐STAT1 mAb (58D6 Ab, 1:1000; #9167, Cell Signaling Technology, Danvers, MA, USA) and mouse anti‐STAT1 mAb (c‐136 Ab, 1:1000; #sc‐464, Santa Cruz Biotechnology, Dallas, TX, USA), overnight at 4°C, and mouse monoclonal anti‐GAPDH (1E6D9 Ab, 1:5000; #60004‐1‐Ig, ProteinTech Group, Rosemont, IL, USA) was used as an internal control. After three washings in 1× TBST, the membrane was probed for 1 hour with HRP‐conjugated goat anti‐mouse IgG (1:5000; #SA00001‐1, ProteinTech Group) or HRP‐conjugated goat anti‐rabbit IgG (1:5000; #SA00001‐2, ProteinTech Group) at room temperature. Secondary Abs were incubated for 1 hour at room temperature before revelation with SuperSignal West Pico Chemiluminescent Substrate (#34077; Thermo Fisher Scientific, Waltham, MA, USA). The images were acquired by Bio‐Rad ChemiDoc Touch (Bio‐Rad, Hercules, CA, USA) and analyzed by Image Lab software (Bio‐Rad).
2.4. Reverse transcription‐PCR
Tumor samples were immediately snap‐frozen in liquid nitrogen after harvesting. Total RNA was extracted using TRIzol reagent (#T9424, Sigma‐Aldrich, St. Louis, MO, USA). Complementary DNA was synthesized from 2 μg total RNA with the ImProm‐II Reverse Transcription System (#A3800, Promega, Madison, WI, USA), according to the manufacturer's directions. Real‐time PCR analysis was carried out with diluted cDNA, and Fast SYBR Green Master Mix (#4385616, Promega) was used. The sequences of the primers were as follows.
CD8A: forward, 5′‐TCCTCCTATACCTCTCCCAAAAC‐3′ – reverse, 5′‐GGAAGACCGGCACGAAGTG‐3′; IFNG: forward, 5′‐TCGGTAACTGACTTGAATGTCCA ‐3′ – reverse, 5′‐TCGCTTCCCTGTTTTAGCTGC‐3′; IDO1: forward, 5′‐TCTCATTTCGTGATGGAGACTGC‐3′ reverse, 5′‐GTGTCCCGTTCTTGCATTTGC‐3′; GAPDH forward: 5′‐GGAGCGAGATCCCTCCAAAAT‐3′ reverse, 5′‐GGCTGTTGTCATACTTCTCATGG‐3′.
Expression relative to the reference gene GAPDH was calculated as , where ΔC t is Ctgene‐Ctctrl. If undetectable, Ct was given a value of 40.
2.5. Immunohistochemistry
After deparaffinization, rehydration, and washing of the 5‐μm sections, endogenous peroxidase was blocked (3% H2O2 for 10 minutes), and microwave antigen retrieval was undertaken in Tris‐EDTA (pH 9.0). The sections were first incubated with blocking serum at room temperature for 30 minutes and with a mouse anti‐IDO1 mAb (4.16H1 Ab, 1:1000; a kind gift from Benoit J. Van den Eynde), a rabbit anti‐CD8 Ab (SP16 Ab, 1:100; #ZA‐0508, ZSGB‐BIO, Beijing, China), a rabbit anti‐CD34 mAb (EP373Y Ab, 1:100; #ab81289, Abcam, Cambridge, UK), a mouse anti‐DC‐LAMP mAb (104G4 Ab, 1:50; #DDX0190A546‐100, Novus Biologicals, Littleton, CO, USA), a rabbit anti‐CD68 polyclonal Ab (1:100; #25747‐1‐AP, ProteinTech Group), a mouse anti‐CD4 Ab (UMAB64 Ab; #ZA‐0418, ZSGB‐BIO, Beijing, China), or a rabbit anti‐NCAM1/CD56 polyclonal Ab (1:400; #14255‐1‐AP, ProteinTech Group) at 4°C overnight. The second incubation was carried out using a goat anti‐mouse/rabbit Ab and visualized with DAB, following counterstaining with hematoxylin after serial rinsing. The observation and images acquisition of these sections were carried out by advanced research microscope (Nikon Eclipse 80i; Nikon, Tokyo, Japan).
2.6. Evaluation of immunostaining parameters
To evaluate IDO1 expression in tumor and inflammatory cells separately, each IHC section was examined by two independent pathologists in the five most representative areas, and the average count was calculated. To reduce the influence of subjective factors, ImageJ computer software (National Institutes of Health, Bethesda, MD, USA) was used to aid in analyzing the images, and the tumor cells and nontumor cells were separated according to morphology. Expression of IDO1 was evaluated quantitatively on the basis of the percentage of positive areas. According to the results of the receiver operating characteristic curve curves, high T‐IDO1 and I‐IDO1 expression were defined as ≥5.80% and ≥6.00%, respectively (Figure S1). CD8+ T lymphocytes were counted in a microscopic field at 400×, the median number (10 counts/high‐power field) of CD8 infiltrations in the cohort as a cut‐off value. Additionally, CD4+ T lymphocytes and NK cells were also counted in another 20 specimens at 400× to evaluate the association between them and IDO1.
2.7. Method using ImageJ
Images were opened at high magnification in ImageJ software, and the open image type was set to RGB color. The option “Color Threshold” was turned on, and “Hue,” “Saturation,” and “Brightness” were adjusted so that threshold color completely covered the ROI. Then the number of pixels in the ROI was analyzed, and the proportion of the ROI area to total area was calculated (the total number of pixels/high magnification image was 2560 × 1920).
2.8. Immunofluorescence
After deparaffinization, rehydration and washing of the sections, microwave antigen retrieval was carried out in Tris‐EDTA (pH 9.0). Sections were first incubated with blocking serum at room temperature for 30 minutes and with the mouse anti‐IDO1 mAb (4.16H1 Ab, 1:1000; a kind gift from Benoit J. Van den Eynde), combined with a rabbit anti‐CD34 mAb (EP373Y Ab 1:100; #ab81289, Abcam), a rabbit anti‐LAMP3 mAb (104G4 Ab, 1:50; #DDX0190A546‐100, Novus Biologicals), or a rabbit anti‐CD68 polyclonal Ab (1:100; #25747‐1‐AP, ProteinTech Group) at 4°C overnight. The second incubation was undertaken using DyLight 488 AffiniPure goat anti‐mouse IgG (1:100; #E032210, EarthOx, Millbrae, CA, USA) and DyLight 594 AffiniPure goat anti‐rabbit IgG (1:100; E032420, EarthOx). Following counterstaining with DAPI after serially rinsing with 1× PBST, images were acquired in a microscopic field at 600×. The observation and images acquisition of these sections were carried out using a confocal laser scanning microscope (Olympus Fluoview FV1000; Olympus, Tokyo, Japan).
2.9. Statistical analyses
All statistical analyses were performed using SPSS statistical software, version 20 (IBM Corporation, Armonk, NY, USA), and a P value (two‐sided) < .05 was considered significant. The χ2 test and Fisher's exact test were used to analyze the association between immune parameters and clinicopathological features. For gene expression analysis, the correlations among the mRNA expression levels of IDO1, CD8A, and IFNG were analyzed using Spearman's rank correlation. Comparisons of IDO1 expression were undertaken using Student's t test. The prognostic significances of IDO1 expression and CD8+ T cell infiltration were evaluated using the Kaplan‐Meier method and were compared with the log‐rank test. A Cox regression model was used to perform univariate analyses, and multivariate analysis was performed on all factors with P values < .05. Overall survival was defined as the length of time from the surgery date to death or until the last follow‐up (censored). Disease‐free survival was defined as the length of time from the surgery date to recurrence, death, or until the last follow‐up (censored).
The authenticity of this article has been validated by uploading the key raw data onto the Research Data Deposit public platform (http://www.researchdata.org.cn), with the approval RDD number as RDDB2018000434.
3. RESULTS
3.1. Clinical profiles of patients
The demographic and clinical characteristics of the patients and tumors are presented in Table 1. A total of 112 patients with adequate clinical data and tumor samples for the evaluation for IDO1 expression and CD8+ T cell infiltration were included. Liver functions of all patients in our cohort were Child‐Pugh A and all patients received completely curative resection based on intraoperative evaluation and postoperative pathology. Post‐recurrent treatments were administered by managing doctors according the National Cancer Center Network Clinical Practice Guidelines.
Table 1.
Clinicopathological features of the cohort of patients with hepatocellular carcinoma
| Variable | Results |
|---|---|
| Age, <50/≥50 y | 48 (43)/64 (57) |
| Gender, male/female | 104 (93)/8 (7) |
| ECOG, 0/1 | 108 (96)/4 (4) |
| HBV, no/yes | 10 (9)/102 (91) |
| AFP, ≤20 ng/mL/>20 ng/mL | 36 (32)/76 (68) |
| Tumor size, cm | 5.76 (1.5‐20)a |
| Tumor number, single/multiple | 83 (74)/29 (26) |
| Vascular invasion, no/yes | 108 (96)/4 (4) |
| Nodal invasion, no/yes | 110 (98)/2 (2) |
| Tumor encapsulation, complete/none | 41 (37)/71 (63) |
| ALBI grade, 1/2 | 86 (77)/26 (23) |
| BCLC stage, 0‐A/B‐C | 81 (72)/31 (28) |
| TNM stage, I/II‐IV | 79 (71)/33 (29) |
| Histologic grade, I‐II/III | 70 (63)/42 (37) |
| Cirrhosis, no/yes | 26 (23)/86 (77) |
| Alive, no/yes | 46 (41)/66 (59) |
| Follow‐up time, mo | 67.2 (28.9‐76.7)b |
Data are shown as n (%) unless otherwise indicated.
Median (range).
Median (interquartile range).
AFP, α‐fetoprotein; ALBI, albumin‐bilirubin; BCLC, Barcelona Clinic Liver Cancer; HBV, hepatitis B virus.
3.2. Expression pattern of IDO1 in HCC tissue samples
Indoleamine 2,3‐dioxygenase 1 expression was present in 97.3% (109/112) of cases within the tumor compartment and showed a predominantly cytoplasmic staining pattern. As shown in Figure 1, three major expression patterns of T‐IDO1 were observed by IHC: focal expression in 91% (102/112) of cases (Figure 1A,B); discrete expression in 6.3% (7/112) of cases (Figure 1C,D); and absence in 2.7% (3/112) of cases (Figure 1E). According to the cut‐off value, T‐IDO1 expression was low for 54 patients (48.2%) and high for 58 patients (51.8%). Placenta was used as a positive control for IDO1 expression (Figure 1F).
Figure 1.
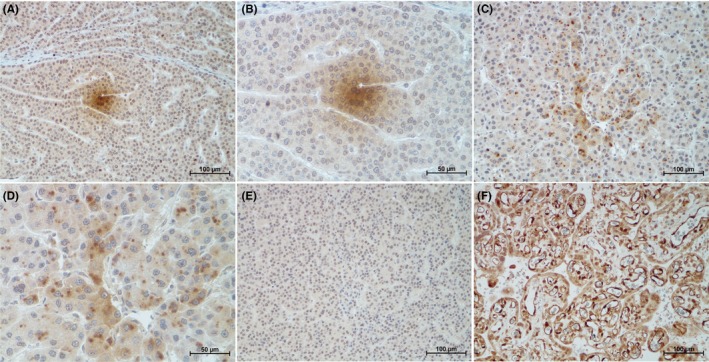
Indoleamine 2,3‐dioxygenase 1 (IDO1) expression pattern in tumor cells of hepatocellular carcinoma (HCC) in formalin‐fixed paraffin‐embedded samples stained with anti‐IDO1 Ab. A,B, Sample displaying pattern 1 showing focal staining in a mass of adjacent tumor cells (brown). (A) 200×; (B) 400×. C,D, Sample displaying pattern 2 showing discrete staining in adjacent or nonadjacent tumor cells (brown). (C) 200×; (D) 400×. E, Sample displaying pattern 3 showing negative expression in HCC cells (brown), 200×. F, Placenta was used as a positive control for IDO1 expression, 200×
3.3. Correlation between IDO1 and clinicopathological features
CD8+ T cell infiltration (Figure S2) and clinicopathological features were submitted to correlation analysis with T‐IDO1 expression. The T‐IDO1‐positive area was larger in samples with abundant CD8+ T cell infiltration than in those with infrequent CD8+ T cell infiltration (P = .043, Figure 2A). In the clinicopathological features, only age was a significant factor affecting T‐IDO1 expression (P = .02; Figure 2B‐D and Table S1).
Figure 2.
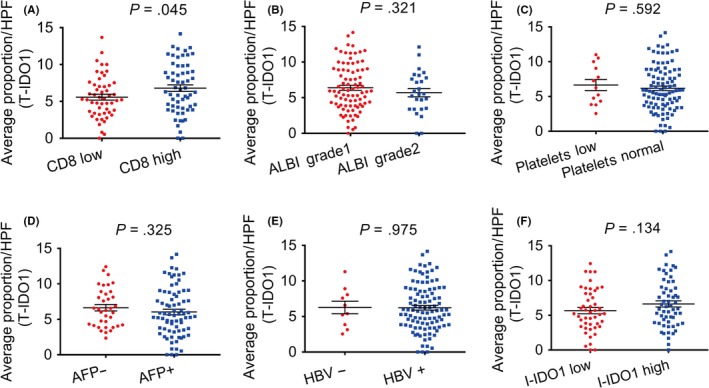
Correlations among the relative expression levels of indoleamine 2,3‐dioxygenase 1 in T cells (T‐IDO1) with CD8, albumin‐bilirubin (ALBI) grade, platelet level, α‐fetoprotein level (AFP), hepatitis B virus (HBV) positivity, and IDO1 expression in inflammatory cells (I‐IDO1). A, Tumors were classified as CD8‐low (n = 52) or CD8‐high (n = 60) and analyzed for the proportion of T‐IDO1. B, Patients were classified as ALBI grade 1 (n = 86) or ALBI grade 2 (n = 26) and analyzed for the proportion of T‐IDO1. C, Patients were classified as platelets low (n = 13) or platelets normal (n = 99) and analyzed for the proportion of T‐IDO1. D, Patients were classified as AFP− (n = 36) or AFP+ (n = 76) and analyzed for the proportion of T‐IDO1. E, Patients were classified as HBV− (n = 10) or HBV+ (n = 102) and analyzed for the proportion of T‐IDO1. C, Tumors were classified as I‐IDO1 low (n = 47) or I‐IDO1 high (n = 58) and analyzed for the proportion of T‐IDO1. HPF, high‐power field
Hepatitis B was the main cause of HCC in our cohort, the T‐IDO1‐positive area showed no significant difference between the HBV (+) and HBV (−) groups (Figure 2E). There was no association between T‐IDO1 and I‐IDO1 (Figure 2F).
3.4. Expression of IDO1 reflects antitumor immunity in HCC
To elucidate the mechanism underlying the association between elevated T‐IDO1 expression and abundant CD8+ T cell infiltration in HCC, the transcription of IDO1, IFNG, and CD8A genes was quantitatively evaluated in 20 surgically excised tumor specimens. Significantly, positive correlations among the transcription levels of CD8A, IDO1, and IFNG were revealed (Figure 3A‐C). To exclude possible bias induced by limited sample size and/or the effects of marker heterogeneity, we further analyzed the levels of IDO1, IFNG, and CD8A mRNA from the TCGA liver cancer dataset obtained by RNA sequencing from whole‐tissue section tumor samples, and positive association among mRNA levels of IDO1, IFNG, and CD8A were also identified (Figure S3; n = 371).31 In vitro experiments further showed that no constitutive IDO1 protein expression was detectable in HCC cell lines until IFN‐γ stimulation, but not type I interferon (Figure 3D, E), and this effect could be abrogated by a JAK2 inhibitor or STAT1 inhibitor, but not by the JAK1/3 inhibitor (Figure 3F, G). Immunohistochemical analysis of CD4 T cells and NK cells was applied to tissues from 20 patients because they are also the major sources of IFN‐γ. No correlation between T‐IDO1 expression and CD4 T cells or NK cells was found (Figure S4).
Figure 3.
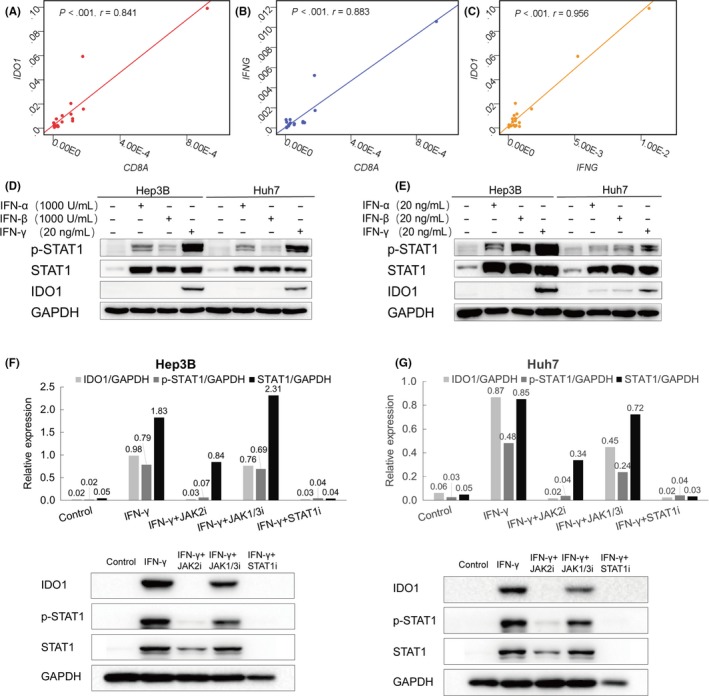
γ‐Interferon (IFN‐γ), but not type I IFN, upregulates indoleamine 2,3‐dioxygenase 1 in T cells (T‐IDO1) through the JAK2‐STAT1 signaling pathway. A‐C, Correlation studies were carried out for IFNG,CD8A, and IDO1 mRNA expression in hepatocellular carcinoma patients’ tumor tissues; GAPDH was used as an internal control (n = 20). D,E, Western blot detection of IDO1 expression in Hep3B and Huh7 cell lines in the presence or absence of 20 ng/mL IFN‐γ, 1000 IU/mL (D) or 20 ng/mL (E) type I IFN for 48 hours. F,G, Western blot detection of IDO1 expression in Hep3B cell lines (F) and Huh7 cell lines (G) in the presence or absence of 20 ng/mL IFN‐γ for 24 hours. This effect could be abrogated by the JAK2 inhibitor (JAK2i) or STAT1 inhibitor (STAT1i) but not by the JAK1/3 inhibitor (JAK1/3i). Total protein (40 μg) was loaded in each lane
3.5. Indoleamine 2,3‐dioxygenase 1 expressed in APCs but not the vascular endothelium of HCC
In our study, except for 7 patients who had too few inflammatory cells in the section to analyze, I‐IDO1 expression was entirely diffused (Figure 4A) and showed no association with clinicopathological features (Figure 4B‐E). We identified the types of IDO1‐positive inflammatory cells according to previous reports, including endothelial cells, macrophages, and DCs by immunofluorescence confocal microscopy. As shown in Figure 4F, IDO1 was expressed by APCs, including macrophages and DCs, but not by endothelial cells in HCC samples. Immunohistochemistry also showed distinctive expression patterns between I‐IDO1 and CD34 (Figure S5) and similar expression patterns between I‐IDO1 and CD68 (Figure S6).
Figure 4.
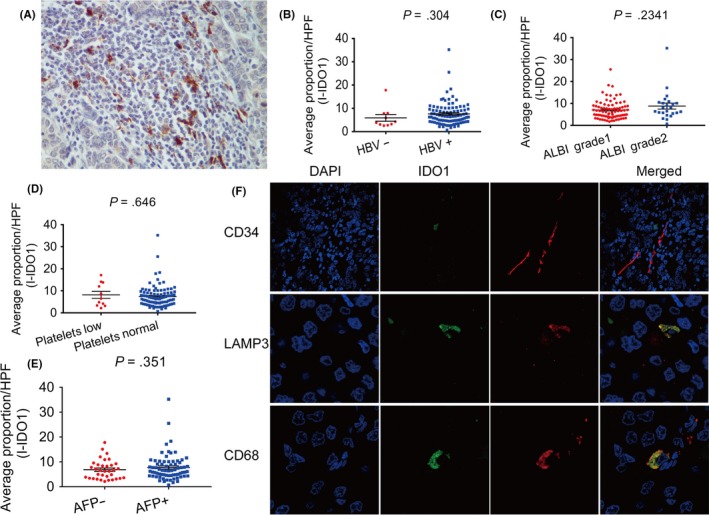
Expression pattern of indoleamine 2,3‐dioxygenase 1 in inflammatory cells (I‐IDO1) and correlations among the relative expression levels of I‐IDO1 with hepatitis B virus (HBV), albumin‐bilirubin (ALBI) grade, platelet level, and α‐fetoprotein level (AFP). A, Samples showing the pattern of IDO1 expression in inflammatory infiltration of hepatocellular carcinoma (brown). B‐E, Correlation analyses between I‐IDO1 expression and (B) HBV, (C) ALBI grade, (D) platelet level, or (E) AFP. F, Resected hepatocellular carcinoma samples were fixed in paraffin for immunofluorescence confocal analysis with primary Ab (red; top, anti‐CD34; middle, anti‐lysosomal associated membrane protein‐3 (LAMP3); bottom, anti‐CD68), anti‐IDO1 (green), and DAPI (blue). HPF, high‐power field
Transcriptional levels of IDO1 and vascular endothelial markers, including CD34, PECAM1, and CEACAM1, were also analyzed in TCGA database, and similarly restricted cochange was observed in HCC samples (Figure S7, n = 371).31
3.6. Associations of IDO1 expression and CD8+ tumor‐infiltrating lymphocytes with HCC patient prognosis
Given that CD8+ T cell infiltration positively correlated with prognosis and T‐IDO1 expression in our study, we next examined whether tumor invasion might be partially restrained in high IDO1 expression HCC. Favorable OS and DFS intervals were highlighted by Kaplan‐Meier curves (P = .003 and P = .045, respectively; Figure 5A,B) for patients with a high T‐IDO1 expression compared with those with low expression. HCC patients with high I‐IDO1 expression also have superior OS and DFS outcomes (P < .001 and P = .002, respectively; Figure 5C,D). In keeping with the previous findings, patients with high levels of CD8+ T cell infiltration had significantly prolonged OS and DFS (P = .004 and P = .027, respectively; Figure 5E,F), confirming the role of the cytotoxic T cell in the eradication of HCC.3 Multivariate analyses showed that T‐IDO1 expression was an independent predictor of OS and DFS (P = .007 and P = .044, respectively; Tables S2, S3).
Figure 5.
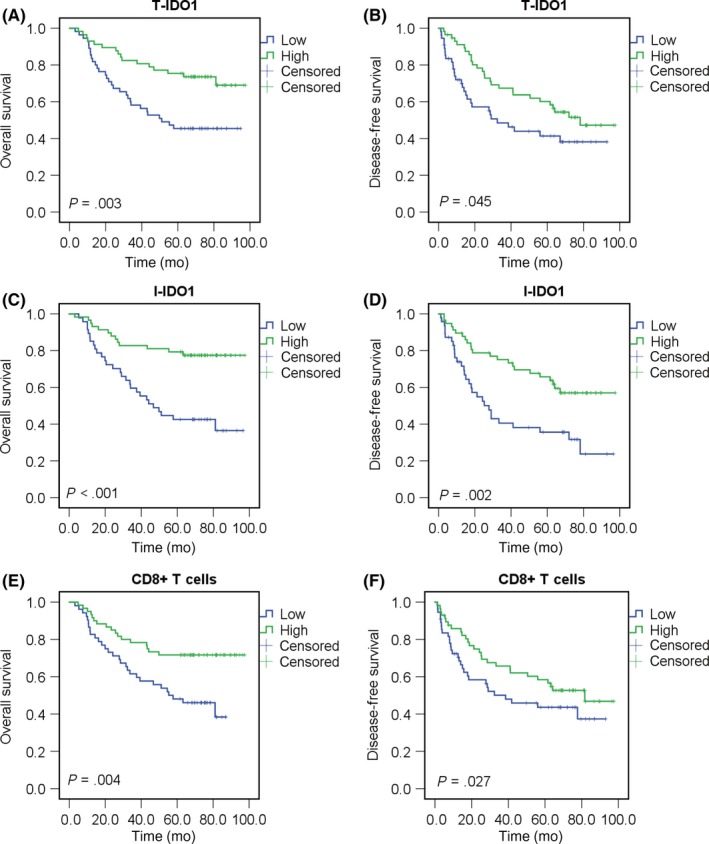
Indoleamine 2,3‐dioxygenase 1 in T cells (T‐IDO1) and inflammatory cells (I‐IDO1) and CD8+ T cell densities correlated with superior overall survival (OS) and disease‐free survival (DFS) of hepatocellular carcinoma (HCC) patients. A,B, Kaplan‐Meier curves for the analysis of OS (A) and DFS (B) in HCC patients (T‐IDO1 low group, n = 55; T‐IDO1 high group, n = 57) according to T‐IDO1 protein levels. P values were calculated by the log‐rank test. C,D, Kaplan‐Meier curves for the analysis of OS (C) and DFS (D) in HCC patients (I‐IDO1 low group, n = 47; I‐IDO1 high group, n = 58) according to I‐IDO1 protein levels. E,F, Kaplan‐Meier curves for the analysis of OS (E) and DFS (F) in HCC patients (CD8 low group, n = 52; CD8 high group, n = 60) according to CD8+ T cell density. P values were calculated by the log‐rank test
4. DISCUSSION
In HCC, previous research has described the prognostic significance of T‐IDO1 expression. Unfortunately, the conclusions are contradictory, and little is known regarding the prognostic value of I‐IDO1 expression; the potential mechanism regulating T‐IDO1 expression must also be clarified.27, 28, 29 Our study explored the potential mechanism regulating T‐IDO1 expression, and the correlations of T‐IDO1 or I‐IDO1 expression with clinicopathological features and survival were also analyzed.
To eliminate the effects of potential subjective factors on outcomes, validated primary Abs, ImageJ image evaluation software, and whole‐tissue sections, but not tissue microarrays, were used in our study. Our IHC data showed that tumors with a high level of T‐IDO1 expression were more likely accompanied by abundant CD8+ T cell infiltration. Substantial evidence has indicated that T‐IDO1 is expressed in response to the presence of T cells producing immune‐stimulating cytokines, such as IFN‐γ.21, 32, 33 In vitro cultured HCC cells with recombinant IFN‐γ, but not type I interferon, led to remarkable IDO1 expression, and the level of IDO1 mRNA significantly correlated with either IFNG or CD8A transcriptional levels in freshly resected HCCs, indicating that IDO1 was inductively expressed by HCC neoplastic cells. This result is in accordance with samples from TCGA database (as of March 2018). To further verify this association, upon IFN‐γ binding to its receptor, p‐STAT1 is the immediate downstream effector was tested in baseline and IFN‐γ‐stimulated HCC cell lysates by western blot. Both STAT1 and p‐STAT1 covaried with IDO1, and this effect could be abrogated by a JAK2 inhibitor or STAT1 inhibitor, but not by the JAK1/3 inhibitor, indicating that the T‐IDO1 expression in HCC was regulated by the IFNγ‐JAK2‐STAT1 signaling pathway. CD4+ T cells and NK cells are also sources of IFN‐γ in the tumor microenvironment, but the results of IHC analyses showed no correlation between T‐IDO1 expression and CD4+ T cell or NK cell infiltration. These findings are consistent with our reported quantitative PCR results in freshly resected HCC tissues, which showed no correlation between IFNG and CD4 or CD56 at the mRNA level.34 This might be due to NK cells seldom infiltrating into HCC, and there is not only Th1 in CD4+ T cells in the HCC microenvironment.35
Notably, T‐IDO1 staining was not proportional to infiltrating CD8+ T cells some of the cases in our study, perhaps due to IDO1 also being constitutively expressed by tumor cells through poorly characterized oncogenic signaling pathways. Previous studies have revealed that intrinsic cellular changes triggered by poorly characterized genetic mutations and epigenetic modifications can also regulate T‐IDO1 expression; for example, either demethylated promoters or low expression of Bin1 could lead to highly inducible IDO1 expression in breast cancer.36, 37, 38 Both intrinsic and extrinsic mechanisms could be independently or jointly determining the T‐IDO1 level in the HCC microenvironment.
Subsequent survival analysis showed that the upregulation of T‐IDO1 is a positive OS and DFS predictor in HCC. Given the positive correlation between CD8 and T‐IDO1 and the role of CD8+ T cells in preventing tumor invasion, T‐IDO1 expression is proposed as a proxy for spontaneous antitumor immune response in HCC. When clinicopathological features were included in the correlation analysis with T‐IDO1, only age correlated with T‐IDO1 expression. Patients with low levels of T‐IDO1 expression were older than patients with high levels, which could be explained by decreased production of cytokines and lower per‐cell cytotoxicity, resulting from innate immunosenescence.39 Thus, overexpression of the immunosuppressive molecule IDO1 by tumors positively correlates with enhanced antitumor immunity, and our seemingly contradictory conclusion has been supported by many previous studies in other tumors.21, 23, 34
Indoleamine 2,3‐dioxygenase 1‐positive cells were also detected in inflammatory infiltration in all cases, and I‐IDO1 expression positively correlated with OS, although it might be not an independent predictor. Extensive mechanistic studies disclosed that I‐IDO1 upregulation elicited by multiplex cytokines existed with inflammatory infiltration; in turn, high I‐IDO1 expression could downregulate inflammatory factors, and tumor cell proliferation was sequentially slowed.40, 41, 42 Consistent with previous studies, local tryptophan deprivation produced by abundant IDO1 could also disturb tumor growth.27, 43 We did not explore the potential mechanism because its inducible expression by APCs has been intensively studied, and it appears that no tumor specificity exists regarding I‐IDO1 expression.40, 44
Expression of I‐IDO1 tends to be higher in HBV+ HCCs than in HBV− HCCs; however, in our study, there was no significant difference between the groups, which might be due to the small number of HBV− HCC patients included in our study. Yoshio and colleagues discovered that IDO1 was mainly induced by IFN‐γ in patients with hepatitis B.45 It was safely deduced that inflammatory cells could be stimulated with cytokines induced by HBV‐infected hepatocytes and then by unregulated IDO1 expression. Hence, I‐IDO1 expression comprehensively reflects the host immune response to the tumor and HBV, supporting that antiviral therapy could improve HCC patient prognosis.46
There were some limitations in this study. A major limitation was its retrospective nature, which incurred the inevitable selection bias. To distinguish tumor and stroma, we did not analyze the entire section but included 5 high‐power fields in a representative region, which might also have contributed to selection bias due to tumor heterogeneity. Because of poor uniformity of the IDO1 IHC Abs and evaluation methods, there is no widely accepted cut‐off value for IDO1 expression in HCC; therefore, we defined one according to the present cohort, which could have led to the discrepancy between our results and other studies. The overwhelming majority cohort in our research is HBV‐associated HCC, which is due to the high incidence of HCC associated with endemic HBV infection in China.47
Our study revealed that IDO1 expression, which is common in HCC, is a positive survival predictor, and its expression is a counter‐regulatory action to suppress the antitumor immune response. Thus, combined inhibition of IDO1 and immune checkpoints to boost host antitumor immune response could be a promising strategy in HCC treatment, and understanding of the mechanism of IDO1 expression would help us explore better combined strategies.7, 23, 48
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
The authors thank Benoit J. Van den Eynde, MD, PhD, for the production of anti‐hIDO1 antibody and two pathologists, Tong Yang and Yan Shen, for the analysis of immunohistochemistry.
Li S, Han X, Lyu N, et al. Mechanism and prognostic value of indoleamine 2,3‐dioxygenase 1 expressed in hepatocellular carcinoma. Cancer Sci. 2018;109:3726–3736. 10.1111/cas.13811
Funding information
This work was supported by grants from the National Natural Science Foundation of China (81771956) and the National Natural Science Foundation of Guangdong (2016A030313282 and 2017A020215149).
REFERENCES
- 1. Berasain C, Castillo J, Perugorria MJ, Latasa MU, Prieto J, Avila MA. Inflammation and liver cancer: new molecular links. Ann N Y Acad Sci. 2009;1155:206‐221. [DOI] [PubMed] [Google Scholar]
- 2. Flecken T, Schmidt N, Hild S, et al. Immunodominance and functional alterations of tumor‐associated antigen‐specific CD8 + T‐cell responses in hepatocellular carcinoma. Hepatology. 2014;59:1415‐1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gabrielson A, Wu Y, Wang H, et al. Intratumoral CD3 and CD8 T‐cell densities associated with relapse‐free survival in HCC. Cancer Immunol Res. 2016;4:419‐430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu K, Kryczek I, Chen L, Zou W, Welling TH. Kupffer cell suppression of CD8+ T cells in human hepatocellular carcinoma is mediated by B7‐H1/programmed death‐1 interactions. Cancer Res. 2009;69:8067‐8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sangro B, Gomez‐Martin C, de la Mata M, et al. A clinical trial of CTLA‐4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J Hepatol. 2013;59:81‐88. [DOI] [PubMed] [Google Scholar]
- 6. El‐Khoueiry AB, Sangro B, Yau T, et al. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open‐label, non‐comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389(10088):2492‐2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brown ZJ, Yu SJ, Heinrich B, et al. Indoleamine 2,3‐dioxygenase provides adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma. Cancer Immunol Immunother. 2018;67:1305‐1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uyttenhove C, Pilotte L, Theate I, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3‐dioxygenase. Nat Med. 2003;9:1269‐1274. [DOI] [PubMed] [Google Scholar]
- 9. Theate I, van Baren N, Pilotte L, et al. Extensive profiling of the expression of the indoleamine 2,3‐dioxygenase 1 protein in normal and tumoral human tissues. Cancer Immunol Res. 2015;3:161‐172. [DOI] [PubMed] [Google Scholar]
- 10. Prendergast GC, Smith C, Thomas S, et al. Indoleamine 2,3‐dioxygenase pathways of pathogenic inflammation and immune escape in cancer. Cancer Immunol Immunother. 2014;63:721‐735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Terness P, Bauer TM, Röse L, et al. Inhibition of allogeneic T cell proliferation by indoleamine 2,3‐dioxygenase‐expressing dendritic cells: mediation of suppression by tryptophan metabolites. J Exp Med. 2002;196:447‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Munn DH, Shafizadeh E, Attwood JT, Bondarev I, Pashine A, Mellor AL. Inhibition of T cell proliferation by macrophage tryptophan catabolism. J Exp Med. 1999;189:1363‐1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fallarino F, Grohmann U, You S, et al. The combined effects of tryptophan starvation and tryptophan catabolites down‐regulate T cell receptor zeta‐chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752‐6761. [DOI] [PubMed] [Google Scholar]
- 14. Van de Velde LA, Guo XJ, Barbaric L, et al. Stress kinase GCN2 controls the proliferative fitness and trafficking of cytotoxic T cells independent of environmental amino acid sensing. Cell Rep. 2016;17:2247‐2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aldajani WA, Salazar F, Sewell HF, Knox A, Ghaemmaghami AM. Expression and regulation of immune‐modulatory enzyme indoleamine 2,3‐dioxygenase (IDO) by human airway epithelial cells and its effect on T cell activation. Oncotarget. 2016;7:57606‐57617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Urakawa H, Nishida Y, Nakashima H, Shimoyama Y, Nakamura S, Ishiguro N. Prognostic value of indoleamine 2,3‐dioxygenase expression in high grade osteosarcoma. Clin Exp Metastasis. 2009;26:1005‐1012. [DOI] [PubMed] [Google Scholar]
- 17. Balachandran VP, Cavnar MJ, Zeng S, et al. Imatinib potentiates antitumor T cell responses in gastrointestinal stromal tumor through the inhibition of Ido. Nat Med. 2011;17:1094‐1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferdinande L, Decaestecker C, Verset L, et al. Clinicopathological significance of indoleamine 2,3‐dioxygenase 1 expression in colorectal cancer. Br J Cancer. 2012;106:141‐147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ino K, Yamamoto E, Shibata K, et al. Inverse correlation between tumoral indoleamine 2,3‐dioxygenase expression and tumor‐infiltrating lymphocytes in endometrial cancer: its association with disease progression and survival. Clin Cancer Res. 2008;14:2310‐2317. [DOI] [PubMed] [Google Scholar]
- 20. Inaba T, Ino K, Kajiyama H, et al. Role of the immunosuppressive enzyme indoleamine 2,3‐dioxygenase in the progression of ovarian carcinoma. Gynecol Oncol. 2009;115:185‐192. [DOI] [PubMed] [Google Scholar]
- 21. Jacquemier J, Bertucci F, Finetti P, et al. High expression of indoleamine 2,3‐dioxygenase in the tumour is associated with medullary features and favourable outcome in basal‐like breast carcinoma. Int J Cancer. 2012;130:96‐104. [DOI] [PubMed] [Google Scholar]
- 22. Soliman H, Rawal B, Fulp J, et al. Analysis of indoleamine 2‐3 dioxygenase (IDO1) expression in breast cancer tissue by immunohistochemistry. Cancer Immunol Immunother. 2013;62:829‐837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spranger S, Spaapen RM, Zha Y, et al. Up‐regulation of PD‐L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med 2013;5:200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heeren AM, van Dijk I, Drai B, et al. Indoleamine 2,3‐dioxygenase expression pattern in the tumor microenvironment predicts clinical outcome in early stage cervical cancer. Front Immunol. 2018;9:1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rubel F, Kern JS, Technau‐Hafsi K, et al. Indoleamine 2,3‐dioxygenase expression in primary cutaneous melanoma correlates with Breslow thickness and is of significant prognostic value for progression‐free survival. J Invest Dermatol. 2018;138:679‐687. [DOI] [PubMed] [Google Scholar]
- 26. Munn DH, Mellor AL. IDO in the tumor microenvironment: inflammation, counter‐regulation, and tolerance. Trends Immunol. 2016;37:193‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishio T, Goto S, Tahara K, Tone S, Kawano K, Kitano S. Immunoactivative role of indoleamine 2,3‐dioxygenase in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2004;19:319‐326. [DOI] [PubMed] [Google Scholar]
- 28. Pan K, Wang H, Chen MS, et al. Expression and prognosis role of indoleamine 2,3‐dioxygenase in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134:1247‐1253. [DOI] [PubMed] [Google Scholar]
- 29. Sideras K, Biermann K, Verheij J, et al. PD‐L1, Galectin‐9 and CD8(+) tumor‐infiltrating lymphocytes are associated with survival in hepatocellular carcinoma. Oncoimmunology. 2017;6:e1273309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson PJ, Berhane S, Kagebayashi C, et al. Assessment of liver function in patients with hepatocellular carcinoma: a new evidence‐based approach‐the ALBI grade. J Clin Oncol. 2015;33:550‐558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakamura T, Shima T, Saeki A, et al. Expression of indoleamine 2, 3‐dioxygenase and the recruitment of Foxp3‐expressing regulatory T cells in the development and progression of uterine cervical cancer. Cancer Sci. 2007;98:874‐881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Choe JY, Yun JY, Jeon YK, et al. Indoleamine 2,3‐dioxygenase (IDO) is frequently expressed in stromal cells of Hodgkin lymphoma and is associated with adverse clinical features: a retrospective cohort study. BMC Cancer. 2014;14:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie QK, Zhao YJ, Pan T, et al. Programmed death ligand 1 as an indicator of pre‐existing adaptive immune responses in human hepatocellular carcinoma. Oncoimmunology. 2016;5:e1181252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yan J, Liu XL, Xiao G, et al. Prevalence and clinical relevance of T‐helper cells, Th17 and Th1, in hepatitis B virus‐related hepatocellular carcinoma. PLoS ONE. 2014;9:e96080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jia Y, Wang H, Wang Y, et al. Low expression of Bin1, along with high expression of IDO in tumor tissue and draining lymph nodes, are predictors of poor prognosis for esophageal squamous cell cancer patients. Int J Cancer. 2015;137:1095‐1106. [DOI] [PubMed] [Google Scholar]
- 37. Noonepalle SK, Gu F, Lee EJ, et al. Promoter methylation modulates indoleamine 2,3‐dioxygenase 1 induction by activated T cells in human breast cancers. Cancer Immunol Res. 2017;5:330‐344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hennequart M, Pilotte L, Cane S, et al. Constitutive IDO1 expression in human tumors is driven by cyclooxygenase‐2 and mediates intrinsic immune resistance. Cancer Immunol Res. 2017;5(8):695‐709. [DOI] [PubMed] [Google Scholar]
- 39. Solana R, Tarazona R, Gayoso I, Lesur O, Dupuis G, Fulop T. Innate immunosenescence: effect of aging on cells and receptors of the innate immune system in humans. Semin Immunol. 2012;24:331‐341. [DOI] [PubMed] [Google Scholar]
- 40. Mondanelli G, Bianchi R, Pallotta MT, et al. A relay pathway between arginine and tryptophan metabolism confers immunosuppressive properties on dendritic cells. Immunity. 2017;46:233‐244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takikawa O. Biochemical and medical aspects of the indoleamine 2,3‐dioxygenase‐initiated L‐tryptophan metabolism. Biochem Biophys Res Commun. 2005;338:12‐19. [DOI] [PubMed] [Google Scholar]
- 42. Ogiso H, Ito H, Kanbe A, et al. The inhibition of indoleamine 2,3‐dioxygenase accelerates early liver regeneration in mice after partial hepatectomy. Dig Dis Sci. 2017;62(9):2386‐2396. [DOI] [PubMed] [Google Scholar]
- 43. Brochez L, Chevolet I, Kruse V. The rationale of indoleamine 2,3‐dioxygenase inhibition for cancer therapy. Eur J Cancer. 2017;76:167‐182. [DOI] [PubMed] [Google Scholar]
- 44. Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol. 2004;4:762‐774. [DOI] [PubMed] [Google Scholar]
- 45. Yoshio S, Sugiyama M, Shoji H, et al. Indoleamine‐2, 3‐dioxygenase as an effector and an indicator of protective immune responses in patients with acute hepatitis B. Hepatology. 2016;63:83‐94. [DOI] [PubMed] [Google Scholar]
- 46. Chen JL, Lin XJ, Zhou Q, Shi M, Li SP, Lao XM. Association of HBV DNA replication with antiviral treatment outcomes in the patients with early‐stage HBV‐related hepatocellular carcinoma undergoing curative resection. Chin J Cancer. 2016;35:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bertuccio P, Turati F, Carioli G, et al. Global trends and predictions in hepatocellular carcinoma mortality. J Hepatol. 2017;67:302‐309. [DOI] [PubMed] [Google Scholar]
- 48. Beatty GL, O'Dwyer PJ, Clark J, et al. First‐in‐Human Phase I Study of the oral inhibitor of indoleamine 2,3‐Dioxygenase‐1 epacadostat (INCB024360) in patients with advanced solid malignancies. Clin Cancer Res. 2017;23(13):3269‐3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


