Abstract
Three new sesquiterpene aryl esters and eight known compounds were isolated from the EtOH extract of the mycelium of Armillaria mellea. The structures of new compounds were established by analysis of their spectroscopic data. Some of the isolates showed cytotoxicity to a variety of cancer cell lines, including MCF-7, H460, HT-29, and CEM.
Keywords: sesquiterpene aryl esters, Armillaria mellea, Tricholomataceae, cytotoxicity
1. Introduction
Armillaria mellea (Tricholomataceae) is a fungus symbiotic with the Chinese medicinal herb “Tianma” (Gastrodia elata Blume). The fruiting bodies of A. mellea have been used in Traditional Chinese Medicine for the treatment of hypertension, headache, insomnia, dizziness, and vertigo. Recently, the cultured mycelium of A. mellea became a health food in Taiwan and China and its tablets are used to treat geriatric patients with palsy, headache, insomnia, dizziness, and neurasthenia [1,2,3,4,5]. Previous chemical studies of A. mellea reported the isolation of a number of sesquiterpene aromatic esters, in which the tricyclic 5-6-4 protoilludane or protoilludene alcohols were esterified with orsellinic acid or its derivatives [1,2,3,6,7,8,9,10,11,12]. Some of the sesquiterpene aryl esters exhibited cytotoxic activities against human cancer cells. Among these sesquiterpene aryl esters, arnamial showed cytotoxicity against MCF-7, CCRF-CEM, HCT-116, and Jurkat T cells, but melledonal C only showed cytotoxic activity against CCRF-CEM cells [11]. Armillaridin was reported to exhibit cytotoxicity against MCF-7, HeLa, K562, and Jurkat T cells [12]. In addition to inhibiting the growth of the cancer cells, armillaridin also enhanced radiosensitivity of human esophageal cancer cells and there might be potential to integrate armillaridin with radiotherapy for esophageal cancer treatment [13]. Moreover, 4-O-methylarmillaridin showed cytotoxicity against MCF-7 and Jurkat T cells and dehydroarmillyl orsellinate showed cytotoxicity against MCF-7 and K562 cells [12]. Recently, armillarikin was reported to inhibit growth and induce apoptosis in human leukaemic K562, U937, and HL-60 cells [14]. In this paper, we describe the isolation and structural elucidation of three new sesquiterpene aryl esters from the mycelium of A. mellea, as well as the cytotoxic activities of the isolated components [15].
2. Results and Discussion
The EtOH extract of the mycelium of A. mellea was partitioned with EtOAc and H2O. The EtOAc layer was chromatographed repeatedly to afford three new compounds, 1–3 (Figure 1), along with eight known compounds: 6′-chloromelleolide F (4) [16], 13-hydroxymelleolide K (5) [17], armillaricin (6) [3], armillaridin (7) [1], armillarikin (8) [2], melleolide F (9) [11], melledonal C (10) [18], and melledonal B (11) [18]. The structures of these new compounds were established by their spectroscopic data and the known compounds were identified by comparison of their NMR data with those reported in the literature.
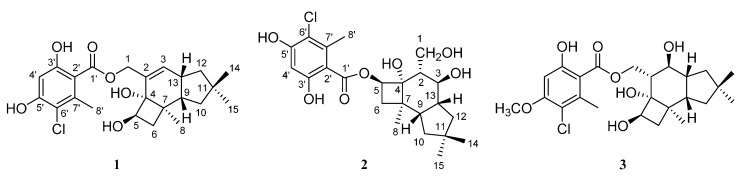
Figure 1.
The structures of 1–3 isolated from the mycelium of Amillaria mellea.
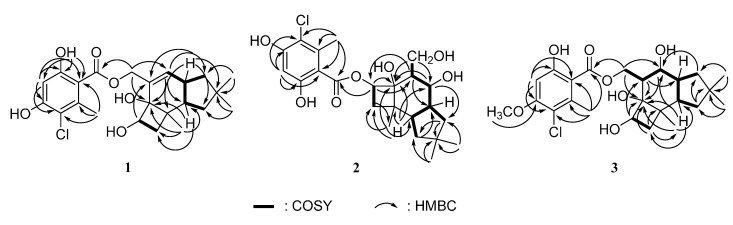
Compound 1 was obtained as a colorless solid. Its ESIMS spectrum showed two peaks at m/z 459 and 461, which were attributed to [M + Na]+ and [M + 2 + Na]+ ions, respectively. The ratio of their intensities was about 3:1, suggesting the presence of a chlorine atom in compound 1. The molecular formula C23H29ClO6 was deduced from FABHRMS and NMR (Table 1) spectra, which indicated that there were nine double bond equivalents. The 13C and DEPT spectra displayed four methyl carbons at δ 19.7, 22.3, 32.1, and 32.3, four methylene carbons, including an oxymethylene group at δ 67.8, five methine carbons, including one oxymethine group at δ 76.9 and two olefinic or aromatic carbons at δ 102.7 and 136.1, and ten quaternary carbons. Four of the quaternary carbons were oxygenated where one carbonyl, one aliphatic, and two aromatic carbons showed signals at δ 170.4, 78.1, 158.2, and 161.8, respectively. The 1H-NMR spectrum of 1 exhibited four methyl singlets at δ 0.95, 0.97, 1.15, and 2.62 and oxymethylene signals at δ 4.93 (dt, J = 12.6, 1.2 Hz) and 5.19 (ddd, J = 12.6, 1.8, 0.6 Hz). Two singlets at δ 5.87 and 6.43 were derived from olefinic or aromatic protons. Moreover, a signal at δ 10.74 revealed the presence of a chelated phenolic hydroxyl group. The one bond 1H–13C connectivities were analyzed by using HSQC data and protons attached to carbons were assigned. In the COSY spectrum of 1, proton H-9 at δ 2.12 showed correlations with H2-10 (δ 1.35 and 1.47) and H-13 (δ 2.73) and proton H-13 also showed correlation with H2-12 (δ 1.83), which indicated a linkage of C-10–C-9–C-13–C-12 (Figure 2). Besides, the linkages of C-3–C-13 and C-5–C-6 were deduced by COSY cross-peaks of H-3/H-13 and H-5/H2-6. Further connectivities were established by long-range HMBC correlations shown in Figure 2. Cross-peaks of H-3/C-4,C-9,C-12 and H-13/C-2,C-7,C-10,C-11 revealed that a cyclopentane ring was fused to a cyclohexene ring. Moreover, HMBC cross-peaks of H-5/C-2,C-4,C-6 and H-9/C-4,C-6 as well as the COSY correlation of H-5 and H2-6 suggested that a cyclobutane ring was fused to the cyclohexene ring. The HMBC data further showed cross-peaks of H3-8/C-4,C-6,C-7,C-9 and H3-14,H3-15/C-10–C-12, which indicated that one and two methyl groups were attached to C-7 and C-11, respectively. Besides, the correlations of H2-1 to C-2, C-3 and C-4 suggested that an oxygenated methylene group was linked to C-2. Based on the above evidence, a protoillud-7-ene skeleton was deduced for compound 1 [6]. In addition to one double bond in the protoilludene moiety, six other unsaturated carbon signals and a total of nine double bond equivalents in 1 revealed that an aromatic ring could be a part of this compound. Furthermore, HMBC cross-peaks of H-8′/C-2′,C-6′,C-7′ and H-4′/C-2′,C-3′,C-5′,C-6′ were observed, which suggested the presence of a 3-chloro-4,6-dihydroxy-2-methylphenyl moiety. In the HMBC spectrum acquired in CDCl3, the proton signal (δ 11.08) of the hydroxyl group chelated to the carbonyl group at the aromatic ring showed correlations with C-2′, C-3′, and C-4′, which indicated that the hydroxyl and the carbonyl groups were located at C-3′ and C-2′, respectively. Hence, the chlorine atom was suggested to be located at C-6′. The linkage of this benzoyl group to the oxygen atom at C-1 was confirmed by the HMBC correlation of H2-1 to C-1′. The relative configuration of 1 was deduced from NOE experiments in CDCl3. The irradiation of H3-8 enhanced the signals of H-5 and H-6a (δ 1.87); however, the irradiation of H-9 enhanced the signals of H-6b (δ 1.30), H-13, and H3-14. Thus, H-6b, H-9, H-13, and H3-14 were on the same face of the protoilludene moiety and H-5, H-6a, and H3-8 were on the other face. In the 13C-NMR spectrum acquired in CDCl3, signals for C-2–C-15 resembled those of a protoilludene-type sesquiterpene, echinocidin B, isolated from a mycelial culture of Echinodontium tsugicola, in which cis junctures of cyclohexene to both cyclobutane and cyclopentane rings were determined [19]. Accordingly, it was deduced that the cyclohexene ring in 1 was also fused to both cyclobutane and cyclopentane rings in a cis-fashion and the relative structure of 1 was established. Compound 1 was given the trivial name melleolide N.
Table 1.
1H- and 13C-NMR data of compounds 1–3 in acetone-d6 a.
| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH b | δC | δH b | δC | δH b | δC | |
| 1 | 4.93 (dt, 12.6, 1.2) 5.19 (ddd, 12.6, 1.8, 0.6) | 67.8 | 3.89 (dd, 10.8, 4.8) 4.03 (dd, 11.4, 3.6) | 62.9 | 4.68 (dd, 11.4, 4.2) 4.71 (dd, 11.4, 4.2) | 66.2 |
| 2 | 132.0 | 2.09 (m) | 46.4 | 2.40 (dt, 10.8, 4.2) | 43.8 | |
| 3 | 5.87 (br s) | 136.1 | 3.72 (t, 11.4) | 69.0 | 3.77 (t, 10.8) | 68.6 |
| 4 | 78.1 | 81.4 | 81.8 | |||
| 5 | 4.33 (t, 8.4) | 76.9 | 5.33 (t, 8.4) | 77.2 | 4.19 (t, 8.4) | 73.4 |
| 6 | 1.38 (dd, 10.8, 9.0) 1.75 (dd, 10.8, 8.4) | 36.4 | 1.85 (dd, 10.8, 9.0) 1.95 (dd, 10.8, 7.8) | 34.5 | 1.54 (dd, 10.2, 8.4) 1.71 (dd, 10.8, 8.4) | 37.0 |
| 7 | 38.0 | 39.0 | 37.7 | |||
| 8 | 1.15 (s) | 22.3 | 1.16 (s) | 22.3 | 1.06 (s) | 22.5 |
| 9 | 2.12 (m) | 45.4 | 2.16 (m) | 48.1 | 2.09 (m) | 48.3 |
| 10 | 1.35 (dd, 12.6, 7.2) 1.46 (t, 12.6) | 42.3 | 1.46 (m) | 44.4 | 1.44 (m) | 44.7 |
| 11 | 38.5 | 36.7 | 36.7 | |||
| 12 | 1.45 (d, 13.2) 1.83 (dd, 13.2, 8.4) | 48.3 | 1.52 (dd, 13.8, 7.8) 1.98 (d, 14.4) | 43.4 | 1.52 (dd, 13.8, 7.8) 1.97 (m) | 43.6 |
| 13 | 2.73 (br t, 7.8) | 40.2 | 2.00 (m) | 47.5 | 1.97 (m) | 47.9 |
| 14 | 0.97 (s) | 32.3 | 0.99 (s) | 32.4 | 0.99 (s) | 32.4 |
| 15 | 0.95 (s) | 32.1 | 1.09 (s) | 32.7 | 1.07 (s) | 32.7 |
| 1′ | 170.4 | 171.1 | 169.0 | |||
| 2′ | 109.0 | 108.3 | 110.4 | |||
| 3′ | 161.8 | 162.4 | 160.9 | |||
| 4′ | 6.43 (s) | 102.7 | 6.45 (s) | 102.6 | 6.45 (s) | 99.6 |
| 5′ | 158.2 | 158.6 | 159.4 | |||
| 6′ | 114.7 | 114.8 | 115.2 | |||
| 7′ | 140.4 | 140.2 | 139.8 | |||
| 8′ | 2.62 (s) | 19.7 | 2.62 (s) | 19.9 | 2.57 (s) | 18.9 |
| 3′-OH | 10.74 (br s) | 10.97 (br s) | ||||
| 5′-OH | 9.42 (br s) | 9.56 (br s) | ||||
| 5′-OCH3 | 3.89 (s) | 55.6 | ||||
a Spectra recorded at 600 MHz for 1H-NMR and 150 MHz for 13C-NMR; b Multiplicities and J values (in Hz) are in parentheses.
Figure 2.
COSY and selected HMBC correlations of 1–3.
Compound 2 gave a molecular formula of C23H31ClO7, which was deduced from its ESIHRMS and NMR (Table 1) spectra. Its 13C and DEPT NMR spectra showed four methyl at δ 19.9, 22.3, 32.4, and 32.7, four methylene, six methine, and nine quaternary carbons. In addition to a carbonyl group with a signal at δ 171.1, six carbons were oxygenated, which included one oxymethylene signal at δ 62.9, two oxymethine signals at δ 69.0 and 77.2, one aliphatic quaternary carbon signal at δ 81.4, and two aromatic carbon signals at δ 158.6 and 162.4. The carbon signals above 100 ppm were almost the same as the signals of 3-chloro-4,6-dihydroxy-2-methylbenzoyl group in 1, suggesting the presence of this benzoyl group in 2. The 1H-NMR spectrum of 2 exhibited four methyl groups at δ 0.99, 1.09 (6H), and 2.62. Besides, it showed two signals at δ 3.89 (dd, J = 10.8, 4.8 Hz) and 4.03 (dd, J = 11.4, 3.6 Hz), which correlated to the carbon signal at δ 62.9 in the HSQC spectrum and were attributed to an oxymethylene group. Moreover, two signals at δ 3.72 (t, J = 11.4 Hz) and 5.33 (t, J = 8.4 Hz) correlating to the carbon signals at δ 69.0 and 77.2, respectively, indicated the presence of two oxymethine groups. A signal at δ 6.45 (s) was also observed and the proton correlated to the carbon resonating at δ 102.6, suggesting that they were in an aromatic ring. Its HMBC spectrum (Figure 2) displayed cross-peaks of H2-1/C-2–C-4, H-3/C-1,C-4,C-12, H-5/C-2,C-4,C-6, H3-8/C-4,C-6,C-7,C-9, and H2-12/C-3,C-10,C-11,C-13–C-15, which revealed a protoilludane skeleton for 2 with C-1, C-3, C-4, and C-5 oxygenated. The 13C-NMR data of the protoilludane moiety in CD3OD were almost the same as those in 5′-methoxy-6′-chloroarmillane [12], which suggested that the structures of the protoilludane moieties in 2 and 5′-methoxy-6′-chloroarmillane were the same. The linkage of 3-chloro-4,6-dihydroxy-2-methylbenzoyl group to the oxygen atom at C-5 was confirmed by the HMBC correlation of H-5 to C-1′. In the NOESY spectrum, the cross-peaks of H3-14/H-9, H-12b (δ 1.52), H-9/H-6b (δ 1.85), and H-6b/H-2 revealed that H-2, H-6b, H-9, H-12b, and H3-14 were on the same face of the protoilludane moiety. Cross-peaks of H-3/H3-15,H-12a (δ 1.98), H3-8/H-5,H-6a (δ 1.95) indicated that H-3, H-5, H-6a, H3-8, H-12a, and H3-15 were on the other face. Furthermore, the large coupling constant of 11.2 Hz for H-3 indicated that H-2 and H-3 were in a trans configuration and the hydroxy-methyl and 3-OH groups were on opposite faces. Thus, the relative structure of 2 was determined and it was named melleolide Q. This compound is similar to 5′-methoxy-6′-chloroarmillane where a methoxyl group instead of a hydroxyl group is located at C-5′ [12].
Compound 3 gave a molecular formula of C24H33ClO7 deduced from FABHRMS and NMR (Table 1) spectra. Its 13C-NMR spectrum displayed 24 signals, including a methoxyl signal at δ 55.6 and a carbonyl signal at δ 169.0, and the signals for C-3, C-4, and C-8–C-15 were similar to those in 2. In the aromatic region, the signals resembled those in 2 except that the signal for C-4′ shifted from δ 102.6 to 99.6. In the HMBC spectrum (Figure 2), the methoxyl protons showed correlation to C-5′, which indicated that the methoxyl group was attached to C-5′ carbon of the aromatic ring. The 1H-NMR spectrum of 3 was also similar to that of 2 except for the signals of H2-1, H-2, H-5, and H2-6 as well as one additional signal at δ 3.89 (s) attributed to a methoxyl group. Signals for H2-1 shifted downfield to δ 4.68 (dd, J = 11.4, 4.2 Hz) and 4.71 (dd, J = 11.4, 4.2 Hz) from δ 3.89 and 4.03; signals for H-2 shifted downfield to δ 2.40 (dt, J = 10.8, 4.2 Hz) from δ 2.09. Moreover, H-5 signal shifted upfield from δ 5.33 to 4.19 (t, J = 8.4 Hz) and the signals of H2-6 shifted upfield from δ 1.85 and 1.95 to δ 1.54 (dd, J = 10.2, 8.4 Hz) and 1.71 (dd, J = 10.8, 8.4 Hz). Therefore, the benzoyloxy group in 3 was suggested to be linked to C-1, which was confirmed by the HMBC correlation of H2-1 to C-1′. The stereochemistry of 3 was established by the NOESY spectrum and the coupling constant of H-2 and H-3. The NOESY cross-peaks of H3-14/H-9,H-12b (δ 1.52), H-9/H-6b (δ 1.54), H-3/H-12a (δ 1.97), and H-5/H3-8,H-6a (δ 1.71) and the large coupling constant of 10.8 Hz for H-3 revealed that its relative configuration was the same as that in 2. Accordingly, the relative structure of 3 was determined and it was named melleolide R.
The isolated compounds from A. mellea were tested in vitro for cytotoxicity to a variety of human cancer cell lines including MCF-7, H460, HT-29, and CEM and the results are summarized in Table 2. Compounds 2–4 and 6–9 showed cytotoxicity to MCF-7 cells, in which compound 2 was most cytotoxic. Compounds 1, 4, and 6–9 exhibited comparable cytotoxicity against H460 cells. Compounds 1 and 6 showed stronger cytotoxicity to HT-29 cells than other tested compounds. Compounds 1, 3, and 5–7 showed comparable cytotoxicity to human leukemia cells. Among all tested compounds, 6 exhibited cytotoxicity to all of these cancer cells. Compounds 1–5 and 7–9 showed selective cytotoxicity and compounds 10 and 11 were inactive to these cancer cell lines.
Table 2.
Cytotoxicity of the isolated compounds from A. mellea against several cancer cell lines a.
| Compound | IC50 (μM) | |||
|---|---|---|---|---|
| MCF-7 | H460 | HT-29 | CEM | |
| 1 | 56.5 ± 4.2 | 5.5 ± 0.6 | 7.1 ± 0.8 | 5.4 ± 0.3 |
| 2 | 1.5 ± 0.1 | 80.0 ± 8.9 | 54.2 ± 4.7 | 10.3 ± 2.3 |
| 3 | 3.7 ± 0.3 | 53.8 ± 6.2 | 18.7 ± 3.2 | 3.4 ± 0.2 |
| 4 | 4.8 ± 0.5 | 4.5 ± 0.4 | 56.7 ± 4.5 | 28.8 ± 1.2 |
| 5 | >100 | >100 | 32.1 ± 3.6 | 5.5 ± 0.6 |
| 6 | 4.8 ± 0.4 | 5.5 ± 0.4 | 4.6 ± 0.3 | 5.8 ± 0.6 |
| 7 | 1.7 ± 0.2 | 4.5 ± 0.3 | 42.1 ± 5.1 | 5.1 ± 0.4 |
| 8 | 4.4 ± 0.8 | 5.7 ± 0.5 | 34.7 ± 4.6 | 44.6 ± 4.4 |
| 9 | 8.3 ± 2.2 | 5.1 ± 0.2 | 58.4 ± 4.9 | 41.2 ± 3.4 |
| 10 | >100 | >100 | 85.6 ± 9.1 | 49.6 ± 5.2 |
| 11 | >100 | >100 | >100 | >100 |
| Dox | 0.27 ± 0.02 | 0.01 ± 0.005 | 0.12 ± 0.01 | 0.09 ± 0.01 |
a MCF-7, human breast cancer; H460, human lung cancer; HT-29, human colon cancer; CEM, human leukaemia. Dox: Doxorubicin as a positive control.
3. Experimental Section
3.1. General Experimental Procedures
Optical rotations were taken on a P-2000 digital polarimeter (JASCO, Tokyo, Japan). UV spectra were measured on a U-3310 spectrophotometer (Hitachi, Tokyo, Japan). IR spectra were recorded on an Avatar 320 FT-IR spectrophotometer (Nicolet, Madison, WI, USA). 1H-, 13C-, and 2D-NMR spectra were recorded on a VNMRS 600 MHz spectrometer (Varian, Palo Alto, CA, USA). ESIMS and HRMS spectra were obtained on LCQ and Quest MAT 95XL spectrometers (Finnigan/Thermo, San Jose, CA, USA), respectively. HPLC was conducted on a model 1100 system (HP, Palo Alto, CA, USA) equipped with a G1311A QuatPump, a G1322A degasser, and a G1315B photodiode array detector set at 254 nm. Semipreparative HPLC was performed using a reversed-phase column (Cosmosil 5C18-MS-II, 5 μm, 10 × 250 mm) at a flow rate of 2.0 mL/min.
3.2. Source of Organism
The strain of the fungus A. mellea (# BCRC 36361) was purchased from the Food Industry Research and Development Institute, Hsinchu, Taiwan.
3.3. Fermentation of Organism
The strain BCRC 36361 was inoculated into 1 L of the medium (1.0% glucose, 1.0% oat powder, 0.1% peptone, 0.1% yeast extract, pH 4.5) in a 2-L Hinton flask at 25 °C on a rotary shaker (120 rpm) for six days. The mycelium was aseptically transferred to a 500-L fermenter containing 400 L of the above medium and incubated at 25 °C for ten days.
3.4. Extraction and Isolation
The mycelium of A. mellea (9.0 kg) was extracted with 95% EtOH (50 L) three times. The 95% EtOH soluble portion was concentrated to give the EtOH extract, which was partitioned with H2O and EtOAc. The EtOAc layer was chromatographed on silica gel column and eluted with n-hexane-EtOAc (20:1 → 0:1) to provide ten fractions (Fr-1–Fr-10). Fraction Fr-3 (n-hexane/EtOAc = 5:1) was concentrated and recrystallized in MeOH to afford armillaricin (6, 114 mg). Fraction Fr-4 (n-hexane/EtOAc = 5:1) was purified by semi-preparative reversed-phase HPLC [H2O/CH3CN (15:85, 0 min) → H2O/CH3CN (0:100, 20 min)] to give armillaridin (7, 365 mg, Rt = 18.53 min). Fraction Fr-7 (n-hexane/EtOAc = 1:1) was repeatedly chromatographed on silica gel (CHCl3/MeOH = 50:1 → 20:1) and Sephadex LH-20 (MeOH) columns to yield armillarikin (8, 676 mg), 2 (42.9 mg), 6′-chloromelleolide F (4, 57.7 mg), and melleolide F (9, 37 mg). Fraction Fr-9 (n-hexane/EtOAc = 0:1) was repeatedly chromatographed on silica gel (CHCl3/MeOH = 30:1 → 20:1), Sephadex LH-20 (H2O/MeOH = 3:7), and RP-18 (H2O/MeOH = 2:8) columns to afford two compounds, melledonal C (10, 3.50 g) and 1 (101 mg), and one sub-fraction Fr-9-1. Sub-fraction Fr-9-1 was further purified by chromatography on a RP-18 column (H2O/MeOH = 2:8) to give 3 (18 mg). Fraction Fr-10 (n-hexane/EtOAc = 0:1) was repeatedly chromatographed on silica gel (CHCl3/MeOH = 30:1 → 15:1), Sephadex LH-20 (MeOH), and RP-18 (H2O/MeOH = 3:7) columns to afford 13-hydroxymelleolide K (5, 595 mg) and melledonal B (11, 118 mg).
3.4.1. Melleolide N (1)
Colorless powder; −25 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 307 (3.65), 261 (3.91), 213 (4.45) nm; IR (KBr) νmax 3528, 1631, 1592, 1461, 1425, 1310, 1243, 1128, 1081 cm−1; 1H- and 13C-NMR data: see Table 1; ESIMS m/z (%) 459 [M + Na]+ (100), 461 (36); FABHRMS m/z 437.1732 [M + H]+ (calcd for C23H30ClO6 [M + H]+, 437.1731).
3.4.2. Melleolide Q (2)
Colorless powder; −70 (c 0.05, MeOH); UV (MeOH) λmax (log ε) 310 (3.44), 263 (3.78), 215 (4.20) nm; IR (KBr) νmax 3400, 2950, 2868, 1655, 1608, 1313, 1241, 1160, 1099, 1029 cm−1; 1H- and 13C-NMR data: see Table 1; ESIMS m/z (%) 477 [M + Na]+ (100), 479 (33); ESIHRMS m/z 453.1684 [M – H]− (calcd for C23H30ClO7 [M − H]−, 453.1675).
3.4.3. Melleolide R (3)
Colorless powder; –10 (c 0.06, MeOH); UV (MeOH) λmax (log ε) 303 (3.61), 258 (3.95), 216 (4.47) nm; IR (KBr) νmax 3500, 2951, 2863, 1648, 1600, 1453, 1366, 1319, 1240, 1101, 1042 cm−1; 1H- and 13C-NMR data: see Table 1; FABMS m/z (%) 469 [M + H]+; FABHRMS m/z 469.1991 [M + H]+ (calcd for C24H34ClO7 [M + H]+, 469.1993).
3.5. Cell Culture
Four cancer cell lines, MCF-7, H460, HT-29, and CEM, were derived from the American Type Culture Collection (Manassas, VA, USA) and were maintained in DMEM or RPMI medium supplemented with 2 mM L-glutamine and 10% heat-inactivated fetal bovine serum (FBS) under standard culture conditions. The cell viability and cell number were determined by the Trypan Blue dye-exclusion method.
3.6. Cancer Cell Cytotoxicity Assay
To assess cell viability, the alamar blue (AB) assay (dye purchased from Biosource International, Nivelles, Belgium) was used as previously described [20]. This involved aspirating medium at the end of each treatment period and adding 100 µL of fresh medium containing 10% v/v AB to control and treated wells. Plates were incubated at 37 °C for 6 h prior to measuring the absorbance at 540 nm and at 595 nm wavelengths using a spectrophotometric plate reader. Experimental data were normalized to control values.
4. Conclusions
Three new sesquiterpene aryl esters, melleolides N (1), Q (2), and R (3), together with eight known compounds 4–11 were isolated from the EtOH extract of the mycelium of Armillaria mellea. Nine isolates showed cytotoxicity to a variety of cancer cell lines, including MCF-7, H460, HT-29, and CEM. Among the isolates, compound 6 exhibited cytotoxicity to all of these cancer cells and compounds 1–5 and 7–9 showed selective cytotoxicity.
Acknowledgments
This work was financially supported by National Science Council, Taiwan (NSC97-2323-B-077-002 and NSC98-2323-B-241-001). Additional financial support at CMU under the Aim for Top University Plan of the Ministry of Education, Taiwan and Taiwan Ministry of Health and Welfare Clinical Trial and Research Center of Excellence (MOHW104-TDU-B-212-113002) was provided.
Author Contributions
Chien-Chih Chen and Chien-Chang Shen conceived and designed the experiments and wrote the paper. Ching-Li Ni performed the experiments. Chien-Chih Chen, Chien-Chang Shen, Yueh-Hsiung Kuo, and Ping-Jyun Sung analyzed the spectroscopic data and determined the chemical structures of the natural products. Jing-Jy Cheng contributed to the bioassays. Chin-Chu Chen contributed to the fermentation of the mycelium.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds 7 and 10 are available from the authors.
References and Notes
- 1.Yang J., Chen Y., Feng X., Yu D., Liang X. Chemical constituents of Armillaria mellea mycelium I. Isolation and characterization of armillarin and armillaridin. Planta Med. 1984;50:288–290. doi: 10.1055/s-2007-969711. [DOI] [PubMed] [Google Scholar]
- 2.Yang J.S., Su Y.L., Wang Y.L., Feng X.Z., Yu D.Q., Cong P.Z., Tamai M., Obuchi T., Kondoh H., Liang X.T. Isolation and structures of two new sesquiterpenoid aromatic esters: Armillarigin and armillarikin. Planta Med. 1989;55:479–481. doi: 10.1055/s-2006-962070. [DOI] [PubMed] [Google Scholar]
- 3.Yang J.S., Chen Y.W., Feng X.Z., Yu D.Q., He C.H., Zheng Q.T., Yang J., Liang X.T. Isolation and structure elucidation of armillaricin. Planta Med. 1989;55:564–565. doi: 10.1055/s-2006-962096. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe N., Obuchi T., Tamai M., Araki H., Omura S., Yang J.S., Yu D.Q., Liang X.T., Huan J.H. A novel N6-substituted adenosine isolated from Mi Huan Jun (Armillaria mellea) as a cerebral-protecting compound. Planta Med. 1990;56:48–52. doi: 10.1055/s-2006-960882. [DOI] [PubMed] [Google Scholar]
- 5.Gao L.W., Li W.Y., Zhao Y.L., Wang J.W. The cultivation, bioactive components and pharmacological effects of Armillaria mellea. Afr. J. Biotechnol. 2009;8:7383–7390. [Google Scholar]
- 6.Midland S.L., Izac R.R., Wing R.M., Zaki A.I., Munnecke D.E., Sims J.J. Melleolide, a new antibiotic from Armillaria mellea. Tetrahedron Lett. 1982;23:2515–2518. doi: 10.1016/S0040-4039(00)87383-9. [DOI] [Google Scholar]
- 7.Arnone A., Cardillo R., Nasini G. Structures of melleolides B–D, three antibacterial sesquiterpenoids from Armillaria mellea. Phytochemistry. 1986;25:471–474. doi: 10.1016/S0031-9422(00)85503-X. [DOI] [Google Scholar]
- 8.Donnelly D.M.X., Quigley P.F., Coveney D.J., Polonsky J. Two new sesquiterpene esters from Armillaria mellea. Phytochemistry. 1987;26:3075–3077. doi: 10.1016/S0031-9422(00)84599-9. [DOI] [Google Scholar]
- 9.Donnelly D.M.X., Hutchinson R.M., Coveney D., Yonemitsu M. Sesquiterpene aryl esters from Armillaria mellea. Phytochemistry. 1990;29:2569–2572. doi: 10.1016/0031-9422(90)85190-Q. [DOI] [Google Scholar]
- 10.Yang J.S., Su Y.L., Wang Y.L., Feng X.Z., Yu D.Q., Liang X.T. Two novel protoilludane norsesquiterpenoid esters, armillasin and armillatin, from Armillaria mellea. Planta Med. 1991;57:478–480. doi: 10.1055/s-2006-960176. [DOI] [PubMed] [Google Scholar]
- 11.Misiek M., Williams J., Schmich K., Hüttel W., Merfort I., Salomon C.E., Aldrich C.C., Hoffmeister D. Structure and cytotoxicity of arnamial and related fungal sesquiterpene aryl esters. J. Nat. Prod. 2009;72:1888–1891. doi: 10.1021/np900314p. [DOI] [PubMed] [Google Scholar]
- 12.Bohnert M., Miethbauer S., Dahse H.M., Ziemen J., Nett M., Hoffmeister D. In vitro cytotoxicity of melleolide antibiotics: Structural and mechanistic aspects. Bioorg. Med. Chem. Lett. 2011;21:2003–2006. doi: 10.1016/j.bmcl.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Chi C.W., Chen C.C., Chen Y.J. Therapeutic and radiosensitizing effects of armillaridin on human esophageal cancer cells. Evid. -Based Complement. Altern. Med. 2013;2013 doi: 10.1155/2013/459271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y.J., Wu S.Y., Chen C.C., Tsao Y.L., Hsu N.C., Chou Y.C., Huang H.L. Armillaria mellea component armillarikin induces apoptosis in human leukemia cells. J. Funct. Foods. 2014;6:196–204. doi: 10.1016/j.jff.2013.10.007. [DOI] [Google Scholar]
- 15.Some contents in this paper were filed for US patent application (Pub. No. US2011/0262561 A1); however, the application was later abandoned due to some concerns.
- 16.Bohnert M., Nützmann H.W., Schroeckh V., Horn F., Dahse H.M., Brakhage A.A., Hoffmeister D. Cytotoxic and antifungal activities of melleolide antibiotics follow dissimilar structure–activity relationships. Phytochemistry. 2014;105:101–108. doi: 10.1016/j.phytochem.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Kobori H., Sekiya A., Suzuki T., Choi J.H., Hirai H., Kawagishi H. Bioactive sesquiterpene aryl esters from the culture broth of Armillaria sp. J. Nat. Prod. 2015;78:163–167. doi: 10.1021/np500322t. [DOI] [PubMed] [Google Scholar]
- 18.Arnone A., Cardillo R., Nasini G., Meille S.V. Secondary mould metabolites. Part 19. Structure elucidation and absolute configuration of melledonals B and C, novel antibacterial sesquiterpenoids from Armillaria mellea. X-ray molecular structure of melledonal C. J. Chem. Soc. Perkin Trans. 1. 1988;3:503–510. doi: 10.1039/p19880000503. [DOI] [Google Scholar]
- 19.Shiono Y., Seto T., Kamata M., Takita C., Suzuki S., Murayama T., Ikeda M. Protoilludane-type sesquiterpenes, echinocidins A and B, from a mycelial culture of Echinodontium tsugicola. Z. Naturforsch. 2004;59:925–929. doi: 10.1002/chin.200449167. [DOI] [Google Scholar]
- 20.Lu M.K., Cheng J.J., Lin C.Y., Chang C.C. Purification, structural elucidation, and anti-inflammatory effect of a water-soluble 1,6-branched 1,3-α-D-galactan from cultured mycelia of Poria cocos. Food Chem. 2010;118:349–356. doi: 10.1016/j.foodchem.2009.04.126. [DOI] [Google Scholar]