Abstract
Three new pigment compounds—terreusinone A (1), pinophilin C (2) and cryptosporioptide A (3) were isolated from a solid culture of Cordyceps gracilioides. The structures of these compounds were determined by extensive spectroscopic analysis including HRESIMS, 1D- and 2D-NMR. The structure of terreusinone A (1) was further confirmed by single-crystal X-ray crystallographic diffraction analysis. In an in vitro activity assay, 1, 2 and 3 exhibited high inhibitory activity against PTP1B, SHP2, CDC25B, LAR and SHP1. Terreusinone A (1) inhibited PTP1B, SHP2, CDC25B, LAR and SHP1 enzyme with IC50 values 12.5, >50, 4.1, 10.6, 5.6 µg/mL, respectively; pinophilin C (2) with IC50 values 6.8, 8.0, 4.5, 4.7, 3.4 µg/mL, respectively; and cryptosporioptide A (3) with IC50 values 7.3, 5.7, 7.6, >50, 4.9 µg/mL, respectively.
Keywords: protein tyrosine phosphatases inhibitors, Cordyceps gracilioides, terreusinone A, pinophilin C, cryptosporioptide A
1. Introduction
Cordyceps is a genus of ascomycete fungi that includes about 400 species, which are parasitic mainly on insects and other arthropods [1]. Cordyceps are rich sources of novel biologically active substances with diverse structural architectures [2], such as the antimalarial erythrostominones [3], antimalarial cordypyridones [4], antitrypanosomal cardinalisamides A–C [5] and opaliferi [6]. Cordyceps gracilioides Kobayasi was a new species in China and the specimen was collected on a coleopteran from the National Natural Conservation of Guniujiang, Shitai County, Anhui Province in September 2000 [7]. Although there has been considerable investigation on the genus Cordyceps, little has been reported about secondary metabolites from C. gracilioides. Here we describe the isolation and structural elucidation of the three new compounds—terreusinone A (1), pinophilin C (2) and cryptosporioptide A (3, Figure 1)—from C. gracilioides and the inhibitory activities against the enzymes PTP1B, SHP2, CDC25B, LAR and SHP1.
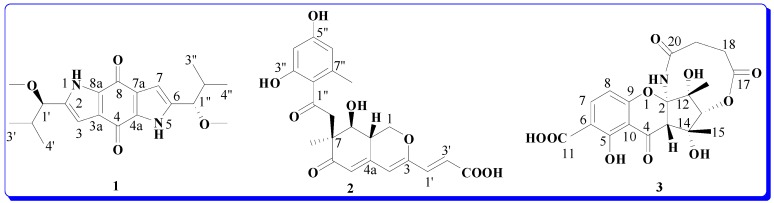
Figure 1.
The structures of compounds 1–3.
2. Results and Discussion
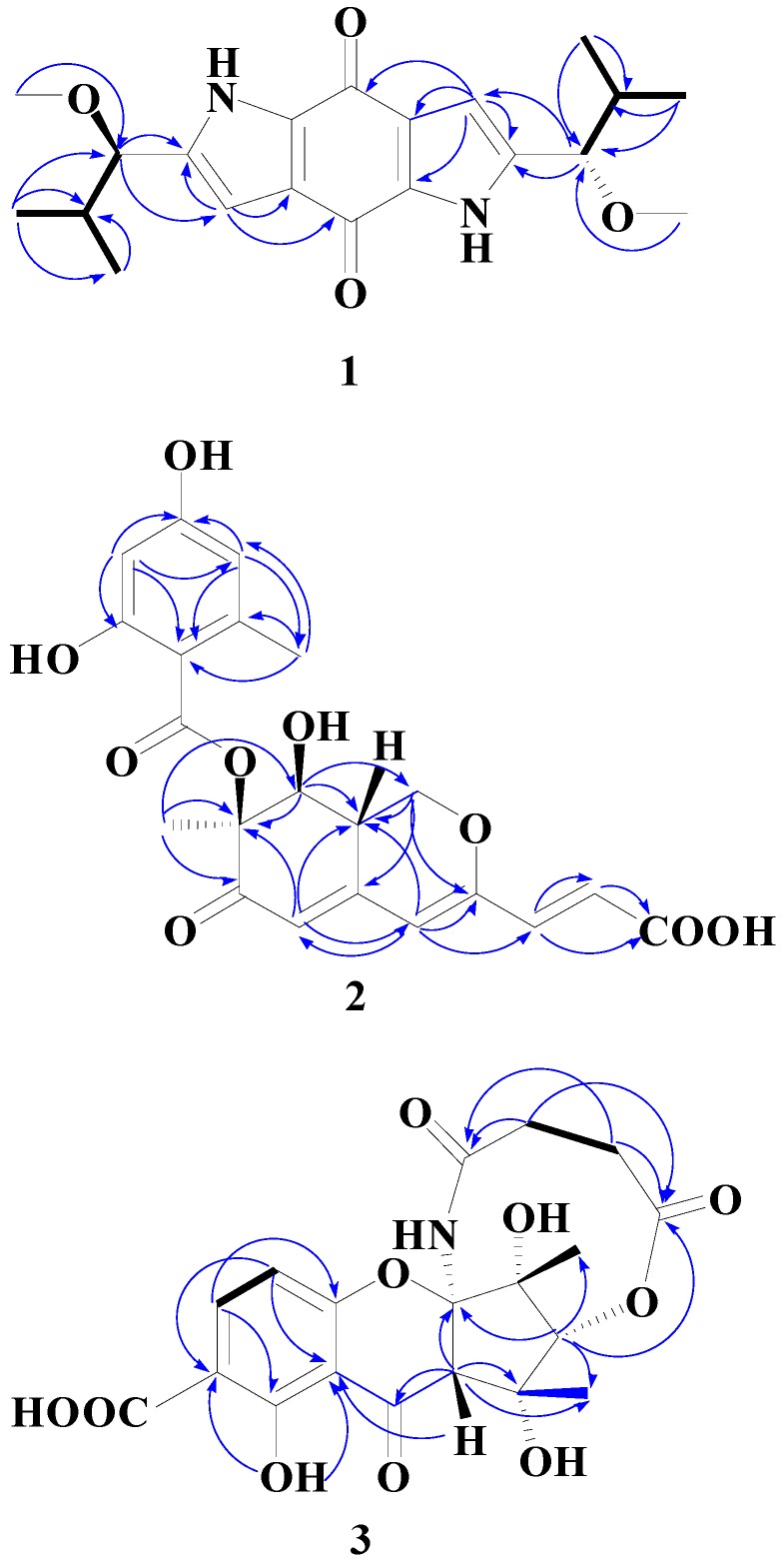
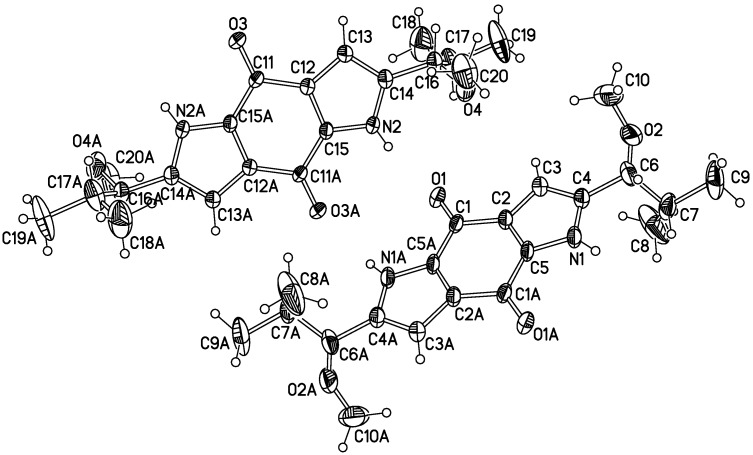
Compound 1 was obtained as red crystals (from MeOH). The high-resolution EIMS exhibited a molecular ion peak at m/z 358.1893, corresponding to the molecular formula of C20H26N2O4 (calcd. 358.1880), indicating nine degrees of unsaturation. The IR absorption spectrum of 1 showed bands characteristic of benzoquinone (1631 cm−1) and pyrrole (3442, 1469 cm−1) functionalities. The 13C-NMR (DEPT) spectrum of 1 showed only 10 signals: three olefinic quaternary carbons (δC 139.1, 126.5, 132.7), one olefinic methine carbons (δC 106.7), two methine carbons (δC 83.0, 33.4), three methyl carbons (δC 18.4, 18.0, 56.8) and one carbonyl group (δC 175.2) for one half of the molecule, suggesting that 1 is a high symmetry dimer. The 1H- and 13C-NMR and HMBC data (Table 1) of 1 were similar to that of terreusinone, which suggested the compound possesses the same dipyrrolobenzoquinone skeleton [8]. The key difference between 1 and terreusinone was that the hydroxyl in terreusinone was replaced by a methoxy in 1. The structure of 1 was further confirmed by long range correlations measured in an HMBC experiment (Figure 2). ROESY spectral data which showed the correlations of H-1' to H-3, H-1 to H-OCH3 indicative of their trans orientations, respectively. The relative configuration was further confirmed by X-ray diffraction (Figure 3). Thus, compound 1 was identified as 2,6-bis(1-methoxy-2-methylpropyl) pyrrolo[2,3-f]indole-4,8(1H,5H)-dione as shoen in Figure 1, and it was named terreusinone A.
Table 1.
NMR Spectroscopic Data for Compound 1 (Terreusinone A) in CD3OD-d6.
| Position | δH a (J in Hz) | δC b, mult | HMBC(H→C#) |
|---|---|---|---|
| 2 (6) | 139.1 (C) | ||
| 3 (7) | 6.42 (s) | 106.7 (CH) | 1' (1''), 8a(4a), 3a(7a), 2(6), 4(8) |
| 3a (7a) | 126.5 (C) | ||
| 4 (8) | 175.2 (C) | ||
| 4a (8a) | 132.7 (C) | ||
| 1' (1'') | 3.86 (d, J = 7.4) | 83.0 (CH) | 2(6),3(7),2' (2''),4' (4'') 1' (1'')-OCH3, 3' (3'') |
| 2' (2'') | 2.03 (m) | 33.4 (CH) | 2(6), 1' (1''), 3' (3''), 4' (4'') |
| 3' (3'') | 1.00 (d, J = 6.6) | 18.4 (CH3) | 1' (1''), 2' (2''), 4' (4'') |
| 4' (4'') | 0.80 (d, J = 6.6) | 18.0 (CH3) | 1' (1''), 2' (2''), 3' (3'') |
| 1' (1'')-OCH3 | 3.26 (s) | 56.8 (CH3) | 1' (1'') |
a 1H-NMR were recorded in CD3OD-d6 at 600 MHz; b 13C-NMR were recorded in CD3OD-d6 at 150 MHz.
Figure 2.
1H-1H COSY correlations (bold lines) and selected HMBC (1H→13C) (single lines) correlations of 1–3.
Figure 3.
X-ray crystal structure of compound 1.
The molecular formula of 2 was deduced from the HRESI-MS [M+H]+ peak at m/z 417.1180 (calcd. for C21H21O9 417.1188), indicating 12 degrees of unsaturation. The IR spectrum indicated the presence of a hydroxy group (3427 cm−1), a conjugated ester (1703 cm−1), and a conjugated carbonyl (1636 cm−1). As shown in Table 2, the 1H- and 13C-NMR spectra suggested that compound 2 had a similar structures to pinophilin A [9] and differed only in C-3' of the side chain, whereby the methyl group in pinophilin A was replaced by a carboxylic acid group in 2 [δC 168.0 (COOH, C-3')]. The azaphilone skeleton was further confirmed by long range correlations measured in an HMBC experiment (Figure 2). The relative configuration of compound 2 was determined by 1H-1H coupling and NOESY correlations. The anti relationship of H-8/H-8a was deduced from the coupling constants (J8, 8a = 9.6 Hz). The syn relationship of H-8a/7-O-benzoyl group was determined from the NOESY correlations between 7-CH3 and H-8. The CD spectrum of compound 2 exhibited Cotton effects due to the interaction between the benzoate and conjugated trienone chromophores at 326 and 301 nm (Δε −15.1 and 5.0), indicating the S configuration [10] at C-7. Thus, compound 2 was identified as (E)-3-((7S,8S,8aS)-7-((2,4-dihydroxy-6-methylbenzoyl)oxy)-8-hydroxyl-7-methyl-6-oxo-6,7,8,8a-tetra-hydro-1H-isochromen-3-yl)acrylic acid as shown in Figure 1, and it was named pinophilin C.
Table 2.
NMR Spectroscopic Data for Compound 2 (Pinophilin C) in CD3OD-d6.
| Position | δH a (J in Hz) | δC b, Mult | HMBC(H→C#) |
|---|---|---|---|
| 1 | 4.81(dd, J = 10.7, 5.3) 3.81(dd, J = 13.6, 10.7) |
68.8 (CH2) | 8a, 4a, 3 |
| 3 | 157.0 (C) | ||
| 4 | 6.04 (s) | 109.6 (CH) | 8a, 5, 3, 1' |
| 4a | 149.2 (C) | ||
| 5 | 5.94 (d, J = 2.0) | 119.5 (CH) | 8a, 7, 4, |
| 6 | 194.2 (C) | ||
| 7 | 84.7 (C) | ||
| 7-Me | 1.67 (s) | 17.0 (CH3) | |
| 8 | 3.62 (d, J = 9.6) | 74.2 (CH) | 7-Me, 8a, 1, 7 |
| 8a | 3.25 (dddd, J = 13.6, 9.6, 5.3, 2.0) | 37.5 (CH) | 8, 4a |
| 1' | 7.05 (d, J = 15.4) | 137.1 (CH) | 4, 2', 3, 3' |
| 2' | 6.33 (d, J = 15.4) | 122.3 (CH) | 1', 3, 3' |
| 3' | 168.0 (COOH ) | ||
| 1'' | 169.8 (C) | ||
| 2' | 105.1 (C ) | ||
| 3'' | 163.9 (C) | ||
| 4' | 6.14 (d, J = 2.4) | 100.4 (CH) | 2'', 6'', 5'', 3'' |
| 5'' | 162.4 (C) | ||
| 6'' | 6.20 (d, J = 2.4) | 111.0 (CH) | 7''-Me, 2'', 4'', 5'' |
| 7'' | 143.1 (C) | ||
| 7''-Me | 23.0 (CH3) |
a 1H-NMR were recorded in CD3OD-d6 at 600 MHz; b 13C-NMR were recorded in CD3OD-d6 at 150 MHz.
Compound 3 was obtained as a yellow solid. The high-resolution EIMS exhibited a molecular ion peak at m/z 421.1012 corresponding to the molecular formula of C19H19NO10 (calcd. 421.1009), indicating 11 degrees of unsaturation. The IR spectrum displayed absorption bands at 3440 (OH), 3030, 2931 (CH), 1745 (C=O) and 1591, 1562 (C=C) cm−1. The 1H-NMR spectrum of 3 (Table 3) showed two ortho-coupled doublets in the aromatic region at δH 7.40 (1H, J = 8.4 Hz, H-7) and 6.42 (1H, J = 8.4 Hz, H-8), which are attributed to a tetrasubstituted benzene ring.
Table 3.
NMR Spectroscopic Data for Compound 3 (Cryptosporioptide A) in CDCl3-d6.
| Position | δH a (J in Hz) | δC b, mult | HMBC(H→C#) |
|---|---|---|---|
| 2 | - | 104.3 (C) | |
| 3 | 3.42 (s) | 55.8 (CH) | 2, 4, 11, 14, 15 |
| 4 | - | 187.8 (C) | |
| 5 | - | 159.0 (C) | |
| 6 | - | 117.1 (C) | |
| 7 | 7.40 (d, J = 8.4) | 140.5 (CH) | 5, 9, 10 |
| 8 | 6.42 (d, J = 8.4) | 108.2 (CH) | 4, 6, 9, 10 |
| 9 | - | 157.1 (C) | |
| 10 | - | 106.0 (C) | |
| 11 | - | 169.8 (COOH) | |
| 12 | - | 78.9 (C) | |
| 13 | 5.65 (s) | 72.5 (CH) | 2, 3, 4, 12, 14, 15, 16, 17 |
| 14 | - | 58.7 (C) | |
| 15 | 1.56 (s) | 18.1 (CH3) | 3, 13, 14, |
| 16 | 1.65 (s) | 28.4 (CH3) | 2, 12, 13 |
| 17 | - | 171.2 (C) | |
| 18 | 2.65 (m) | 28.8 (CH2) | 17 |
| 19 | 2.66 (m) | 28.8 (CH2) | 20 |
| 20 | - | 177.2 (C) | |
| 5-OH | 11.75 (s) | - | 5, 6, 10 |
| N–H | 14.03 (s) | - |
a 1H-NMR were recorded in CDCl3-d6 at 600 MHz; b 13C-NMR were recorded in CDCl3-d6 at 150 MHz.
It also showed δH 5.65 (1H, s, H-13), 3.42 (1H, s, H-3), 2.65 (2H, m, H-18), 2.66 (2H, m, H-18), 1.56 (3H, s, H-15), 1.65 (3H, s, H-16). The most downfield signals appeared at δH 14.03 (1H, s, NH) and 11.75 (1H, s, OH). The unusual downfield shift of the amide hydrogen could be due to an anisotropic effect of oxygen of the chromone moiety. The 13C-NMR (DEPT) spectrum of 3 (Table 3) showed signals corresponding to 19 signals: two methyls (δC 28.4, 18.1), two methylenes (δC 28.8, 28.8), four methines (δC 140.5, 108.2, 72.5, 55.8) and 11 quaternary carbons (δC 187.8, 177.2, 171.2, 169.8, 159.0, 157.1, 117.1, 106.0, 104.3, 78.9, 58.7). The 1H- and 13C-NMR and HMBC experiment results (Table 3) of 3 were similar to those of cryptosporioptide [11]. The distinct differences between 3 and cryptosporioptide were that a methoxy group [δC 52.5 (OCH3)] in cryptosporioptide was replaced by an OH in 3 and one methylene (δC 41.1, C-18) in cryptosporioptide was replaced by two methylenes [δC 28.8, 28.8 (C-18, 19)] in 3. The polyketide skeleton was further confirmed by long range correlations measured in an HMBC experiment (Figure 2). The syn relationship of 15-Me/16-Me/H-3/H-13 was determined from NOESY correlations between 15-Me, 16-Me, H-3, and H-13. The absolute configuration of C-2, 3, 12, 13, 14 in 3 was further determined by comparing the circular dichroism (CD) and [α]D spectra with cryptosporioptide (see Supporting Information). The (CD) and [α]D of 3 were very similar to cryptosporioptide, therefore, we may assign the absolute configuration of compound 3 as (+)-(2R, 3S, 12R, 13R, 14R)-5,12,14-trihydroxy-12,14-dimethyl-4,17,20-trioxo-3,7,8, 13,18,19-octahydro-21H-2,13-methanochromeno[3,2-d][1,6]oxazecine-11-carboxylic acid, named cryptosporioptide A (3) as shown in Figure 1.
Compounds 1–3 were examined for their inhibitory activity against the enzymes PTP1B, SHP2, CDC25B, LAR and SHP1 using an in vitro assay. The results showed that 1 and 2 have significant inhibitory activity against CDC25B and SHP1, with IC50 values of 4.1, 3.4 µg/mL, respectively (Table 4).
Table 4.
The activity of enzyme inhibition of compounds 1–3.
| Compounds | IC50 Values (µg/mL) | ||||
|---|---|---|---|---|---|
| PTP1B | SHP2 | CDC25B | LAR | SHP1 | |
| 1 | 12.5 | >50 | 4.1 | 10.6 | 5.6 |
| 2 | 6.8 | 8.0 | 4.5 | 4.7 | 3.4 |
| 3 | 7.3 | 5.7 | 7.6 | >50 | 4.9 |
| Sodium orthovanadate (positive control) | 38.3 | 122.2 | 0.93 | 163.0 | 4.3 |
3. Experimental Section
3.1. General Information
The 1H- and 13C-NMR spectra were recorded on a Bruker AM-600 spectrometer (Faellanden, Switzerland) at 600 and 150.9 MHz using TMS as internal standard. Optical rotations were obtained on a Perkin-Elmer 341 spectropolarimeter (Milpitas, CA, USA). IR spectra were recorded on a Perkin-Elmer 577 spectrometer (Waltham, MA, USA). HRESIMS data were measured on a Bruker FT-ICR-MS mass spectrometer (Billerica, MA, USA). UV Spectra were recorded on a UV-210 spectrometer (Kyoto, Japan). Column chromatography (CC): silica gel (200~300 mesh, Yantai Zhi Fu Chemical Co. Ltd., Yantai, China), TLC: silica gel GF254 plates (Yantai Zhi Fu Chemical Co. Ltd.) and Sephadex LH-20 gel (25~100 µm, GE Healthcare Co. Ltd., Uppsala, Sweden).
3.2. Fungal Material and Cultivation Conditions
The Cordyceps gracilioides Kobayasi was isolated from Coleoptera larvae collected in Guniujiang (Anhui Province, China), and assigned the accession number ACCC47758 in the culture collection at the College of Life Science, Key Laboratory of Medicinal Chemistry and Molecular Diagnosis of Ministry of Education, Hebei University. The fungal strain was cultured on slants of potato dextrose agar (PDA) at 28 °C for 7 days, and then inoculated into a 500 mL Erlenmeyer flask containing 100 mL of PDA medium (20.0 g of glucose, 200.0 g of potato (peeled), 3.0 g of KH2PO4, 1.5 g of MgSO4, 0.1 g of citric acid, and 10.0 mg of thiamin hydrochloride, in 1 L of deionized H2O). The final pH of the medium was adjusted to 6.5 before sterilization. After 7 days of incubation at 28 °C on rotary shakers at 150 rpm, 25 mL of culture liquid were transferred as seed into a 500 mL Erlenmeyer flask containing 200.0 g rice medium, and the fermentation was carried out in an incubator for 40 days.
3.3. Extraction and Isolation
The culture broth (30 L) was extracted three times with ethyl acetate (30 L, soaking each time for 2 days), and the organic layer was concentrated in vacuo to yield a brown oily residue (70 g). This residue was subjected to silica gel column chromatography (CC) with elution using a gradient of petroleum ether/EtOAc (100:0, 98:2, 95:5, 90:10, 60:10, 30:10, 10:10 (v/v)) to obtain seven fractions 1–7. Fraction 5 (150 mg) eluted with petroleum ether/EtOAc (60:10) (v/v) was repeatedly purified by CC (silica gel; petroleum ether/EtOAc 10:1 (v/v)) and Sephadex LH-20 chromatography (CHCl3/MeOH, 1:1 (v/v)) to afford 2 (20 mg) and 3 (30 mg) as yellow amorphous powders. Fraction 6 (100 mg) eluted with petroleum ether/EtOAc (30:10) (v/v) was repeatedly purified by CC (silica gel; petroleum ether/EtOAc 5:1 (v/v), Sephadex LH-20 chromatography (CHCl3/MeOH, 1:1 (v/v)) and Sephadex LH-20 (methanol) to give 1 (15 mg) as red crystals.
3.4. Analytical Data
3.4.1. Terreusinone A (1)
Red crystals. HREIMS m/z 358.1893 (calcd. for C20H26N2O4 358.1880); [α]D21.5 = −7° (c, 0.001, MeOH); UV (MeOH) λmax (logε): 357 (3.93), 287 (4.10), 250 (4.58) nm; IR (KBr)vmax: 3442, 3125, 2960, 1631, 1555, 1469, 1384, 1228, 1162, 1088, 993, 752, 670 cm−1; 1H- and 13C-NMR: Table 1. X-Ray Crystallographic Analysis of 1. Upon crystallization from MeOH by the vapor-diffusion method, red crystals of 1 were obtained. A crystal (0.29 mm × 0.28 mm × 0.20 mm) was separated from the sample and mounted on a glass fiber, and data were collected with a Bruker-SMART-1000-CCD diffractometer and graphite-monochromated MoKa radiation (λ = 0.71073 Å) at 296(2) K. Crystal data: C20H26N2O4, M = 358.43, Monoclinic space group: P-1; Unit cell dimensions a = 8.3255(10) Å, b = 9.9479(12) Å, c = 12.4585(15) Å. V = 968.9(2) Å3, Z = 2, D = 1.229 mg/m3. F (000) = 384, µ = 0.086 mm−1.The structure was solved by direct methods with SHELXL-97 and refined by full-matrix least-squares difference Fourier techniques. All non-H-atoms were refined with anisotropic displacement parameters, and all H-atoms were placed in idealized positions and refined as riding atoms with the relative isotropic parameters. Absorption corrections were applied with the Siemens area detector absorption program (SADABS). The 5740 measurements yielded 4139 independent reflections after equivalent data were averaged, and Lorentz and polarization corrections were applied. The final refinement gave Rf = 0.1278 and Rw = 0.4121 (I > 2s (I)). Crystallographic data for the structure of 1 has been deposited in the Cambridge Crystallographic Data Centre (deposition number: CCDC 1041522). Copies of these data can be obtained, free of charge, on application to the CCDC through http://www.ccdc.cam.ac.uk/conts/retrieving.html (or from the Cambridge Crystallographic Data Centre, 12, Union Road, Cambridge CB2 1EZ, UK; Fax (Internat.): +44-1223/336-033; E-Mail: deposit@ccdc.cam.ac.uk).
3.4.2. Pinophilin C (2)
Yellow solid. HRESI-MS [M+H]+ m/z 417.1180 (calcd. for C21H21O9 417.1188); [α]D21.6 = −146° (c, 0.002, MeOH); IR(KBr) νmax: 3427, 2962, 2933, 2875, 2374, 1703, 1636, 1589, 1489, 1447, 1382, 1200, 1172, 1103, 995, 616 cm−1; UV (MeOH) λmax (logε): 347 (4.36), 266 (4.23), 216 (4.45) nm; 1H- and 13C-NMR: Table 2.
3.4.3. Cryptosporioptide A (3)
Yellow solid. HREIMS m/z 421.1012 (calcd.for C19H19NO10 421.1009); [α]D21.5 = +20.1° (c, 0.003, MeOH); IR(KBr) νmax: 3440, 3030, 2931, 1745, 1591, 1562 cm−1; UV (MeOH) λmax (log ε): 388 (3.59), 203 (3.61) nm; 1H- and 13C-NMR: Table 3.
3.5. PTP Assay
Protein tyrosine phosphatases (PTPs) constitute a large family of signaling enzymes that control the cellular levels of protein tyrosine phosphorylation. Several “classical” PTPs are attractive therapeutic targets, including PTP1B for obesity and Type II diabetes, SHP1, and SHP2 and CDC25B for cancer [12]. GST-ΔPTP1B, GST-ΔSHP2, GST-ΔSHP1, GST-ΔLAR and GST-ΔCDC25B fusion proteins which only carry the PTP activity domain were expressed in Escherichia coli DH5α and affinity purified with glutathione Sepharose (Amersham Biosciences, Piscataway, NJ, USA). p-Nitrophenyl phosphate (pNPP) was used in the enzymatic reactions to determine the intrinsic catalytic activities of the PTPs. All assays using pNPP as the substrate were performed in a buffered reaction medium containing 10 mM of sodium acetate (NaAc), 1 mM of ethylenediaminetetraacetic acid and 1 mM of dithiothreitol at 37 °C, pH 5.5. Sodium orthovanadate was used as the positive control. Every experiment was performed in triplicate. The inhibitory activity measurements of PTPs were performed with different concentrations (0, 2.5, 5, 10, 20, 40 µg mL−1) of inhibitor and 20 µg mL−1 of PTPs. The reaction mixture was incubated at 37 °C for 15 min and the catalytic activity was detected by monitoring the absorbance at 405 nm immediately after adding 12.5 mM of pNPP. IC50 (the concentration of an inhibitor that is required for 50% inhibition of an enzyme) data were derived from the experimental results [13,14,15,16,17].
4. Conclusions
In summary we have isolated three new pigments—terreusinone A (1), pinophilin C (2) and cryptosporioptide A (3)—from cultures of the insect fungus Cordyceps gracilioides. We also demonstrate for the first time that 1 and 2 have significant inhibitory activity against CDC25B and SHP1.
Acknowledgments
This work was supported by National Natural Science Foundation of China (31171885 and 31371957), the Programs Foundation of Ministry of Education of China (20121301110006), key Projects in the Hebei Province Science & Technology (13226508D) and support by The Program for Changjiang Scholars and Innovative Research Team in University (IRT1124). We also thank Zhi-Hui Zheng from New Drug Research and Development Center, North China Pharmaceutical Group Corporation, China for his help with the activity assays.
Supplementary Materials
Supplementary materials can be assessed at: http://www.mdpi.com/1420-3049/20/04/5825/s1.
Author Contributions
Du-Qiang Luo conceived and designed the study. Pei-Yao Wei, Lin-Xia Liu, Chuan Chen and Bao-Zhong Shi performed the isolation and identification experiments. Ting Liu performed the biological evaluation. Pei-Yao Wei, Lin-Xia Liu and Du-Qiang Luo wrote the paper. Lin-Xia Liu and Du-Qiang Luo reviewed and edited the manuscript. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds 1–3 are available from the authors.
References
- 1.Sung G.H., Hywel-Jones N.L., Sung J.M., Luangsa-Ard J.J., Shrestha B., Spatafora J.W. Phylogenetic classification of cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007;57:5–59. doi: 10.3114/sim.2007.57.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isaka M., Kittakoop P., Kirtikara K., Hywel-Jones N.L., Thebtaranonth Y. Bioactive substances from insect pathogenic Fungi. Acc. Chem. Res. 2005;38:813–823. doi: 10.1021/ar040247r. [DOI] [PubMed] [Google Scholar]
- 3.Isaka M., Tanticharoen M., Kongsaree P., Thebtaranonth Y. Structures of Cordypyridones A–D, antimalarial N-hydroxy- and N-methoxy-2-pyridones from the insect pathogenic fungus Cordyceps nipponica. J. Org. Chem. 2001;66:4803–4808. doi: 10.1021/jo0100906. [DOI] [PubMed] [Google Scholar]
- 4.Kittakoop P., Punya J., Kongsaeree P., Lertwerawat Y., Jintasirikul A., Tanticharoen M., Thebtaranonth Y. Bioactive naphthoquinones from Cordyceps unilateralis. Phytochemistry. 1999;52:453–457. doi: 10.1016/S0031-9422(99)00272-1. [DOI] [Google Scholar]
- 5.Umeyama A., Takahashi K., Grudniewska A., Shimizu M., Hayashi S., Kato M., Okamoto Y., Suenaga M., Ban S., Kumada T., et al. In vitro antitrypanosomal activity of the cyclodepsipeptides, cardinalisamides A–C, from the insect pathogenic fungus Cordyceps cardinalis NBRC 103832. J. Antibiot. 2014;67:163–166. doi: 10.1038/ja.2013.93. [DOI] [PubMed] [Google Scholar]
- 6.Grudniewska A., Hayashi S., Shimizu M., Kato M., Suenaga M., Imagawa H., Ito T., Asakawa Y., Ban S., Kumada T., et al. Opaliferin, a new polyketide from cultures of entomopathogenic fungus Cordyceps sp. NBRC 106954. Org. Lett. 2014;16:4695–4697. doi: 10.1021/ol502216j. [DOI] [PubMed] [Google Scholar]
- 7.Fan M.Z., Li C.R., Chen Y.Y., Li Z.Z. Cordyceps Gracilioides, a new record from China. Mycosystema. 2001;20:273–274. [Google Scholar]
- 8.Lee S.M., Li X.F., Jiang H., Cheng J.G., Seong S., Choi H.D., Son B.W. Terreusinone, a novel UV-A protecting dipyrroloquinone from the marine algicolous fungus Aspergillus terreus. Tetrahedron Lett. 2003;44:7707–7710. doi: 10.1016/j.tetlet.2003.08.101. [DOI] [Google Scholar]
- 9.Myobatake Y., Takeuchi T., Kuramochi K., Kuriyama I., Ishido T., Hirano K., Sugawara F., Yoshida H., Mizushina Y. Pinophilins A and B, inhibitors of mammalian A-, B-, and Y-family DNA polymerases and human cancer cell proliferation. J. Nat. Prod. 2012;75:135–141. doi: 10.1021/np200523b. [DOI] [PubMed] [Google Scholar]
- 10.Takeshi I., Nobuko O., Koohei N., Ken-ichi K. Isolation and structures of new azaphilone derivatives, falconensins E–G, from Emericella falconensis and absolute configurations of falconensins A–G. Chem. Pharm. Bull. 1996;44:2213–2217. doi: 10.1248/cpb.44.2213. [DOI] [Google Scholar]
- 11.Saleem M., Tousif M.I., Riaz N., Ahmed I., Schulz B., Ashraf M., Nasar R., Pescitelli G., Hussain H., Jabbar A., et al. Cryptosporioptide: A bioactive polyketide produced by an endophytic fungus Cryptosporiopsis sp. Phytochemistry. 2013;93:199–202. doi: 10.1016/j.phytochem.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 12.Barr A.J. Protein tyrosine phosphatases as drug targets: Strategies and challenges of inhibitor development. Future Med. Chem. 2010;2:1563–1576. doi: 10.4155/fmc.10.241. [DOI] [PubMed] [Google Scholar]
- 13.Chen L., Sung S.S., Yip M.L., Lawrence H., Ren Y., Guida W.C., Sebti S.M., Lawrence N.J., Wu J. Discovery of a novel Shp2 protein tyrosine phosphatase inhibitor. Mol. Pharmacol. 2006;70:562–570. doi: 10.1124/mol.106.025536. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z., Chai Q., Li Y.Y., Shen Q., Ma L.P., Zhang L.N., Wang X., Sheng L., Li J.Y., Li J., et al. Discovery of novel PTP1B inhibitors with antihyperglycemic activity. Acta Pharmacol. Sin. 2010;31:1005–1012. doi: 10.1038/aps.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sohn J., Rudolph J. Temperature dependence of binding and catalysis for the Cdc25B Phosphatase. Biophys. Chem. 2007;125:549–555. doi: 10.1016/j.bpc.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y.L., Hollfelder F., Gordon S.J., Chen L., Keng Y.F., Wu L., Herschlag D., Zhang Z.Y. Impaired Transition State Complementarity in the Hydrolysis of O-Arylphosphorothioates by Protein-Tyrosine Phosphatases. Biochemistry. 1999;38:12111–12123. doi: 10.1021/bi990836i. [DOI] [PubMed] [Google Scholar]
- 17.Valencia A.M., Ohva J.L., Bodega G., Chiloeches A., Lopez-Ruiz P., Prieto J.C., Susini C., Colas B. Identification of a protein-tyrosine phosphatase (SHPl) different from that associated with acid phosphatase in rat prostate. FEBS Lett. 1997;406:42–48. doi: 10.1016/S0014-5793(97)00235-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.