Abstract
Four new steroid saponins 1–4 were isolated from the rhizomes of Anemarrhena asphodeloides (Asparagaceae), as well as four known saponins: anemarsaponin B (5) timosaponin D (6), timosaponin E1 (7) anemarsaponin B II (8). Their structures were established through UV and NMR as well as MS data. All the compounds were evaluated for cytotoxicity against HepG2 and SGC7901 human cancer lines. Compounds 3 and 7 displayed medium antiproliferative activities on HepG2 and SGC7901 cells, with IC50 values of 43.90 and 57.90 μM, respectively.
Keywords: Asparagaceae, furostanol saponin, spirostanol saponin, MTT, cytotoxicity
1. Introduction
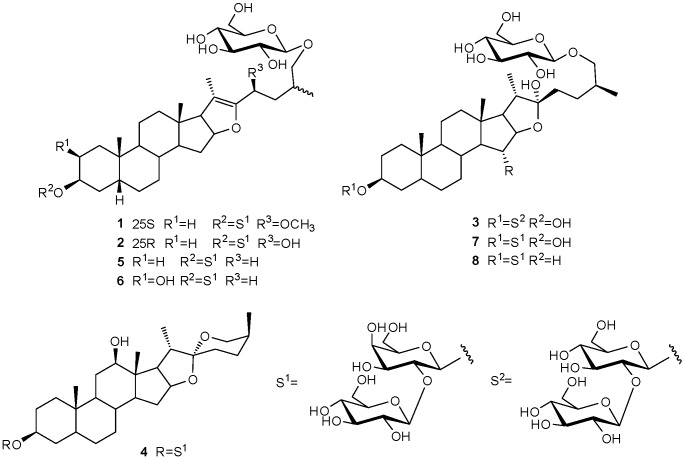
The rhizomes of Anemarrhena asphodeloides (Asparagaceae) have been used as traditional Chinese medicine for centuries. Phytochemical studies on Anemarrhena asphodeloides have led to the identification of a series of compounds, such as steroidal saponins, flavonoids, phenylpropanoids and alkaloids [1]. The most remarkable of the bioactive ingredients are the steroidal saponins. Recently, steroidal saponin studies were focused on a range of bioactivities, such as anti-inflammatory [2], antiplatelet [3,4], and especially anti-tumor properties [5,6]. To explore its active components, we carried out a series of studies on the rhizomes of Anemarrhena asphodeloides. Our experiments have led to the isolations of four new steroidal saponins 1–4, which we have named anemarsaponin P–S, along with four known ones, including anemarsaponin B (5) [7], timosaponin D (6) [8], timosaponin E1 (7) [9], anemarsaponin B II (8) [10] (Figure 1).
Figure 1.
Structures of compounds 1–8 from Anemarrhena asphodeloides.
The cytotoxicity of the isolated compounds has also been evaluated by the MTT method. This paper therefore reports the isolation, structural elucidation, and antiproliferative activities of steroidal saponins from Anemarrhena asphodeloides.
2. Results
2.1. Structure Elucidation
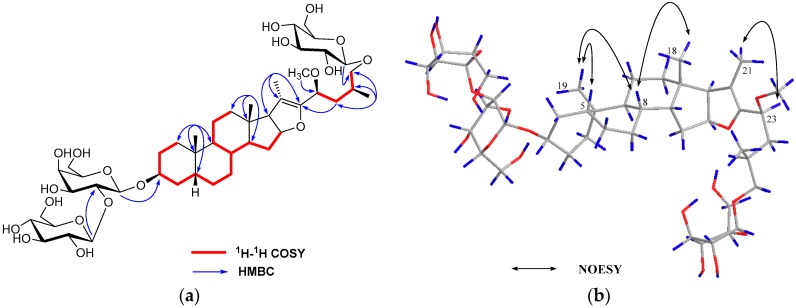
Anemarsaponin P (1) was obtained as an amorphous power and its molecular formula was C46H76O19 according to the HR-ESI-MS data (m/z 955.4644 [M + Na]+) (Figure S6). The corresponding 1H-NMR (Figure S7) data of the aglycone portion (Table 1) showed four methyl signals at δH 0.66 (3H, s), 0.99 (3H, s), 1.73 (3H, s) and 1.12 (3H, d, J = 6.8 Hz), three anomeric protons at δH 4.83 (1H, d, J = 8.0 Hz), 5.28 (1H, d, J = 7.6 Hz) and 4.92 (1H, d, J = 8.0 Hz) and a methoxy group at δH 3.32 (3H, s). The 13C-NMR spectrum (Figure S8) showed four methyl groups at δC 14.6, 24.1, 11.6 and 17.9. Characteristic signals at 109.2 (C-20), 150.4 (C-22) and the secondary carbon signal at δC 75.5 (C-26) indicated that compound 1 was a Δ20(22)-unsaturated furostanol saponin [11]. Comparison of the 1H- and 13C-NMR spectra in 1 with those of anemarsaponin B (5) revealed the ring A–E portions and glycoside moiety of C-3 of the former were consistent with those of 5. On the other hand, remarkable differences were indicated by the carbon signals from the ring F portion (C-22~C-27). The HMBC correlations between the methoxy signal (δH 3.32) and C-23 (δC 73.7) indicated that methoxy group should be placed in C-23, which was proved by the HMBC correlations from H-24a (δH 2.07) to C-22 (δC 150.4) (Figure 2a). The key NOESY correlations between H-23 and H-21/H-27 were indicative of α-orientation for H-23 (Figure 2b). Therefore, the methoxy group at C-23 had a β-orientation. On the basis of above features, a 23S configuration was established by reference to notation of R,S-configuration. The absolute configuration of 25S in 1 was established by the chemical shift of H2-26 (δH 3.56 and 4.15 ppm, Δδ = 0.59) (Δδ ≥ 0.57 ppm) [11]. Thus, compound 1 was inferred as (23S)-3β,26-dihydroxy-23β-methoxyl-5β-furost-20(22)-en. The absolute configurations of the β-glucose and β-galactose [12] were determined as D by GC analysis of their hydrolyzed forms. The linkage of the sugar in 1 was proved by long-range HMBC correlations between δH 4.83 (H-1′) and δC 75.5 (C-26, aglycone), δH 4.92 (H-1′′) and δC 75.3 (C-3, aglycone) and δH 5.28 (H-1′′′) and δC 81.8 (C-2′′, 3-O-β-d-galactose) (Figure 2a, Table 2). Thus, the structure of compound 1 was deduced to be (23S,25S)-26-O-β-d-glucopyranosyl-3β,26-dihydroxy-23β-methoxyl-5β-furost-20(22)-en-3β-yl-O-β-d-glucopyranosyl-(1→2)-β-d-galactopyranoside.
Table 1.
1H-NMR and 13C-NMR data for aglycone moiety of compounds 1–4 (pyridine-d5).
| No. | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δH mult (J, Hz) | δC | δH mult (J, Hz) | δC | δH mult (J, Hz) | δC | δH mult (J, Hz) | δC | |
| 1a | 1.49 m | 31.0 | 1.48 m | 31.0 | 1.48 a | 31.6 | 1.47 a | 31.0 |
| 1b | 1.87 a | 1.83 a | 1.88 a | 1.80 a | ||||
| 2a | 1.21 a | 27.0 | 1.23 a | 27.0 | 1.45 a | 27.3 | 1.51 a | 26.7 |
| 2b | 1.84 a | 1.87 a | 1.92 a | 1.95 m | ||||
| 3 | 4.28 m | 75.3 | 4.28 m | 75.3 | 4.30 a | 75.7 | 4.31 m | 75.5 |
| 4a | 1.45 a | 31.0 | 1.52 m | 31.0 | 1.80 a | 31.2 | 1.81 a | 31.0 |
| 4b | 1.81 a | 1.89 a | 1.82 a | 1.85 a | ||||
| 5 | 2.15 m | 37.0 | 2.18 m | 37.0 | 2.20 m | 37.4 | 2.16 m | 36.8 |
| 6a | 0.92 m | 26.8 | 0.98 m | 26.9 | 1.25 a | 27.6 | 1.21 m | 27.1 |
| 6b | 1.82 a | 1.20 a | 1.87 a | 1.83 a | ||||
| 7a | 1.51 a | 1.51 a | 1.50 a | 0.93 m | ||||
| 7b | 1.98 m | 26.9 | 2.00 m | 26.9 | 2.31 m | 27.5 | 1.28 a | 26.7 |
| 8 | 1.38 a | 35.1 | 1.40 a | 35.2 | 1.86 a | 36.9 | 1.54 a | 34.7 |
| 9 | 1.22 a | 40.2 | 1.27 a | 40.2 | 1.42 a | 40.9 | 1.46 a | 39.4 |
| 10 | 35.3 | 35.3 | 35.9 | 35.3 | ||||
| 11a | 1.15 m | 21.3 | 1.15 a | 21.3 | 1.22 a | 21.7 | 1.49 a | 31.4 |
| 11b | 1.32 m | 1.32 m | 1.35 m | 1.78 m | ||||
| 12a | 1.17 a | 1.15 a | 1.20 a | |||||
| 12b | 1.70 m | 40.1 | 1.72 d (9.2) | 40.1 | 1.68 m | 41.6 | 3.53 dd (4.0, 10.0) | 79.5 |
| 13 | 44.0 | 43.9 | 41.8 | 46.7 | ||||
| 14 | 0.81 m | 54.8 | 0.80 m | 54.8 | 1.53 a | 61.4 | 1.10 m | 55.3 |
| 15a | 1.04 a | 34.5 | 1.42 a | 34.5 | 4.40 a | 79.5 | 1.58 m | 31.9 |
| 15b | 2.05 m | 2.07 dd (5.6, 12.4) | 2.09 dd (5.4, 12.0) | |||||
| 16 | 4.87 m | 84.8 | 4.85 m | 84.7 | 5.05 dd (4, 8.8) | 91.9 | 4.66 t (5.4) | 81.4 |
| 17 | 2.55 d (10.0) | 65.0 | 2.50 d (10.4) | 65.1 | 2.18 a | 61.9 | 2.22 a | 62.8 |
| 18 | 0.66 s | 14.6 | 0.70 s | 14.6 | 0.95 s | 18.5 | 1.08 s | 11.2 |
| 19 | 0.99 s | 24.1 | 0.99 s | 24.0 | 1.04 s | 24.7 | 1.00 s | 24.0 |
| 20 | 109.2 | 105.1 | 2.26 m | 41.4 | 2.18 m | 43.6 | ||
| 21 | 1.73 s | 11.6 | 1.75 s | 11.7 | 1.30 d (6.8) | 16.9 | 1.48 d (6.4) | 14.3 |
| 22 | 150.4 | 154.4 | 110.8 | 110.0 | ||||
| 23 | 4.22 m | 73.7 | 4.92 m | 63.9 | 2.09 a
1.97 m |
37.6 | 2.18 a
1.36 m |
26.3 |
| 24a | 1.87 a | 37.3 | 1.80 br.d (12.4) | 39.7 | 1.70 m | 28.9 | 1.30 m | 26.5 |
| 24b | 2.07 a | 2.40 m | 2.07 a | 1.50 a | ||||
| 25 | 2.21 m | 31.1 | 2.48 br.d (10.4) | 31.0 | 1.92 a | 34.9 | 1.62 a | 27.6 |
| 26a | 3.56 m | 75.5 | 3.81 br.d (10.4) | 75.6 | 3.48 dd (6.8, 9.2) | 75.9 | 3.40 br.d (10.8) | 65.1 |
| 26b | 4.15 a | 4.07 m | 4.08 m | 4.13 m | ||||
| 27 | 1.12 d (6.8) | 17.9 | 1.18 d (6.4) | 17.8 | 1.01 d (6.8) | 17.9 | 1.09 d (6.8) | 16.3 |
| OCH3 | 3.32 s | 56.0 | ||||||
a Overlapped signals.
Figure 2.
Key HMBC and 1H-1H COSY correlations (a) of compound 1; Key NOESY correlations (b) of compound 1.
Table 2.
1H-NMR and 13C-NMR data for sugar portions of compounds 1–4 (pyridine-d5).
| No. | 1 | 2 | 3 | 4 | ||||
|---|---|---|---|---|---|---|---|---|
| δH (mult, J, Hz) | δC | δH (mult, J, Hz) | δC | δH (mult, J, Hz) | δC | δH (mult, J, Hz) | δC | |
| Glc-1 | 4.83 d (8.0) | 105.3 | 4.89 d (7.6) | 105.1 | 4.81 d (7.6) | 105.7 | ||
| 2 | 4.01 m | 75.3 | 4.07 a | 75.2 | 4.03 a | 75.7 | ||
| 3 | 3.84 a | 78.5 | 3.97 m | 78.6 | 4.24 br.d (8.8) | 79.1 | ||
| 4 | 4.20 a | 71.7 | 4.22 t (8.4) | 71.8 | 4.23 br.d (8.8) | 72.2 | ||
| 5 | 4.21 a | 78.6 | 4.24 m | 78.7 | 3.95 a | 78.9 | ||
| 6 | 4.36 m 4.45 d (8.0) |
62.9 | 4.39 dd (2.8, 12.4) 4.55 dd (5.6, 12.4) |
63.0 | 4.32 dd (3.2, 14.8) 4.50 br.d (14.8) |
63.3 | ||
| Gal/Glc-1 | 4.92 d (8.0) | 102.6 | 4.93 d (7.6) | 102.6 | 4.94 d (7.6) | 102.3 | 4.93 d (8.0) | 102.6 |
| 2 | 4.68 t (7.6) | 81.8 | 4.69 t (11.2) | 82.0 | 4.26 m | 83.6 | 4.70 a | 81.9 |
| 3 | 4.08 t (7.6) | 77.0 | 4.10 d (11.2) | 76.9 | 4.32a | 78.6 | 4.27 dd (3.2, 9.6) | 75.3 |
| 4 | 4.57 dd (2.2, 8.4) | 69.9 | 4.58 a | 69.9 | 4.33 br.d (9.6) | 72.3 | 4.59 d (3.2) | 69.9 |
| 5 | 4.03 a | 76.7 | 4.04 m | 76.6 | 3.87 m | 78.7 | 4.03 t (8.4) | 76.6 |
| 6 | 4.43 br.d (9.2) 4.39 dd (3.6, 9.2) |
62.2 | 4.40 dd (1.8, 12.0) 4.45 br. d (12.0) |
62.2 | 4.40 d (10.4) 4.54 br.d (10.4) |
63.4 | 4.40 dd (1.2, 8.4) 4.45 dd (2.4, 8.4) |
62.2 |
| Glc-1 | 5.28 d (7.6) | 106.1 | 5.30 d (8.0) | 106.2 | 5.39 d (8.0) | 106.4 | 5.30 d (7.6) | 106.2 |
| 2 | 4.35 m | 75.6 | 4.31 t (8.4) | 75.6 | 4.09 a | 77.5 | 4.10 m | 77.0 |
| 3 | 4.18 a | 78.1 | 4.20 t (8.4) | 78.1 | 4.28 a | 78.4 | 4.22 t (11.2) | 78.1 |
| 4 | 4.30 br.d (9.6) | 71.8 | 4.32 m | 71.9 | 4.18 br.d (10.0) | 72.1 | 4.34 t (11.2) | 71.8 |
| 5 | 3.84 m | 78.5 | 3.87 m | 78.4 | 4.26 a | 79.0 | 3.86 m | 78.5 |
| 6 | 4.53 dd (1.2, 10.8) 4.47 m |
62.9 | 4.36 dd (4.0, 9.6) 4.48 br.d (9.6) |
62.9 | 4.38 d (8.8) 4.57 br.d (8.8) |
63.2 | 4.47 dd (3.6, 8.4) 4.53 dd (1.2, 8.4) |
62.8 |
a Overlapped signals.
Anemarsaponin Q (2) was an amorphous power and its molecular formula was C45H74O19 according to HR-ESI-MS at m/z 941.4714 [M + Na]+ (Figure S9). The 13C-NMR spectrum data (Figure S11) (Table 1) showed four methyl groups at δC 14.6, 24.0, 11.7 and 17.8. In addition, the carbon signals at δC 105.1 (C-20), 154.4 (C-22) and the secondary C-26 carbon signal (δC 75.6) indicated that compound 2 was Δ20(22)-unsaturated furostanol saponin [11]. The 13C-NMR data of compound 2 was very similar to those of 1, except for the absence of a methoxy in 2. Significant differences were observed between 1 and 2 for C-22~C-27. The C-23 that appeared at δC 73.7 of 1 was instead an upfield-shifted carbon at δC 63.9 in compound 2. The above data suggested that a hydroxy group existed at C-23 of 2, which was supported by the correlations from H-23 to H-24, H-24 to H-25, and H-25 to H-27 in the 1H-1H COSY spectrum of 2 (Figure S1).
The NOESY cross-peaks between H-23 and H-21 indicated the α-orientation of H-23 and the β-orientation of hydroxyl group for C-23 (Figure S2). Being similar to compound 1, the absolute configuration of 23S was confirmed by reference to the notation of R,S-configuration. The 25R configuration in 2 was proved by the protons of H2-26 (δH 3.81 and 4.07 ppm, Δδ = 0.26) (Δδ ≤ 0.48 ppm) [11]. Compound 2 afforded d-glucose and d-galactose, identified by GC analysis of their acid hydrolysis derivatives. Compound 2 was thus established as (23S,25R)-26-O-β-d-glucopyranosyl-3β,23β,26-trihydroxy-5β-furost-20(22)-en-3β-yl-O-β-d-glucopyranosyl-(1→2)-β-d-galactopyranoside.
Anemarsaponin R (3) (Figure S12). had a molecular formula C45H76O20, based on HR-ESI-MS at m/z 959.4840 [M + Na]+. In the 13C-NMR spectrum (Figure S14) of 3 (Table 1) four methyl groups (δC 18.5, 24.7, 16.9 and 17.9) and quaternary carbon (δC 110.8) suggested that compound 3 was a furostanol saponin [11]. Comparison of 13C-NMR data indicated the same skeleton in 3 and timosaponin E1 (7). The tiny differences between them were seen in the sugar moiety of C-3. Instead of glucose and galactose in 7, two glucoses were identified in 3 by the signals at δC 102.3, 83.6, 78.6, 72.1, 78.7, 63.4 and δC 106.4, 77.5, 78.4, 72.3, 79.0, 63.2 (Table 2). In addition, the existence of glucose was also confirmed by the coupling constant of 9.6 Hz in H-4 for compound 3 instead of the typical one of galactose [13,14]. Meanwhile, the 13C-NMR data of the sugar residue matched those of anemarsaponin C [15]. Their sugar linkages were established by the existence of long-range HMBC correlations between δH 4.81 (H-1′) and δC 75.9 (C-26, aglycone), δH 4.94 (H-1′′) and δC 75.7 (C-3, aglycone) and δH 5.39 (H-1′′′) and δC 83.6 (C-2′′, 3-O-β-d-glucose) (Figure S3). The sugars of 3 were also only assignable to d-glucose by GC analysis of their chiral derivatives. The NOESY cross-peaks between the signals of H-23a and H-20, H-15 and H-18 indicated the α-orientations of the hydroxyl groups at C-15 and C-22 (Figure S4). In addition, the NOESY correlations between H-3 and H-1′′, H-5 and H-19 indicated the β-orientations of OH-3, H-5 (Figure S4). Thus, the structure of compound 3 was established as (25S)-26-O-β-d-glucopyranosyl-3β,15α,22α,26-tetrahydroxy-5β-furost-3β-yl-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranoside.
Anemarsaponin S (4) had a molecular formula C46H76O19, on the basis of HR-ESI-MS at m/z 779.4193 [M + Na]+ (Figure S15). In the 1H-NMR spectrum (Figure S16) of 4 (Table 1) the signals at δH 1.08 (3H, s), 1.00 (3H, s), 1.48 (3H, d, J = 6.4 Hz) and 1.09 (3H, d, J = 6.8 Hz) were assignable to four methyl groups. Correspondingly, the signals at δC 11.2, 24.0, 14.3 and 16.3 also demonstrated the existences of four methyl groups. In addition, the signal at δC 110.0 was assignable to the characteristic C-22 carbon signal of a spirostanol saponin. Comparison of the carbon signals revealed multiple similarities between 4 and (25R)-5β-spirostane-3β,12β-diol-3-O-β-d-glucopyranosyl-(1→2)-β-d-galacto-pyranoside [16], excluding the F ring. In the comparison of the F ring signals (δC 32.2, 29.6, 31.0, 67.2, 17.7), apparent differences were observed at δC 26.3, 26.5, 27.6, 65.1, 16.3 of 4. Meanwhile, the 13C-NMR data of F ring in 1 were consistent with markogenin 3-O-d-glucopyranosyl-(1→2)-β-d-galactopyranoside [17]. Certainly, both the shift of H3-27 (δH 1.09, d, J = 6.8 Hz) and H2-26 proton chemistry shift (δH 3.40 and 4.13) directly confirmed a 25S configuration in 4 [18]. Therefore, compound 4 was formulated as (25S)-3β,12β-dihydroxy-5β-spirostane-3β-yl-O-β-d-glucopyranosyl-(1→2)-β-d-galactopyranoside.
By comparison of NMR data with those reported, the four known compounds were established as anemarsaponin B (5) [7], timosaponin D (6) [8], timosaponin E1 (7) [9], anemarsaponin B II (8) [10].
2.3. Cytotoxic Activity
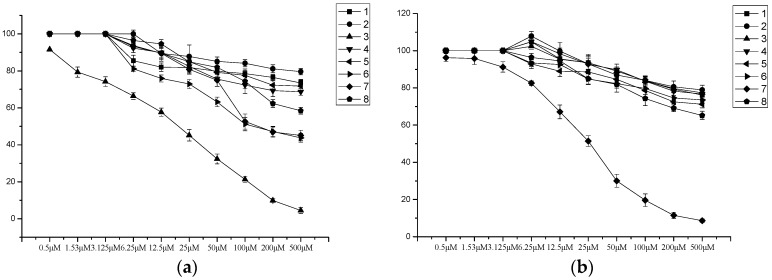
Anemarsaponins P-S (compounds 1–4), anemarsaponin B (5), timosaponin D (6), timosaponin E1 (7) and anemarsaponin B II (8) were evaluated for their in vitro cytotoxic activities against two human tumor cell lines (HepG2 and SGC7901) through the MTT method. Among them, anemarsaponin R (3) showed medium cytotoxicity against HepG2 cells, with an IC50 value of 43.90 μM. Timosaponin E1 (7) exhibited medium cytotoxicity against SGC7901 cells, with an IC50 value of 57.90 μM. The other compounds did not show significant cytotoxicity (IC50 > 100 μM) (Table 3). The dose dependent of cell viabilityon HepG2 (a) and SGC7901 (b) for compounds (Figure 3).
Table 3.
Cytotoxicities of compounds 1–8.
| Compounds | IC50 (μM) | Compounds | IC50 (μM) | ||
|---|---|---|---|---|---|
| HepG2 | SGC7901 | HepG2 | SGC7901 | ||
| 1 | >100 | >100 | 6 | >100 | >100 |
| 2 | >100 | >100 | 7 | >100 | 57.90 ± 2.88 |
| 3 | 43.90 ± 3.36 | >100 | 8 | >100 | >100 |
| 4 | >100 | >100 | doxorubicin | 8.20 ± 1.25 | 6.25 ± 2.18 |
| 5 | >100 | >100 | |||
Figure 3.
(a) Inhibition of HepG2 cell proliferation by the tested compounds; (b) Inhibition of SGC7901 cell proliferation by the tested compounds.
3. Experimental Section
3.1. General Experimental Procedures
Optical rotations were obtained on a P-2000 polarimeter (JASCO, Tokyo, Japan). IR spectra were obtained on a FTIR-8400S instrument (Shimadzu, Kyoto, Japan). UV spectra were obtained on a Shimadzu UV-1601 instrument. NMR spectra were obtained on DPX 400 NMR instrument (Bruker, Rheinstetten, Germany). HR-ESI-MS were recorded on a Xevo-TOF-MS™ instrument (Waters, Milford, MA, USA). HPLC was performed by using a Waters 515 HPLC system coupled with a Waters 2414 refractive index detector. A Waters XBridge preparative C18 column (19 × 250 mm, 10 μm) was used. Macroporous absorption resin D-101 (Cangzhou Bon Adsorber Technology Co., Ltd., Cangzhou, China), MPLC (C-610, Büchi, Flawil Switzerland) and ODS silica gel (YMC Ltd., Kyoto, Japan) were used for column chromatography. MeOH and EtOH were analytical grade and purchased from Tian-Jin Fu Yu Co., Ltd. (Tianjin, China). MeCN was HPLC grade and obtained from J&K Scientific Ltd. (Beijing, China).
3.2. Plant Material
The rhizomes of Anemarrhena asphodeloides Bunge were collected at Bozhou Country, Anhui Province, People’s Republic of China, in September 2014, and identified by Ruifeng Fan from Heilongjiang University of Chinese Medicine. A voucher specimen (20140930) has been deposited in the laboratory.
3.3. Extration and Isolation
The dried slice of rhizomes from Anemarrhena asphodeloides (56 kg) were extracted three times with hot water (280 L) under reflux. The solutions were combined and evaporated to give a residue (22.09 kg). The crude was suspended with EtOAc, n-BuOH, H2O, respectively. The n-BuOH elute was concentrated under vacuum to yield the n-BuOH–soluble fraction.
A part of the fraction (120 g) was chromatographed by MPLC, eluted with a stepwise gradient of MeOH–H2O and finally with MeOH, giving 20 subfractions. Fr.5 (3.1 g) was subjected to ODS column chromatography, eluted with a gradient of MeOH–H2O (4:6 to 1:0) to yield Fr.5-1~Fr.5-4. Fr.5-4 (0.8 g) was separated by preparative HPLC (5.0 mL/min, 15% MeCN–H2O) and yielded compound 4 (4 mg). Fr.17 (0.2 g) was purified with HPLC (5.0 mL/min, 27% MeCN–H2O) to give compound 7 (6 mg). Fr.18 (25 g) was chromatographed over an ODS column to afford Fr.18-1~Fr.18-5. Compound 5 (2.9 g) was crystallized from Fr.18-3. Fr.18-5 was further purified by HPLC (5.0 mL/min, 30% MeCN–H2O) to afford compounds 1 (5 mg), 2 (28 mg) and 3 (8 mg). Fr.19 (2 g) was purified by HPLC (5.0 mL/min, 38% MeCN–H2O) to give compounds 6 (7 mg) and 8 (57 mg).
Anemarsaponin P (1). White amorphous power; −67 (c 2.0, MeOH); UV (MeOH) γmax 217 nm; CD Δε +1.5 (231 nm); IR (KBr) νmax 3454, 2930, 1646, 1405, 1070 cm−1; 1H- and 13C-NMR data (Figures S7 and S8), see Table 1 and Table 2; HR-ESI-MS m/z 955.4644 [M + Na]+ (calc. for C46H76O19 955.4878).
Anemarsaponin Q (2). White amorphous power; −7.1 (c 1.7, MeOH); UV (MeOH) γmax 211 nm; CD Δε +1.7 (232 nm); IR (KBr) νmax 2418, 2931, 1067, 1033 cm−1; 1H- and 13C-NMR data (Figures S10 and S11), see Table 1 and Table 2; HR-ESI-MS m/z 941.4714 [M + Na]+ (calc. for C45H74O19 941.4722).
Anemarsaponin R (3). White amorphous power; −38.0 (c 2.0, MeOH); IR (KBr) νmax 3465, 2935, 1397, 1080 cm−1; 1Hp and 13C-NMR data (Figures S13 and S14), see Table 1 and Table 2; HR-ESI-MS m/z 959.4840 [M + Na]+ (calc. for C45H76O20 959.4828).
Anemarsaponin S (4). White amorphous power; −5.0 (c 2.0, MeOH); IR (KBr) νmax 3428, 2930, 1397, 1070 cm−1; 1H- and 13C-NMR data (Figures S16 and S17), see Table 1 and Table 2; HR-ESI-MS m/z 779.4193 [M + H]+ (calc. for C39H64O14 779.4194).
Anemarsaponin B (5). White amorphous power; 1H-NMR (400 MHz, Pyridine) δ: 0.70 (3H, s, H-18), 1.00 (3H, s, H-19), 1.64 (3H, s, H-21), 1.04 (3H, d, J = 6.4 Hz, H-27), 4.95 (1H, d, J = 8.0 Hz, H-1′′), 4.85 (1H, d, J = 7.6 Hz, H-1′′′), 5.31 (1H, d, J = 7.6 Hz, H-1′); 13C-NMR (100 MHz, Pyridine) δ: 31.0 (C-1), 27.0 (C-2), 75.3 (C-3), 31.0 (C-4), 37.0 (C-5), 26.9 (C-6), 26.9 (C-7), 35.2 (C-8), 40.2 (C-9), 35.3 (C-10), 21.3 (C-11), 40.1 (C-12), 43.9 (C-13), 54.8 (C-14), 31.4 (C-15), 84.6 (C-16), 64.7 (C-17), 14.4 (C-18), 24.0 (C-19), 103.6 (C-20), 11.9 (C-21), 152.4 (C-22), 34.5 (C-23), 23.7 (C-24), 33.7 (C-25), 75.3 (C-26), 17.2 (C-27), 102.6 (C-1′′), 82.0 (C-2′′), 77.0 (C-3′′), 69.9 (C-4′′), 76.7 (C-5′′), 62.2 (C-6′′), 106.2 (C-1′′′), 75.6 (C-2′′′), 78.1 (C-3′′′), 71.8 (C-4′′′), 78.5 (C-5′′′), 62.8 (C-6′′′), 105.2 (C-1′), 75.3 (C-2′), 78.6 (C-3′), 71.8 (C-4′), 78.7 (C-5′), 62.9 (C-6′).
Timosaponin D (6). White amorphous power; 1H-NMR (400 MHz, Pyridine) δ: 0.70 (3H, s, H-18), 1.00 (3H, s, H-19), 1.64 (3H, s, H-21), 1.05 (3H, d, J = 6.4 Hz, H-27), 4.99 (1H, d, J = 7.6 Hz, H-1′′), 4.82 (1H, d, J = 7.6 Hz, H-1′′′), 5.28 (1H, d, J = 7.6 Hz, H-1′); 13C-NMR (100 MHz, Pyridine) δ: 40.1 (C-1), 67.2 (C-2), 82.0 (C-3), 31.9 (C-4), 36.6 (C-5), 26.3 (C-6), 26.9 (C-7), 35.3 (C-8), 41.4 (C-9), 37.1 (C-10), 21.5 (C-11), 40.6 (C-12), 43.8 (C-13), 54.7 (C-14), 31.4 (C-15), 84.6 (C-16), 64.7 (C-17), 14.4 (C-18), 23.9 (C-19), 103.4 (C-20), 11.8 (C-21), 152.4 (C-22), 34.4 (C-23), 23.7 (C-24), 33.7 (C-25), 75.3 (C-26), 17.2 (C-27), 103.6 (C-1′′), 81.8 (C-2′′), 75.3 (C-3′′), 69.8 (C-4′′), 77.0 (C-5′′), 62.1 (C-6′′), 106.2 (C-1′′′), 77.0 (C-2′′′), 78.1 (C-3′′′), 71.8 (C-4′′′), 78.6 (C-5′′′), 62.9 (C-6′′′), 105.2 (C-1′), 75.3 (C-2′), 78.6 (C-3′), 71.8 (C-4′), 78.6 (C-5′), 62.9 (C-6′).
Timosaponin E1 (7). White amorphous power; 1H-NMR (400 MHz, Pyridine) δ: 0.81 (3H, s, H-18), 1.00 (3H, s, H-19), 1.28 (3H, d, J = 6.8 Hz, H-21), 1.01 (3H, d, J = 6.8 Hz, H-27), 4.81 (1H, d, J = 7.6 Hz, H-1′′), 4.96 (1H, d, J = 7.6 Hz, H-1′′′), 5.26 (1H, d, J = 7.6 Hz, H-1′); 13C-NMR (100 MHz, Pyridine) δ: 31.0 (C-1), 27.2 (C-2), 75.5 (C-3), 31.0 (C-4), 37.3 (C-5), 27.0 (C-6), 26.5 (C-7), 36.4 (C-8), 40.5 (C-9), 35.4 (C-10), 21.4 (C-11), 41.3 (C-12), 41.6 (C-13), 60.8 (C-14), 79.1 (C-15), 91.4 (C-16), 61.4 (C-17), 18.0 (C-18), 24.1 (C-19), 40.9 (C-20), 16.5 (C-21), 110.4 (C-22), 37.1 (C-23), 28.4 (C-24), 34.5 (C-25), 75.5 (C-26), 17.5 (C-27), 103.3 (C-1′′), 81.8 (C-2′′), 75.3 (C-3′′), 69.9 (C-4′′), 76.8 (C-5′′), 62.1 (C-6′′), 106.2 (C-1′′′), 77.0 (C-2′′′), 78.1 (C-3′′′), 71.7 (C-4′′′), 78.4 (C-5′′′), 62.9 (C-6′′′), 105.2 (C-1′), 75.3 (C-2′), 78.7 (C-3′), 71.8 (C-4′), 78.5 (C-5′), 62.9 (C-6′).
Anemarsaponin BII (8). White amorphous power; 1H-NMR (400 MHz, Pyridine) δ: 0.81 (3H, s, H-18), 0.99 (3H, s, H-19), 1.32 (3H, d, J = 6.8 Hz, H-21), 1.02 (3H, d, J = 6.8 Hz, H-27), 4.82 (1H, d, J = 8.0 Hz, H-1′′), 4.93 (1H, d, J = 7.6 Hz, H-1′′′), 5.30 (1H, d, J = 7.6 Hz, H-1′); 13C-NMR (100 MHz, Pyridine) δ: 31.4 (C-1), 27.5 (C-2), 75.7 (C-3), 31.4 (C-4), 37.4 (C-5), 27.5 (C-6), 27.3 (C-7), 36.0 (C-8), 40.7 (C-9), 35.7 (C-10), 21.6 (C-11), 40.9 (C-12), 41.7 (C-13), 56.9 (C-14), 32.9 (C-15), 81.7 (C-16), 64.5 (C-17), 17.2 (C-18), 24.5 (C-19), 41.1 (C-20), 17.0 (C-21), 111.1 (C-22), 37.6 (C-23), 28.8 (C-24), 34.9 (C-25), 75.9 (C-26), 17.9 (C-27), 103.0 (C-1′′), 82.3 (C-2′′), 77.4 (C-3′′), 70.3 (C-4′′), 77.1 (C-5′′), 63.2 (C-6′′), 106.6 (C-1′′′), 76.0 (C-2′′′), 78.5 (C-3′′′), 72.2 (C-4′′′), 78.9 (C-5′′′), 63.2 (C-6′′′), 105.6 (C-1′), 75.7 (C-2′), 79.1 (C-3′), 72.2 (C-4′), 79.0 (C-5′), 62.6 (C-6′).
3.4. Acid Hydrolysis and GC Analysis
The hydrolysis and GC analysis of the four new compounds were carried out for the chiral sugar derivatives. Compounds 1–4 (2 mg) were heated with 5 mL 2 M HCl at 90 °C for 3 h. The mixtures were extracted with EtOAc (5 mL) for three times. The sugar residue was dispersed with 1 mL pyridine and reacted with l-cysteine methyl ester hydrochloride (1.5 mg) at 60 °C for 1 h. Then 150 μL of HMDS-TMCS (hexamethyldisilazane–trimethylchlorosilane, 3:1) was added into the mixture that was further reacted at 60 °C for 30 min. The supernatant of the mixture was evaporated to dryness with a N2 stream. The residue was separated with H2O (0.1 mL) and n-hexane (0.1 mL), and the supernatant layer (1 μL) was analyzed by GC. The configurations of the sugar portion for compounds 1–4 were determined by comparison the retention times of their derivatives with those of standard d-glucose (tR = 15.68 min) and d-galactose (tR = 13.47 min) [19].
3.5. Cytotoxic Activity
The isolated compounds were evaluated for their in vitro antiproliferative activities by the MTT method. Doxorubicin was used as positive control (Table 3). Two cell lines, HepG2 and SGC7901, were obtained from the Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). They were cultured in RPMI 1640 supplemented with 10% FBS, 100 IU/mL penicillin, 100 μg/mL streptomycin in 5% CO2 at 37 °C. Cells were cultured in 96-well plates for 24 h with 100 μL complete medium, followed by treating with compounds at different concentrations. 20 μL MTT (5 mg/mL in PBS) was added in the 96-well plates for another 4 h. The solutions were assayed at 490 nm using a VICTOR-X3 ELISA instrument (PerkinElmer, Waltham, MA, USA), after the precipitates were dissolved in DMSO [20]. The cytotoxicities of compounds against HepG2 and SGC7901 were calculated and expressed as IC50 values.
4. Conclusions
As described in the introduction, in recent years steroidal saponins have become a research hotspot due to their multiple and strong bioactivities, especially their cytotoxic activities against a series of tumor cell lines [5,6,21]. In this paper, four new saponins were isolated from Anemarrhena asphodeloides their strictures elucidated, and their antiproliferative activities against HepG2 and SGC7901 were evaluated. Obvious differences were observed between the antiproliferative activities of compounds 3, 7 and the others. The above results represent a contribution to the discovery of new active ingredients and lead compounds and provide an experimental and scientific basis for drug design and drug discovery.
Acknowledgments
This project was supported by Major State Basic Research Development Program (973 Program)of China (2013CB531800) and National Natural Science Foundation of China (No. 81274103).
Abbreviations
The following abbreviations are used in this manuscript:
| IR | Infrared |
| NMR | Nuclear magnetic resonance |
| UV | Ultraviolet |
| HR-ESI-MS | High-resolution electrospray ionization mass spectrometry |
| HMBC | Heteronuclear multiple bond correlation |
| HSQC | Heteronuclear multiple quantum coherence |
| NOESY | Nuclear overhauser effect |
| 1H-1H COSY | Correlation spectroscopy |
| MeOH | Methanol |
| HPLC | High performance liquid chromatography |
| MPLC | Middle pressure liquid chromatography |
| MeCN | Acetonitrile |
| GC | Gas chromatography |
| EtOAc | Ethyl acetate |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2-H-tetrazolium bromide |
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/21/8/1075/s1.
Author Contributions
B.-Y.Y. and H.-X.K. designed the experiments; J.Z. performed the experiments; J.Z. wrote the paper. Y.L. modified the paper; All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Not available.
References
- 1.Wang Y.L., Dan Y., Yang D.W., Hu Y.L., Zhang L., Zhang C.H., Zhu H., Cui Z.H., Li M.H., Liu Y.Z. The genus Anemarrhena Bunge: A review on ethnopharmacology, phytochemistry and pharmacology. J. Ethnopharmacol. 2014;153:42–46. doi: 10.1016/j.jep.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Lim S.M., Jeong J.J., Kang J.D., Kim K.A., Choi H.S., Kim D.H. Timosaponin AIII and its metabolite sarsasapogenin ameliorate colitis in mice by inhibiting NF-κB and MAPK activation and restoring Th17/Treg cell balance. Int. Immunopharm. 2015;25:493–503. doi: 10.1016/j.intimp.2015.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Lu W.Q., Qiu Y., Li T.J., Tao X., Sun L.N., Chen W.S. Antiplatelet and antithrombotic activities of timosaponin B-II, an extract of Anemarrhena asphodeloides. Clin. Exp. Pharmacol. Physiol. 2011;38:380–384. doi: 10.1111/j.1440-1681.2011.05530.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang J.Y., Meng Z.Y., Ma D.H., Xu S.X., Kodama H. Effect of six steroidal saponins isolated from anemarrhenae rhizoma on platelet aggregation and hemolysis in human blood. Clin. Chim. Acta. 1999;289:78–88. doi: 10.1016/S0009-8981(99)00160-6. [DOI] [PubMed] [Google Scholar]
- 5.Kang Y.J., Chung H.J., Nam J.W., Park H.J., Seo E.K., Kim Y.S., Lee D., Lee S.K. Cytotoxic and Antineoplastic Activity of Timosaponin A III for Human Colon Cancer Cells. J. Nat. Prod. 2011;74:701–706. doi: 10.1021/np1007735. [DOI] [PubMed] [Google Scholar]
- 6.Ni Y., Wang Y., Liu M., Chen H.M., Gong X.G. Mitochondrial ROS burst as an early sign in sarsasapogenin induced apoptosis in HepG2 cells. Cell. Biol. Int. 2008;32:337–343. doi: 10.1016/j.cellbi.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Dong J.X., Han G.Y. Studies on the active constituents of Anemarrhena asphodeloides Bge. Acta Pharm. Sin. 1996;27:26–32. [PubMed] [Google Scholar]
- 8.Meng Z.Y., Zhou X.M., Xu S.X. A New Steroidal Saponin from Anemarrhena asphodeloides Bge. J. Shenyang Pharm. Univ. 1998;15:254–256. [Google Scholar]
- 9.Meng Z.Y., Xu S.X., Meng L.H. Timosaponin E1 and E2. Acta Pharm. Sin. 1998;33:693–696. [PubMed] [Google Scholar]
- 10.Qin L.P., Han T., Cao D.P., Zhang Q.Y., Nian H., Rahman K., Zheng H.C. Antiosteoporotic chemical constituents from Er-Xian Decoction, a traditional Chinese herbal formula. J. Ethnopharmacol. 2008;118:271–279. doi: 10.1016/j.jep.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal P.K. Assigning stereo diversity of the 27-Me group of furostane-type steroidal saponins via NMR chemical shifts. Steroids. 2005;70:715–724. doi: 10.1016/j.steroids.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Kostova I., Dinchev D. Saponins in Tribulusterrestris-chemistry and bioactivity. Phytochemistry. 2005;4:111–137. doi: 10.1007/s11101-005-2833-x. [DOI] [Google Scholar]
- 13.Ma B.P., Dong J.X., Yan X.Z. Studies on the furostanol saponins from Anemarrhena asphodeloides Bunge. Acta Pharm. Sin. 1996;31:271–277. [Google Scholar]
- 14.Cheng S.B., Zhang Y.F., Wang Y., Wang Y. Steroidal saponins from Allii Macrostemonis Bulbus. Chin. Tradit. Herb. Drugs. 2013;44:1078–1081. [Google Scholar]
- 15.Yukiko M., Nana A., Chisato H., Fumito T., Yoshihiro M. Steroidal glycosides from the bulbs of Besseraelegans and their cytotoxic activities. Phytochemistry. 2013;96:244–256. doi: 10.1016/j.phytochem.2013.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Setsuo S., Satoshi N., Koki I. New Steroidal Saponins from the Rhizome of Anemarrhena asphodeloides Bunge (Liliaceae) Chem. Pharm. Bull. 1994;42:2342–2345. doi: 10.1248/cpb.42.2342. [DOI] [PubMed] [Google Scholar]
- 17.Nagumo S., Kishi S., Inoue T., Nagai M. Saponins of Anemarrhenae Rhizoma. Yakugaku Zasshi. 1991;111:306–310. doi: 10.1248/yakushi1947.111.6_306. [DOI] [PubMed] [Google Scholar]
- 18.Agrawal P.K. 25R/25S stereochemistry of spirostane-type steroidal sapogenins and steroidal saponins via chemical shift of geminal protons of ring-F. Magn. Reson. Chem. 2003;41:965–968. doi: 10.1002/mrc.1278. [DOI] [Google Scholar]
- 19.Yang B.Y., Guo R., Li T., Wu J.J., Zhang J., Liu Y., Wang Q.H., Kuang H.X. New anti-inflammatory withanolides from the leaves of Datura metel L. Steroids. 2014;87:26–34. doi: 10.1016/j.steroids.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Hussain R.F., Nouri A.M.E., Oliver R.T.D. A new approach for measurement of cytotoxicity using colorimetric assay. J. Immunol. Methods. 1993;160:89–96. doi: 10.1016/0022-1759(93)90012-V. [DOI] [PubMed] [Google Scholar]
- 21.Guo C.R., Li L., Yang X.L., Meng Z.Q., Li F., Zhang C.F., Yang Z.L. Protective effects of timosaponin B II on high glucose-induced apoptosis in human umbilical vein endothelial cells. Environ. Toxicol. Pharmacol. 2014;37:37–44. doi: 10.1016/j.etap.2013.11.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.