Abstract
Three new diterpenes, uprolide N (1), uprolide O (2), uprolide P (3) and a known one, dolabellane (4), were isolated from the CH2Cl2-MeOH extract of the gorgonian octocoral Eunicea succinea, collected from Bocas del Toro, on the Caribbean coast of Panama. Their structures were determined using spectroscopic analyses, including 1D and 2D NMR and high-resolution mass spectrometry (HRMS) together with molecular modeling studies. Compounds 1–3 displayed anti-inflammatory properties by inhibiting production of Tumor Necrosis Factor (TNF) and Interleukin (IL)-6 induced by lipopolysaccharide (LPS) in murine macrophages.
Keywords: Eunicea succinea, cembranolide diterpenes, uprolides, anti-inflammatory
1. Introduction
Gorgonian octocorals are a well-known source of natural products [1,2]. Many of these secondary metabolites are important to the survival strategies of these corals in that they have ecological roles as defensive substances against predators and pathogens and also enhance their ability to compete during reproduction and for space in which to grow [3]. Octocoral metabolites have also proven to be a good resource for drug discovery showing a wide range of pharmacological properties [1,2]. Among octocoral metabolites, diterpenes are a group of compounds frequently found, and these have shown biological activities in different therapeutic areas including antibacterial, antiviral, antifungal, antitumor, anti-arthritic, calcium-antagonistic, anti-inflammatory and antiprotozoal [1,2].
Octocorals from the genus Eunicea (family Plexauridae) were among the first marine invertebrates subjected to chemical studies [4]. These animals have continued to be a prolific source of chemically complex diterpenoids with diverse architectures, such as the cembradienes, cembrane lactones, dolabellanes, cubitanes, dilophols and fuscols [1]. Furthermore, previous work on Eunicea species resulted in the isolation of a group of highly functionalized α-methylene-γ-lactone cembranolides, including a series of compounds that were characterized by the presence of a Δ6 olefin known as the uprolides [5,6,7,8,9,10].
As part of our drug discovery program at INDICASAT, we collected several gorgonian octocorals from the Caribbean and Pacific Coasts of Panama. The extracts of these organisms were subsequently evaluated using a series of bioassays including antimicrobial, anticancer, anti-parasitic and anti-inflammatory. Herein we describe the isolation and structural determination of three new compounds, uprolide N (1), uprolide O (2) and uprolide P (3), from the octocoral Eunicea succinea, which possessed in vitro anti-inflammatory effects by inhibiting the production of TNF and IL-6 in LPS-induced macrophages.
2. Results and Discussion
2.1. Isolation and Structural Elucidation
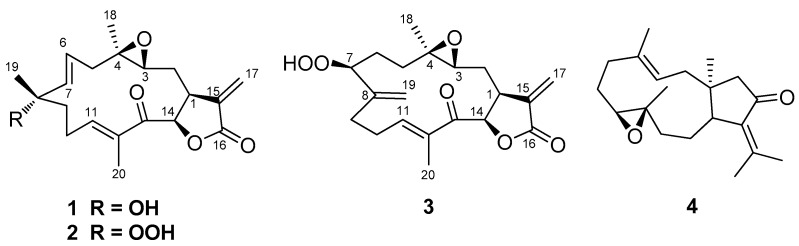
The octocoral Eunicea succinea was collected at −4.5 m by hand using SCUBA in the Isla Bastimentos National Park on the Caribbean coast of Panama. The sample was extracted with a methanol-dichloromethane mixture, and the extract was fractionated using silica gel column chromatography, and subsequently, high performance liquid chromatographic (HPLC) separation to yield compounds 1–4 (Figure 1).
Figure 1.
Structures of compounds 1–4.
The HRESITOF-MS data, collected in positive ion mode for compound 1, showed a pseudomolecular ion peak [M + H]+ at m/z 347.1871. This mass corresponded with the molecular formula C20H26O5 and was consistent with the carbon and proton count from the 13C-NMR and 1H-NMR experiments (Table 1, Figures S1 and S2). DEPT and HSQC experiments revealed six quaternary carbons, six methines, five methylenes and three methyl groups (Figures S3 and S5). Their chemical shifts and multiplicities were consistent with an exocyclic olefin at δC 136.1 (C-15) and 120.0 (CH2-17), four sp3 carbons bearing oxygen (two at δC 73.1 (C-8) and 72.5 (CH-14), and two forming an epoxide at δC 60.9 (C-4) and 56.3 (CH-3)), one ester carbonyl at δC 169.4 (C-16), and a disubstituted olefin at δC 140.7 (CH-7) and 124.2 (CH-6). An α,β-unsaturated ketone moiety was implicated by shifts at δC 195.2 (C-13), 147.8 (CH-11) and 137.2 (C-12). Eight degrees of unsaturation were inferred from the molecular formula: five were accounted for by the two carbonyls and three double bonds. Therefore, compound 1 was deduced to be tricyclic.
Table 1.
1H- and 13C-NMR data in ppm for uprolides N, O and P (1–3) measured in CDCl3 at 400 MHz.
| Position | Uprolide N (1) | Uprolide O (2) | Uprolide P (3) | |||
|---|---|---|---|---|---|---|
| δC | δH (J in Hz) | δC | δH (J in Hz) | δC | δH (J in Hz) | |
| 1 | 41.7, CH | 3.17, m | 41.6, CH | 3.17, m | 40.8, CH | 3.26, m |
| 2 | 27.7,CH2 | 1.74, m | 27.6, CH2 | 1.74, m | 29.1, CH2 | 1.95, m 1.69, m |
| 3 | 56.3, CH | 2.65, dd (4.0, 9.9) | 56.2, CH | 2.72, m | 56.6, CH | 2.47, dd (4.7, 9.5) |
| 4 | 60.9, C | 60.8, C | 59.2, C | |||
|
5a
5b |
39.2, CH2 | 2.58, m 2.43, m | 39.1, CH2 | 2.51, m 2.65, m | 31.9, CH2 | 1.24, m |
| 6 | 124.2, CH | 5.34, ddd (4.7, 9.8, 15.3) | 128.3, CH | 5.45, ddd (4.3, 10.2, 16.5) | 30.8, CH2 | 1.84, m 1.61, m |
| 7 | 140.7, CH | 5.65, d (15.3) | 136.5, CH | 5.63, d (16.5) | 87.3, CH | 4.28, br t (6.9) |
| 8 | 73.1, C | 84.4, C | 145.7, C | |||
| 9 | 41.6, CH2 | 1.97, m | 36.9, CH2 | 2.01, m | 30.2, CH2 | 2.40, m 2.34, m |
| 10 | 25.3, CH2 | 2.40, m | 24.8, CH2 | 2.45, m | 28.3, CH2 | 2.72, m 2.51, m |
| 11 | 147.8, CH | 6.81, br t (6.6) | 147.2, CH | 6.76, br t (5.9) | 145.4, CH | 6.91 br dd (4.8, 9.1) |
| 12 | 137.2, C | 137.5, C | 138.0, C | |||
| 13 | 195.2, C | 195.2, C | 195.5, C | |||
| 14 | 72.5, CH | 5.80, br d (7.3) | 72.9, CH | 5.80, d (8.0) | 73.3, CH | 5.82, d (8.0) |
| 15 | 136.1, C | 136.1, C | 136.8, C | |||
| 16 | 169.4, C | 169.4, C | 169.3, C | |||
|
17a 17b |
120.0, CH2 | 6.28, d (3.3) 5.44, d (3.3) |
120.0, CH2 | 6.28, d (3.3) 5.44, d (3.3) |
120.8, CH2 | 6.28, d (2.2) 5.47, d (2.2) |
| 18 | 18.2, CH3 | 1.37, s | 18.2, CH3 | 1.36, s | 18.0, CH3 | 1.32, s |
|
19a 19b |
25.9, CH3 | 1.34, s | 19.5, CH3 | 1.35, s | 113.7, CH2 | 5.25, s 5.15, s |
| 20 | 11.4, CH3 | 1.83, s | 11.3, CH3 | 1.82, s | 11.2, CH3 | 1.86, s |
Four spin systems were developed on the basis of COSY experiments (Figure 2 and Figure S4). In spin system 1i, a deshielded doublet proton at δH 5.80, attached to an oxygen bearing carbon (C-14, δC 72.5), was adjacent to a midfield methine (C-1, δH 3.17, δC 41.7) with shifts consistent with its allylic nature. In turn, this latter resonance was adjacent to a methylene group (C-2, δH 1.74, δC 27.7) that was further coupled to an α-epoxy proton (C-3, δH 2.65, δC 56.3). In spin system 1ii, a doublet vinyl proton (C-7, δH 5.65, δC 140.7) was coupled to its olefinic partner (C-6, δH 5.34, δC 124.2) with a J value of 15.3 Hz, indicating the trans nature of this double bond [6,8]. The latter resonance was also coupled to an adjacent methylene group (C-5, δH 2.58/2.43, δC 39.2). The spin system 1iii was comprised of a significantly deshielded olefin proton (C-11, δH 6.81, δC 147.8) that was adjacent to a CH2-CH2 moiety (C-10, δH 2.40, δC 25.3; C-9, δH 1.97, δC 41.6). The final spin system 1iv was comprised of two olefinic protons at δH 6.28 (d, J = 3.3 Hz, H-17a) and 5.44 (d, J = 3.3 Hz, H-17b), and were assigned to an exocyclic methylene. In addition, the 1H-NMR spectrum for compound 1 showed signals for three methyl groups, two attached to sp3 quaternary carbons at δH 1.37 (s, H3-18) and 1.34 (s, H3-19) and one attached to a double bond at δH 1.83 (s, H3-20).
Figure 2.
Correlation spectroscopy (COSY), heteronuclear multiple bond correlation (HMBC) and selected 1D nuclear Overhauser effect correlations for compound 1.
The connectivity of these four spin systems with their intervening quaternary carbon atoms and proximate methyl groups was determined using 2,3J HMBC experiments (Figure 2 and Figure S6). An α-methylene-γ-lactone scaffold was indicated by long range H–C coupling from the exocyclic methylene protons (H2-17) to the ester carbonyl at δC 169.4 (C-16) and methine carbon C-1. A further long range H–C coupling from methine proton H-14 to the same ester carbonyl clarified this bond connection, the 5-membered ring nature of the lactone, and the interconnection of spin systems 1i and 1iv. The singlet methyl group at δH 1.37 (H3-18), a shift consistent with its placement on a carbon bearing oxygen, showed HMBC correlations to C-3, C-4 and C-5, thereby connecting partial structures 1i and 1ii and defining the epoxide ring. Similarly, the methyl group at δH 1.34 (H3-19), also at a shift consistent with its placement on a carbon bearing oxygen, tied together spin systems 1ii and 1iii. Finally, the third singlet methyl group at δH 1.83 (H3-20), a shift consistent with its placement on an olefinic bond, showed HMBC correlations to C-11, C-12 and carbonyl carbon C-13, thereby defining the terminus of spin system 1iii as belonging to a disubstituted enone system. Because all atoms of 1 were accounted for and placed into a linear array, but required the presence of one additional ring, a final bond connection was made between C-13 and C-14, thereby completing the planar structure of compound 1 as a novel cembrenolide diterpene.
A literature search for related cembrenolides indicated that compound 1 was structurally similar to uproeuniolide, a diterpene isolated from Eunicea succinea [7], uprolide K, which was isolated from Eunicea pinta [8] and peunicin, isolated from a Eunicea succinea-mammosa complex [11]. Comparisons with the spectroscopic data reported for these compounds supported our assignments for compound 1.
The relative configuration of compound 1 was determined by 1D NOE experiments (Figure 2 and Figures S7–S11), and showed correlations between H-1 and H-3, H-14 and H3-18, indicating they were in the same side of the molecule, assigned as the α-plane. Correlations between H-14 and H-11 revealed these latter protons were also in an α-orientation. Moreover, H-7 showed correlations with H3-18 and H-11 but not with H-6, confirming their orientation in the α-plane, as well as the E geometry of the C-6/C-7 double bond. On the other hand H-6 showed a correlation with H3-19, confirming its orientation on the β-plane. Finally, H-11 showed correlations with H-3, H-7 and H-14, but not with H3-20, confirming its orientation into the α-plane and the E-geometry of the C-11/C-12 double bond.
To validate the stereo-configuration of compound 1, molecular modeling studies were undertaken. A conformational hybrid search method recommended for macrocyclic systems [12], implemented in Macromodel [13], allowed the detection of several conformers for compound 1 (Figure 3). The C-8 epimer was also considered in this study to corroborate the configuration at this carbon atom. The relative stabilities for each conformer were obtained using the MMFF94s force field calculation. To emulate the experimental conditions, the effects of chloroform were considered implicitly during the calculations. The two lower energy conformations for each C-8 epimer of uprolide N (1) are represented in Figure 3. The lowest energy conformer 1a of uprolide N, which is 1.4 kcal mol-1 more stable than 1b, showed a spatial disposition for the Me-19 and C6-C7 double bond very similar to the one of the 3D model of uprolide-A acetate obtained through X-ray diffraction [6]. The calculated inter-proton distances for the lowest energy conformer (1a) of uprolide N (Figure 3) agreed with the experimentally determined nOe effects (Figures S7–S11) for this compound, but they disagreed with values for conformer 1b. In fact, the observations of a strong nOe between H-6 and H-19 and between H-3 and H-11 in compound 1 agree with the distances calculated between both pairs of protons in the lowest energy conformation 1a. In the case of conformer 1b, the distances between H-6 and H-19 and between H-3 and H-11 (4.0 and 4.7Å respectively) are too large and do not agree with the strong nOe’s observed between these proton pairs in compound 1. Consequently, compound 1 should correspond to the configuration and conformation represented in 1a, reinforcing the assignment of structure (1) for this compound.
Figure 3.
The two lower energy conformers for each C-8 epimer of uprolide N (1). Relative energies are expressed in Kcal∙mol−1. Selected distances (in blue) are given in Å.
The HRESITOF-MS spectra of compound 2 showed a pseudomolecular ion peak [M + Na]+ at m/z 385.1615, consistent with a molecular formula of C20H26O6 which is one O-atom more than compound 1. This was consistent with the data obtained from the 13C- and 1H-NMR spectra (Figures S12 and S13), and indicated that compound 2 also had 8 degrees of unsaturation. Five of these were accounted for by two carbonyls (δC 169.4 (C-16) and 195.2 (C-13)) and three carbon-carbon double bonds (δC 147.2 (CH-11), 137.5 (C-12), 136.5 (CH-7), 136.1 (C-15), 128.3 (CH-6), 120.0 (CH2-17)), indicating that compound 2 was also tricyclic. Indeed, the NMR data for compounds 1 and 2 were highly similar (Table 1), indicating a high degree of structural relatedness (Figure 2 and Figure 4). The major differences between the two compounds were found at C-8 and its surrounding carbon atoms. The presence of an additional oxygen in the molecular formula of compound 2 combined with the chemical shift at C-8, which appeared more deshielded (δC 84.4) than that in compound 1 (δC 73.1), was consistent with the presence of a hydroperoxide group at this position (Figure 4) [8].
Figure 4.
1H-1H-COSY and 1H-13C-HMBC correlations for compounds 2 and 3.
The relative stereochemistry of 2 was determined by 1D NOE experiments (Figures S18–S23) wherein H-1 correlated with H-14, and H3-18 and H-14 correlated with H-1 and H-11, indicating their same relative disposition, assigned to the α-plane. Irradiation on H-7 showed correlations with H-11 but not with H-6. Irradiation on H-6 showed no correlation with H-7 confirming the trans orientation of this double bond. A correlation between H-6 and H3-19 indicated the β-orientation of these protons.
The HRESITOF-MS spectra of compound 3 showed a pseudomolecular [M + Na]+ ion at m/z 385.1620; this was consistent with the molecular formula C20H26O6. As in compounds 1 and 2, eight degrees of unsaturation were deduced from the molecular formula; five were accounted for by two carbonyls and three double bonds, indicating compound 3 also had three rings. The 1H-NMR data for compound 3 were highly comparable to compounds 1 and 2, except in the region of C-5 to C-9 and C-19 (Table 1, Figures S24 and S25). The differences were that a new exocyclic olefin was present at C-8 to C-19 (δH 5.25 (s, H-19a), 5.15 (s, H-19b); δC 145.7 (C-8), 113.7 (CH2-19)), and a new oxygen-bearing carbon appeared at δC 87.3, C-7 (δH 4.28, br t, J = 6.9 Hz, H-7). By COSY and 2,3J HMBC experiments (Figure 4, Figures S27 and S29), this latter resonance could be connected to higher field methylene groups at C-6 (δH 1.84 and 1.61; δC 30.8) and C-5 (δH 1.24; δC 31.9). The location of oxidation was further revealed by a 3J HMBC correlation observed for H2-19 to C-7. Considering the molecular formula of 3 which showed one more O-atom than compound 1, and the relatively deshielded shift for C-7 (δC 87.3) [8], it became clear that compound 3 was the alternate allylic oxidation product from a presumed biosynthetic precursor containing a C-7-C-8 double bond such as peunicin [11]. The structure of compound 3 resembles that of uprolide C, previously described from Eunicea mammosa [6,8]. Comparisons with the spectroscopic data for this latter compound supported our assignments for compound 3.
The relative stereochemistry of compound 3 was confirmed by 1D NOE experiments (Figures S30–S36). Irradiation of H-1 showed correlations with, H-14 and H3-18, indicating that they are all on the same α-plane. Also, correlations observed between H-14 and H-11 and between H-11 and H-7 indicated these protons were α-oriented, and therefore, the hydroperoxy group on C-7 must be β-orientated.
To date, more than 30 uprolide diterpenes have been isolated from several Eunicea species. This family is characterized by the presence of an α-methylene-γ-lactone moiety attached to a highly functionalized 14-membered carbocycle which forms the cembranolide scaffold. The major differences among the uprolides are the stereochemical assignments of C-1, C-8, C-12, C-13 and C-14, the degree of oxidation at C-13, and the presence of a hydroxyl group, a hydroperoxy group or an exocyclic double bond at C-8. Additionally, uprolides D-G possess an unusual 4,7-oxa-bridge functionality.
2.2. Anti-inflammatory Effect
Inflammation is the biological response of a host towards harmful stimuli, such as pathogens, physical injury or damaged cells. The inflammatory response initiates with the recruitment of immune cells to injured areas. These cells produce inflammatory mediators including cytokines, chemokines and reactive nitrogen and oxygen species. These mediators are critical for resolving inflammation and for tissue repair; however they can also cause tissue damage. Immune cells, such as macrophages, are equipped with various pattern recognition receptors (PRRs) that recognize many structurally different molecules, including lipopolysaccharide (LPS), lipoproteins, lipids, nucleic acids and proteins [14]. The activation of macrophages by LPS leads to the production of high levels of pro-inflammatory cytokines such as TNF-α and IL-6.
Inhibition of secretion of pro-inflammatory cytokines is frequently used to indicate anti-inflammatory activity. Herein, we evaluated the ability of compounds 1–3 to inhibit the production of TNF-α and IL-6 in primary murine macrophages stimulated with LPS. Compounds 1–3 inhibited the production of TNF and IL-6 in a dose-dependent manner (Figure 5A,B). IC50 values for both mediators are presented in Table 2 (also in Figure S46). The effect of these compounds was not attributed to cellular death because cell viability was not affected at the inhibitory concentrations (Figure 5C and Table 2). However, 60%–70% of cell death was observed after the exposure of cells to 14 μM concentrations of compounds 1 and 3 (data not shown). A cytotoxic effect on cancer cell lines has been demonstrated for other uprolides [15]. In the absence of LPS compounds were unable to induce the production of TNF and IL-6 (Figure S47).
Figure 5.
Uprolides compounds inhibit the production of TNF (A) and IL-6 (B) induced by LPS in macrophages. Peritoneal macrophages from C57B1/6 mice were treated with different concentrations of compounds 1, 2 or 3 (uprolides N, O or P) 1 h before stimulation with 100 μg/mL of LPS. The supernatants were harvested after 6 h, and cytokine concentrations were determined by ELISA. Results are represented as Mean ± S.E.M. from two independent experiments performed in duplicate. **, p < 0.01, ***, p < 0.001 relative to the LPS stimulus alone. (C) The cell viability was assessed using a MTT assay after collecting the supernatant. Results are represented as Mean ± S.E.M. from stimuli performed in duplicate and are representative of two different experiments. C represents the control plus 0.5% of DMSO without the stimulus.
Table 2.
Anti-inflammatory activity of the new uprolides N-P (1–3) and dolabellane (4).
| Mediator | IC50 ± S.D. (μM) | |||
|---|---|---|---|---|
| Uprolide N (1) | Uprolide O (2) | Uprolide P (3) | Dolabellane (4) | |
| TNF | 1.39 ± 0.48 | 2.73 ± 0.03 | 2.27 ±0.91 | n.a. |
| IL-6 | 3.26 ± 0.32 | 4.22 ± 1.52 | 2.60 ±1.47 | n.a. |
Values represent average of IC50 from four independent experiments ± S.D. n.a. non-active.
The anti-inflammatory activity of certain families of cembranoids has been attributed to the presence of the α-methylene-γ-lactone group [16], a feature that is also present in compounds 1–3. Although the lactone group is commonly present in other uprolides previously described, the anti-inflammatory activity of these compounds was not evaluated. This is the first report showing this effect and thus identifies the uprolides as possessing anti-inflammatory properties; however, further studies are necessary to characterize the mechanism of action of these compounds. Compound 4 did not exhibit inhibitory effect on the activation of macrophages by LPS (Table 2).
3. Experimental Section
3.1. General Procedures
Optical rotations were measured in CHCl3 using either a P-2000 digital polarimeter (JASCO, Easton, MD, USA) or an Autopol® III automatic polarimeter (Rudolph, Hackettstown, NJ, USA). UV spectra were recorded using a model UV-2401 (PC) UV-Vis spectrophotometer (Shimadzu, Columbia, MD, USA). IR spectra were recorded using a Platinum ATR ALPHA instrument (Bruker, Billerica, MA, USA). The 1H-, 13C- and 2D-NMR spectra were recorded on a Eclipse 400 MHz spectrometer (JEOL, Peabody, MA, USA). The chemical shifts were calibrated internally based on the signal for the residual solvent in which the sample was dissolved (CDCl3, δH 7.26, δC 77.0). Accurate mass APCI spectra were acquired on a JEOL LC-mate mass spectrometer (Peabody, MA, USA) and HRESITOF-MS were acquired on a microTOF-QIII spectrometer (Bruker Daltonics, Billerica, MA, USA). HPLC purification was carried out using an 1100 HPLC system (Agilent, Santa Clara, CA, USA) equipped with a quaternary pump, an Agilent diode array detector 1200 Series and a normal phase silica gel column (Sphereclone, 250 mm × 10.0 mm, 5 μm, Phenomenex®, Torrance, CA, USA). The flash chromatographic separations were performed using grade 60 silica gel (70–230 mesh, 60 Å, Sigma-Aldrich, St. Louis, MO, USA) and silica gel (Sigma-Aldrich, St. Louis, MO, USA, grade 9385, 230–400 mesh, 60 Å, Sigma). Merck TLC sheets (silica gel 60 F254) were used to perform analytical TLC (aluminum-supported, layer-thickness 200 μm).
3.2. Animal Material
One specimen of the octocoral Eunicea succinea (Order Gorgonacea, Family Plexauridae) was collected at −4.5 m by hand using SCUBA in the Bastimentos National Park, which is located in the Caribbean off the coast of Bocas del Toro, Panama, in November 2009. The coral specimen was identified as Eunicea succinea (Pallas 1766) based on its morphology. A reference specimen is deposited at INDICASAT’s CBDD under the number GLBO-241109-02.
3.3. Extraction and Isolation
The organism (442 g) was minced and exhaustively extracted at room temperature with 1 × 400 mL of CH2Cl2 and then with 6 × 400 mL of 1:1 CH2Cl2/MeOH. The organic extracts were combined and evaporated in vacuo to yield a dark-green oily residue (16.0 g). The CH2Cl2–MeOH extract was fractionated using column chromatography on silica gel with a stepwise gradient of 0%–100 % of EtOAc in hexanes followed by 0%–100% of MeOH in EtOAc to yield 10 fractions (A–J). Fraction D (2.4 g) was chromatographed on silica gel with a stepwise gradient of CH2Cl2-acetone (100:0, 98:2, 90:10, 80:20, 0:100) to obtain 150 fractions collected in 18-mL vials. The fractions were analyzed and combined based on their TLC profile to yield 13 fractions (D1–D13). Fraction D10 (136.7 mg) was purified by HPLC (normal phase, gradient from 20% to 100% of EtOAc in hexanes at 1 mL/min, for 75 min, followed by 5 min of 100% EtOAc) to yield uprolides O (2) (1.3 mg, tR 53.6 min) and N (1) (1.3 mg, tR 76.4 min). Fraction D7 (647.1 mg) was fractionated using column chromatography on silica gel with a gradient of CH2Cl2–acetone (100:0, 97:3, 95:5, 91:9, 80:20, 0:100) to yield 12 subfractions (I-XII). Subfraction III was purified by HPLC (normal phase, gradient from 0% to 100% of EtOAc in hexanes at 1 mL/min, in 120 min) to yield dolabellane 4 (1.3 mg, tR 48.1 min). Fraction E (300 mg) was purified by HPLC (normal phase, isocratic gradient 46:54% Hx-EtOAc, at 1 mL/min) to yield uprolide O (7.6 mg, tR 35 min) and uprolide P (3) (5.0 mg, tR 31.8 min).
3.4. Compound Characterization
Uprolide N (1): colorless glassy solid; −26.0° (0.8 c, CHCl3); UV (MeOH) λmax 241 nm (ε 7711); IR 3533, 3093, 1764, 1664, 1377, 1262 cm−1; for 1H- and 13C-NMR data see Table 1; HRESITOF-MS m/z 347.1871 [M + H]+ (calcd for C20H27O5, 347.1853).
Uprolide O (2): colorless glassy solid; −23.3° (0.45 c, CHCl3); UV (MeOH) λmax 243 nm (ε 9926); IR 3510, 3127, 1766, 1664, 1461, 1377, 1251 cm−1; for 1H- and 13C-NMR data see Table 1; HRESITOF-MS m/z 385.1615 [M + Na]+ (calcd for C20H26O6Na, 385.1622).
Uprolide P (3): colorless glassy solid; −32.6° (0.31 c, CHCl3); UV (MeOH) λmax 242 nm (ε 10014); IR 3514, 3162, 1763, 1666, 1457, 1377, 1263 cm−1; for 1H- and 13C-NMR data see Table 1; HRESITOF-MS m/z 385.1620 [M + Na]+ (calcd for C20H26O6Na, 385.1622).
Dolabellane (4) [17]: colorless glassy solid, 1H-NMR (CDCl3, 400 MHz) 5.42 (1H, dd, J = 11.6. 11.8 Hz, H-3), 2.91 (1H, d, J = 12.6 Hz, H-7), 2.68 (1H, d, J = 12.6, H-11), 2.40 (1H, br d, J = 12.6, H-14), 2.26 (3H, s, H3-20), 2.16 (1H, dd, J = 11.6, 11.8, H-2), 2.12 (1H, d, J = 18.6, H-14), 2.04 (1H, br m, H-5), 1.93 (3H, s, H3-19), 1.68 (1H, m, H-6), 1.63 (1H, dd, J = 11.8, 4.7, H-2), 1.57 (3H, s, H3-16), 1.50 (1H, m, H-6), 1.38 (1H, m, H-5), 1.36 (3H, s, H3-17), 1.19 (3H, s, H3-15) . 13C-NMR (CDCl3, 100 MHz) 206.5 (C=O, C-13), 149.2 (C, C-18), 13 7.7 (C, C-4), 136.0 (C, C-12), 124.7 (CH, C-3), 65.8 (CH, C-7), 60.5 (C, C-8), 54.4 (CH2, C-14), 42.1 (CH, C-11), 41.1 (C, C-1), 39.8 (CH2, C-2), 38.0 (CH2, C-9), 36.9 (CH2, C-5), 27.5 (CH2, C-6), 25.0 (CH3, C-19), 23.4 (CH3, C-15), 22.9 (CH2, C-10), 21.8 (CH3, C-20), 17.7 (CH3, C-17), 15.7 (CH3, C-16); APCI-MS m/z 303.25 [M + H]+; -LRAPCI-MS m/z 303.2255 [M + H]+ (calcd for C20H31O2, 303.2324).
3.5. Molecular Modeling Studies
A conformational search was performed using the default protocol for macrocycle sampling in Macromodel. This method uses a hybrid search method that employs 5000 cycles of MD-simulated annealing followed by 5000 cycles of LLMOD (large-scale low mode) search steps. The simulated annealing stage produces a set of nonredundant candidate structures that are used as seeds for the conformational search. The torsion sampling option called “enhanced” was selected. Each conformation was minimized using a MMFF94 force field, and no energy cutoff was applied to discard unreasonable conformations. The structural drawings were produced in Spartan08 (Wavefunction, Inc., Irvine, CA, USA).
3.6. Cell Culture and Cytokines Determination
The peritoneal macrophages from C57B1/6 mice were obtained five days after an i.p. instillation of 2 mL of thioglycollate 3%, through peritoneal washing with chilled RPMI. The cells were seeded in RPMI with 10% FCS at 2 × 105 cells/well in 96-well plates. The plates were incubated for 2 h at 37 °C under 5% CO2 atmosphere. The non-adherent cells were removed by washing, and the adherent cells were stimulated with 100 ng/mL of LPS with or without different concentrations of compounds 1, 2 or 3 (14, 7, 3.5 or 1.75 μM). The supernatants were collected 6 h after the stimulus, and the concentrations of TNF and IL-6 were determined by ELISA (DuoSet kit, R & D System, Inc. Minneapolis, MN, USA), according to the manufacturer′s protocol.
3.7. Cytotoxicity Assay
After the removing the supernatants, 100 μL of MTT (Sigma) (0.5 mg/mL) in RPMI was added to each well, and the cells were incubated overnight at 37 °C. The MTT is a water soluble tetrazolium salt, which is reduced by the activity of succinate dehydrogenase of living cells mitochondria to an insoluble purple product (formazan crystals). The supernatants were removed, and the formazan crystals were dissolved in 100 μL of 0.04 M HCl in isopropanol. The color was analyzed at 570 nm on an ELISA plate reader. The percentage of viable cells was calculated as % viability = (OD sample/OD control) × 100%. The non-stimulated cells cultured in medium plus 10% FCS represent the control. As compounds were diluted in DMSO, we determined the sensitivity of macrophages to several concentrations of this diluent (2%, 1%, 0.5%, 0.25% and 0.125%). Only concentrations above 2% showed significant toxicity. Therefore, we performed the rest of the experiments in the presence of 0.5% DMSO. At this concentration the cell viability was 99.1% ± 9.0% (mean ± SD).
3.8. Statistical Analysis
The results were analyzed using the statistical software package GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA). The statistical analysis was performed using a one-way ANOVA followed by Dunnet’s multiple comparisons post-test. The differences between groups were considered significant when p < 0.05. The 50% inhibitory concentration (IC50) was calculated by adjusting a sigmoidal dose-response curve following the standard procedure in GraphPad Prism 5.
4. Conclusions
In this work, we isolated three new uprolides N-P (1–3); these compounds are the first members of this family that have shown anti-inflammatory activity, indicating that this group of compounds should be further explored as a source of new anti-inflammatory lead compounds.
Acknowledgments
We gratefully acknowledge the Government of Panama (Ministerio de Ambiente) for granting permission to collect the coral used in this study. This work was partially supported by the National Secretariat for Science and Technology of Panama (SENACYT, grant numbers COL08-061 and COL09-047), the Fogarty International Center’s International Cooperative Biodiversity Groups program (grant number TW006634), and the Global Environmental Fund (GEF ID 4780–UNDP 81860). We thank Carlos Guevara for his assistance during the collections trips. Daniel Torres-Mendoza and Yisett González thank SENACYT-IFARHU for financial support.
Supplementary Materials
NMR, MS and IR spectra for uprolides N (1), O (2) and P (3). The following are available online at http://www.mdpi.com/1420-3049/21/6/819/s1.
Author Contributions
D.T.M. designed and performed the experiments, analyzed the data and wrote the paper. Y.G. performed the experiments and analyzed data. J.F.G.R. designed, performed and analyzed some NMR experiments. H.M.G collected and identified the coral samples and wrote the paper. J.L.L.P. performed the molecular modeling calculations and wrote the paper. P.L.F and M.G. and W.H.G. designed experiments, analyzed the data and wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds 1–3 are available from the authors.
References
- 1.Berrue F., Kerr R.G. Diterpenes from gorgonian corals. Nat. Prod. Rep. 2009;26:681–710. doi: 10.1039/b821918b. [DOI] [PubMed] [Google Scholar]
- 2.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H.G., Prinsep M.R. Marine Natural Products. Nat. Prod. Rep. 2016;33:382–431. doi: 10.1039/C5NP00156K. [DOI] [PubMed] [Google Scholar]
- 3.Sammarco P.W., Coll J.C. Chemical adaptations in the Octocorallia: Evolutionary considerations. Mar. Ecol. Prog. Ser. 1992;88:93–104. doi: 10.3354/meps088093. [DOI] [Google Scholar]
- 4.Weinheimer A., Middlebrook R.E., Bledsoe J.O., Marsico W.E., Karns T.K.B. Eunicin, an oxa-bridged cembranolide of marine origin. Chem. Comm. 1968:384–385. doi: 10.1039/c19680000384. [DOI] [Google Scholar]
- 5.Rodríguez A.D., Soto J.J., Piña I.C. Uprolides D-G, 2. A rare family of 4,7-oxa-bridged cembranolides from the Caribbean gorgonian Eunicea mammosa. J. Nat. Prod. 1995;58:1209–1216. doi: 10.1021/np50122a008. [DOI] [PubMed] [Google Scholar]
- 6.Rodríguez A.D., Piña I.C., Soto J.J., Rojas D.R., Barnes C.L. Isolation and structure of the uprolides. I. Thirteen new cytotoxic cembranolides from the Caribbean gorgonian Eunicea mammosa. Can. J. Chem. 1995;73:643–654. doi: 10.1139/v95-083. [DOI] [Google Scholar]
- 7.Rodríguez A.D., Acosta A.L. New Cembranolides from the Gorgonian Eunicea succinea. J. Nat. Prod. 1998;61:40–45. doi: 10.1021/np970354c. [DOI] [PubMed] [Google Scholar]
- 8.Shi Y-P., Rodríguez A.D., Barnes C.L., Sánchez J.A., Raptis R.G., Baran P. New Terpenoid Constituents from Eunicea pinta. J. Nat. Prod. 2002;65:1232–1241. doi: 10.1021/np0200820. [DOI] [PubMed] [Google Scholar]
- 9.Rodríguez A.D., Dhasmana H. Further Bioactive Cembranolide Diterpenes from the Gorgonian Eunicea succinea. J. Nat. Prod. 1993;56:564–570. doi: 10.1021/np50094a017. [DOI] [PubMed] [Google Scholar]
- 10.Wei X., Rodríguez A.D., Baran P., Raptis R.G., Sánchez J.A., Ortega-Barría E., González J. Antiplasmodial cembradiene diterpenoids from a Southwestern Caribbean gorgonian octocoral of the genus Eunicea. Tetrahedron. 2004;60:11813–11819. doi: 10.1016/j.tet.2004.09.108. [DOI] [Google Scholar]
- 11.Chang C.Y., Ciereszko L.S., Hossain M.B., van Der Helm D. Structure of Peunicin. Acta Cryst. 1980;B36:731–733. doi: 10.1107/S0567740880004323. [DOI] [Google Scholar]
- 12.Kolossváry I., Guida W.C. Low Mode Search. An Efficient, Automated Computational Method for Conformational Analysis: Application to Cyclic and Acyclic Alkanes and Cyclic Peptides. J. Am. Chem. Soc. 1996;118:5011–5019. doi: 10.1021/ja952478m. [DOI] [Google Scholar]
- 13.Mohamadi F., Richards N.G.J., Guida W.C., Liskamp R., Lipton M., Caufield C., Chang G., Hendrickson T., Still W.C. MacroModel-An Integrated Software System for Modeling Organic and Bioorganic Molecules Using Molecular Mechanics. J. Comput. Chem. 1990;11:440–467. doi: 10.1002/jcc.540110405. [DOI] [Google Scholar]
- 14.Kawai T., Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 15.Mariottini G.L., Pane L. Cytotoxic and Cytolytic Cnidarian Venoms. A Review on Health Implications and Possible Therapeutic applications. Toxins. 2014;6:108–151. doi: 10.3390/toxins6010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin W.-Y., Lu Y., Chen B.-W., Huang C-Y., Su J-H., Wen Z-H., Dai C.-F., Kuo Y.-H., Sheu J.-H. Sarcocrassocolides M-O, Bioactive Cembranoids from the Dongsha Atoll Soft Coral Sarcophyton crassocaule. Mar. Drugs. 2012;10:617–626. doi: 10.3390/md10030617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Look S.A., Fenical W. New Bicyclic Diterpenoids from the Caribbean Gorgonian Octocoral Eunicea calyculata. J. Org. Chem. 1982;47:4129–4134. doi: 10.1021/jo00142a024. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.