Abstract
Bis-naphtho-γ-pyrones (BNPs) are an important group of aromatic polyketides derived from fungi, and asperpyrone-type BNPs are produced primarily by Aspergillus species. The fungal strain Aspergillus niger SCSIO Jcsw6F30, isolated from a marine alga, Sargassum sp., and identified according to its morphological traits and the internal transcribed spacer (ITS) region sequence, was studied for BNPs secondary metabolisms. After HPLC/MS analysis of crude extract of the fermentation broth, 11 asperpyrone-type BNPs were obtained directly and quickly by chromatographic separation in the extract, and those isolated asperpyrone-type BNPs were structurally identified by NMR and MS analyses. All of the BNPs showed weak cytotoxicities against 10 human tumor cells (IC50 > 30 μM). However, three of them, aurasperone F (3), aurasperone C (6) and asperpyrone A (8), exhibited obvious COX-2–inhibitory activities, with the IC50 values being 11.1, 4.2, and 6.4 μM, respectively. This is the first time the COX-2–inhibitory activities of BNPs have been reported.
Keywords: bis-naphtho-γ-pyrones, Aspergillus niger, COX-2–inhibitory, HPLC/MS
1. Introduction
Endophytic fungi from various marine algal hosts are increasingly being considered as sources of pharmaceutical bioactive compounds, especially Aspergillus and Penicillium, as the most commonly isolated fungal endophytes [1]. Aspergillus niger, probably the most common of the black aspergilla, exhibits a remarkably productive metabolism, which has made it one of the most important microorganisms used for industrial fermentations [2].
Bis-naphtho-γ-pyrones (BNPs), also called dimeric naphtho-γ-pyrones or bis-naphthopyran-4-ones, are an important group of aromatic polyketides derived from fungi. They have a variety of biological activities including antioxidant, antitumor, antimicrobial, and tyrosine kinase and HIV-1 integrase inhibition properties, demonstrating their potential applications in medicine and agriculture. About 60 BNPs have been reported from filamentous fungi in the past few decades, such as Aspergillus sp. and Fusarium sp. [3]. BNPs can be divided into three types, named chaetochromin, asperpyrone, and nigerone types, according to the diaryl bond connection. It is interesting that both asperpyrone and nigerone types of BNPs are produced primarily by Aspergillus species, where chaetochromin types are not distributed [4].
Cyclooxygenase (COX) enzymes are the pharmacological targets of nonsteroidal anti-inflammatory drugs (NSAIDs). COX-2, as a well-established target, is an inducible enzyme in which expression is activated by cytokines, mitogens, endotoxins and tumor promoters. The anti-inflammatory and analgesic properties of traditional NSAIDs are primarily due to the inhibition of COX-2 [5].
During our continuous studies of screening for COX-2 inhibitors in marine-derived fungi, BNPs, as the main secondary metabolites of a marine alga-derived Aspergillus fungus, showed obvious COX-2–inhibitory activities. In the present paper, 11 BNPs (1–11) (Figure 1) were obtained quickly by chromatographic separation, in the fermentation broth of the strain Aspergillus niger SCSIO Jcsw6F30, guided by HPLC/MS analysis. Those asperpyrone-type BNPs were structurally identified by NMR and MS analyses and tested for their cytotoxic activities against several human tumor cells and for their COX-2–inhibitory activities.
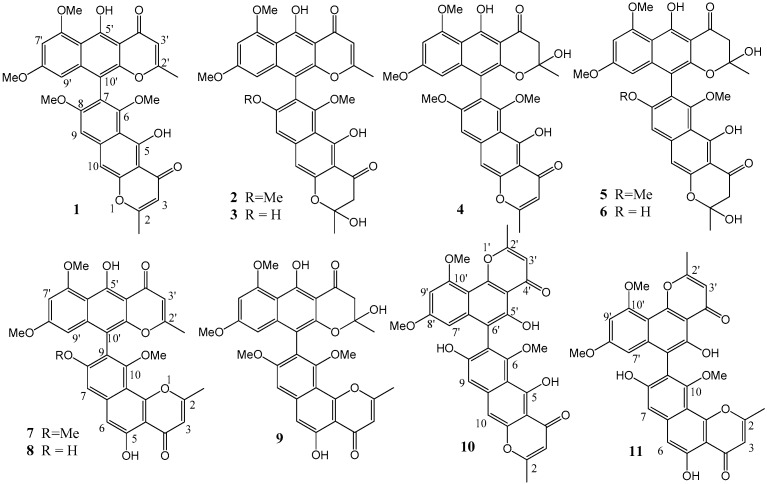
Figure 1.
Structures of compounds 1–11.
2. Results
2.1. Characterization and Identification of Isolated Strain SCSIO Jcsw6F30
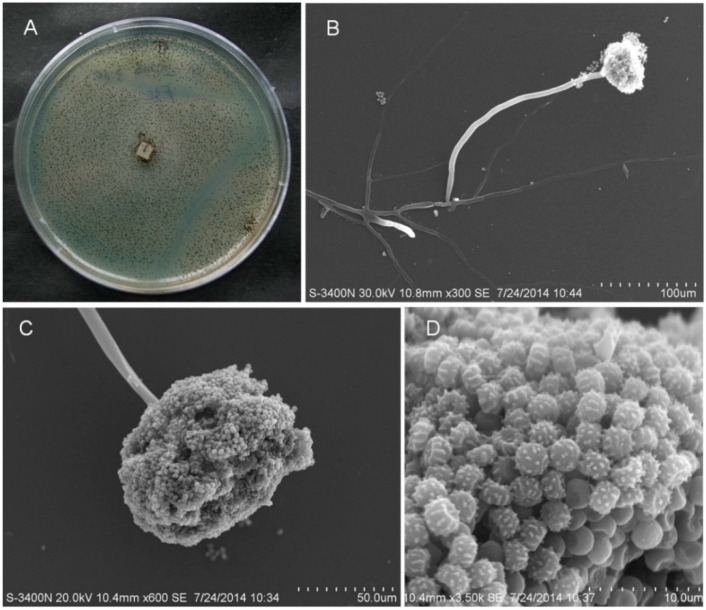
Fungal strain SCSIO Jcsw6F30 was isolated and purified from a marine alga, Scagassum sp., After 7 days of growth on Medium B agar medium at 25 °C; colonies were about 90 mm in diameter, and showed good sporulation. The spores were black, whereas the mycelia were white (Figure 2A).
Figure 2.
Colony appearance and micromorphology of A. niger SCSIO Jcsw6F30. (A) Colony appearance after seven days at 25 °C (MB); (B,C) Conidiophores after seven days at 25 °C under SEM; (D) Conidia as seen using SEM.
Scanning electron microscopy (SEM) revealed that the mycelia were composed of branched, septate, smooth-walled hyphae 2.6 μm to 3.7 μm wide (mean = 3.4 μm). Conidial heads were radiated and subglobose. Stipes were 243 μm to 485 μm long (mean = 364 μm), were 5.9 μm to 9.2 μm wide (mean = 7.1 μm), and had smooth walls. Conidia were black and wheel-shaped when mature, measuring 1.5–2.4 (mean = 1.9) × 2.8–3.5 (mean = 3.1) µm, and smooth or roughened (Figure 2B–D). A teleomorphic state was not observed.
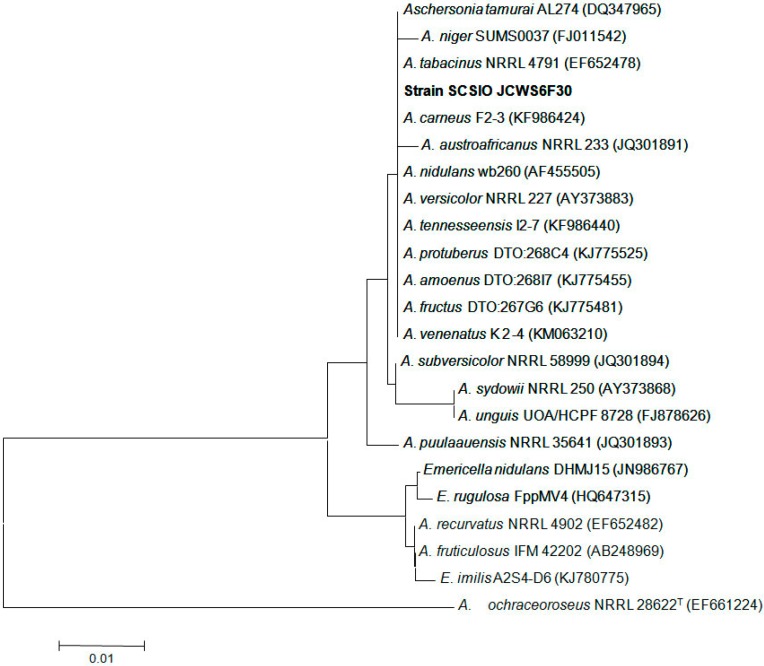
The ITS1-5.8S-ITS2 sequence region (521 basepairs, accession number KT119567) of strain SCSIO Jcsw6F30 was amplified by PCR and sequenced. A phylogenetic tree was constructed, using the neighbor-joining method based on the similarity of a 461 bp consensus length of the ITS1-5.8S-ITS2 sequence (Figure 3). Strain SCSIO Jcsw6F30 was found to belong to a clade related to A. cameus F2-3, A. tabacinus NRRL 4791 and A. niger SUMS0037 et al. in the tree, with sequence identities of 99.8%, 99.6% and 99.2%, respectively. The properties of the culture and the morphology of SCSIO Jcsw6F30 were consistent with those of A. niger as previously reported [6]. The ITS phylogenetic analyses confirmed that the fungus strain SCSIO Jcsw6F30 belonged to A. niger, and was designated as A. niger.
Figure 3.
The neighbor-joining tree based on ITS1-5.8S-ITS2 sequences, showing the phylogenetic relationship between strain SCSIO Jcsw6F30 and related species. GenBank accession numbers are given in parentheses. Bar: 1% sequence divergence.
2.2. HPLC/MS Analysis of Crude Extract of the Fermentation Broth
Naphtho-γ-pyrones present very characteristic UV/vis spectra, two strong absorbance peaks at 215–230 nm, 270–285 nm, and two weak absorbance peaks at 330–340, 395–410 nm, because of their fully conjugated system. The absorption spectra mainly depend on the isomeric form and the polymerization degree [2]. Moreover, monomeric or dimeric naphtho-γ-pyrones could be determined by MS data.
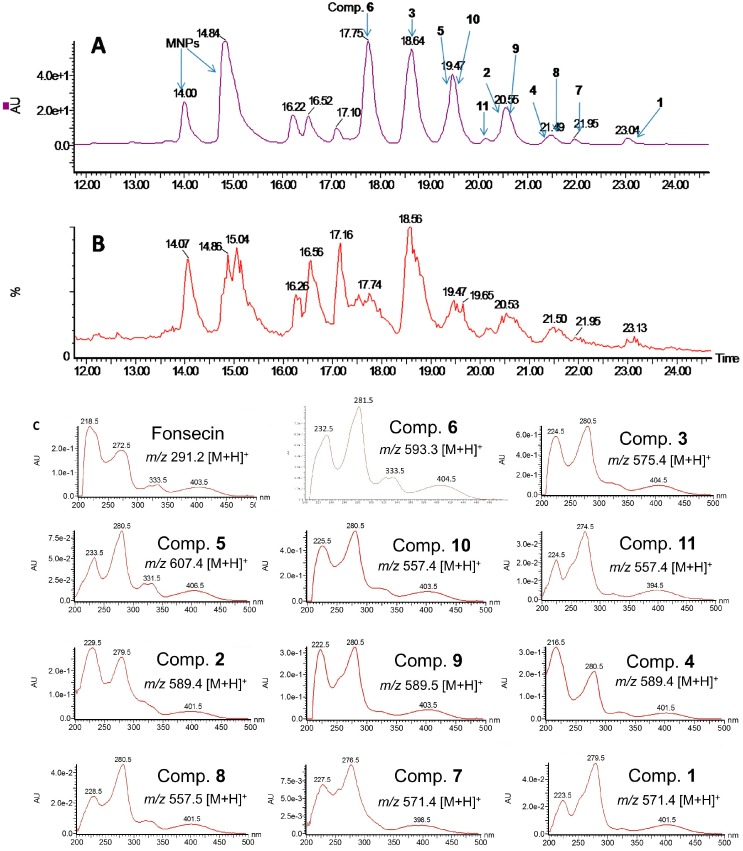
The HPLC/MS data showed at least 11 BNPs signals (Rt 17.5–23.5 min), according to the characteristic UV/vis spectra and molecular ion peaks (m/z 557–607), although their exact structures remained unknown (Figure 4). For example, the UV spectrum [232.5 (71%), 281.5 (100%), 333.5 (23%), 404.5 (17%)] and molecular ion peaks (m/z 593.3 [M + H]+, 615.5 [M + Na]+) of the HPLC peak at Rt 17.75 min suggest it is a BNP compound with the molecular formula C31H28O12. The HPLC peak at Rt 14.00 min showed a similar UV spectrum; however, the molecular ion peaks (m/z 291.2 [M + H]+) suggest they are monomeric naphtho-γ-pyrones (MNPs) (Figure 4). The molecular formulas of 11 BNPs were suggested to be C32H26O10 (1), C32H28O11 (2), C31H26O11 (3), C32H28O11 (4), C32H30O12 (5), C31H28O12 (6), C32H26O10 (7), C31H24O10 (8), C32H28O11 (9), C31H24O10 (10), and C31H24O10 (11), respectively, according to the molecular weight and the molecular rules of BNPs.
Figure 4.
HPLC/MS data of the extract of the fermentation broth. (A) Main part of the HPLC (diode array) chromatogram of the extract; (B) Main part of the total ion chromatogram (TIC) of the extract; (C) All of the 11 BNPs (1–11) had similar UV absorption spectra (UV spectra are placed in order of retention time in chromatogram A), together with their molecular ion given by MS.
2.3. Structural Elucidation
The 1H-NMR spectra of the isolated BNPs were obtained on a Bruker AVANCE-500 spectrometer. The 1H-NMR characteristics of those BNPs showed they were asperpyrone-type BNPs with different linkages. The diagnostic signals at δH 14.86 (s) and 15.27 (s) for the two phenolic OH groups (5, 5’-OH) in its 1H-NMR spectrum suggested that compound 1 was linked by two linear monomeric aromatic polyketide moieties via C-7/C-10’, like the linkages of compounds 2–6. Compound 7 was suggested to have C-9/C-10’ linkages with angular and linear monomeric moieties, according to the two phenolic OH signals at δH 12.86 (s) and 15.27 (s), as well as compounds 8–9 [7]. Compounds 10 and 11 were considered as C-7/C-6’ and C-9/C-6’ linkages because of their different chemical shift of two phenolic OH groups (δH 14.80 and 13.44 of 10, δH 12.74 and 13.47 of 11) [8]. The AB system of the methylene protons (such as δH 3.03 and 3.07, J = 17.0 Hz, in 2) in some 1H-NMR spectra suggested that the C2/C3 or C2’/C3’ double bonds might be hydrogenated in those compounds (compounds 2–6 and 9) [9].
Compounds 1–11 were identified as aurasperone A (1) [7], fonsecinone D (2) [10], aurasperone F (3) [11], fonsecinone B (4) [12], aurasperone B (5) [10], aurasperone C (6) [11], fonsecinone A (7) [7], asperpyrone A (8) [13], fonsecinone C (9) [12], asperpyrone D (10) [8], asperpyrone E (11) [14] by molecular formula given by LC/MS and comparison with their 1H NMR (Supplementary Materials Table S1) with those reported.
2.4. Bioactivities
These 11 BNPs displayed very weak cytotoxicities (IC50 > 30 μM) against 10 human cancer cell lines (K562, A549, Du145, H1975, MCF-7, Huh-7, HL7702, HL60, HeLa, and Molt-4). Aurasperone F (3) was showed to be the relative strongest cytotoxic BNP compound among them, with the best inhibitory rates of 38.8%, 41.0%, 44.9%, 46.6%, and 49.3% against HeLa, MCF-7, Molt-4, Huh-7, and H1975, respectively, at the concentration of 30 μM. However, three of these BNPs, aurasperone F (3), aurasperone C (6), and asperpyrone A (8), with a C-8 phenolic OH group in the structure, exhibited obvious COX-2–inhibitory activities, with the IC50 values being 11.1, 4.2, and 6.4 μM, respectively (Supplementary Materials Table S2).
3. Discussion
BNPs, known as mycotoxins, have been reported with a variety of biological activities including antioxidant, antitumor, antimicrobial, and tyrosine kinase and HIV-1 integrase inhibition properties. Although the activities of three BNPs are much weaker than that of nonsteroidal anti-inflammatory drugs (NSAIDs) Celecoxib, a highly selective COX-2 inhibitor, this is the first time the COX-2–inhibitory activities of BNPs have been reported, presenting a new type of natural product as potential COX-2 inhibitors. Three BNPs with obvious COX-2–inhibitory activities are characterized as C-8 free hydroxyl group and C-10’ (linear naphtho-γ-pyrone) linkages. However, the complete structure-activity relationship is yet to be determined in a future study. The weak cytotoxicities reported in this paper also contribute to these BNPs being potential anti-inflammatory agents. BNP compounds should be screened comprehensively or structurally modified to be optimized as COX-2 inhibitors or anti-inflammatory agents.
BNPs could be produced in large amounts by industrial fungi, such as Aspergillus niger [2]. Aspergillus niger SCSIO Jcsw6F30, obtained from marine alga Sargassum sp., could be considered as a good industrial fungus to produce asperpyrone-type BNPs, according to our research. The LC/MS analysis, together with the quick isolation and structural identification of BNPs by 1H-NMR described in this paper, also shows benefits of the industrial application of asperpyrone-type BNPs.
4. Materials and Methods
4.1. Cultural and Morphological Properties of Strain SCSIO Jcsw6F30
The fungal strain SCSIO Jcsw6F30 was isolated from a marine alga Scagassum sp. collected near the Yongxing Island, South China Sea (in July 2012), and grown on Medium B agar at 25 °C [15]. This strain was stored on Medium B agar slants at 4 °C and then deposited at the Key Laboratory of Tropical Marine Bio-Resources and Ecology, CAS (Guangzhou, China).
The cultural properties of strain SCSIO Jcsw6F30 were described after culturing on Medium B agar medium at 25 °C for 7 days. The morphological features of the spores and mycelia in the 7 d cultures grown on Medium B medium (consisting of 15 g malt extract, 10 g sea salt, 15 g agar, 1000 mL distilled water, pH 7.4–7.8) were examined. The samples were observed with a Hitachi S-3400N scanning electron microscope using a previously described cover technique [16,17].
4.2. ITS Region Sequence and Phylogenetic Analysis
The mycelia of strain SCSIO Jcsw6F30 cultured in Sabouraud’s Dextrose Broth (consisting of 40 g dextrose, 10 g peptone, 2.5 g NaCl, and 1000 mL distilled water, pH 5.6) were sampled and powdered in a mixer mill after liquid nitrogen was added. DNA was isolated through the Hpure Fungal DNA Kit (Genebase Bioscience Co., Guangzhou, China) according to the manufacturer’s protocol. The ITS region of strain SCSIO Jcsw6F30 was amplified by polymerase chain reaction with the primer pair ITS1–ITS4. The amplified product was purified with a TIANgel mini purification kit (TianGen Biotech, Beijing, China). Pure PCR product was submitted for sequencing together with the primer ITS1 to a commercial service (Shanghai Majorbio Bio-pharm Technology Co., Ltd., Shanghai, China). The derived ITS region sequence was compared against the GenBank database (NCBI) through BLAST-Algorithmus. Similarity analysis was performed using ClustalW program. The phylogenetic tree of strain SCSIO Jcsw6F30 was constructed using neighbor-joining method, as previously described [18]. Aspergillus ochraceoroseus NRRL 28622T was used as an outgroup. The nucleotide sequence of the ITS region reported in this article was assigned the GenBank accession number KT119567.
4.3. Fermentation and Extraction
The strain was cultured on Medium B agar plates and then incubated for seven days previously described [19]. Seed medium (consisting of 6.25 g maltose, 6.25 g malt extract, 1 g yeast extract, 6.25 g peptone, 1.25 g potassium dihydrogen phosphate, and 1000 mL distilled water, pH 7.0) in 500 mL Erlenmeyer flasks was inoculated with strain Jcsw6F30 and then incubated at 25 °C for 3 days on a rotating shaker (120 rpm). Production medium (the same as the seed medium) in 500 mL flasks was inoculated with 10% seed solution. The flasks were incubated at 28 °C statically. After 7 days, broth from 100 flasks (15 L) was harvested to isolate substances. The broth (15 L) was extracted with 10 L ethyl acetate stirring three times for 30 min.
4.4. HPLC/MS Analysis and Isolation of the Compounds
First 10 Mg crude extract was prepared in 1 mL acetonitrile for HPLC/MS analysis. Two microliters of the extract were subjected to HPLC (a WATERS e 2695 separation module with a WATERS 2996 photodiode array detector). A reverse phase C18 column (2.1 × 150 mm, 3.5 μm, sunfire™, Waters) was employed. The analysis condition was set to a flow rate of 0.3 mL·min−1, with a water:acetonitrile step gradient used in the separation method and consisting of 5% acetonitrile for 2 min, gradient to 100% acetonitrile for 25 min, 100% acetonitrile for 5 min, gradient to 5% for 5 min and 5% acetonitrile for 3 min.
MS data were obtained with Waters Quattro ZQ mass spectrometer coupled to the HPLC system. MS parameters: ESI, alternative ion polarity mode, capillary voltage, 3.5 kv, Cone voltage, 45 kv, Desolvation temperature, 300 °C, Desolvation gas flow, 500 L/h, mode, full scan: m/z range, 100–1500.
The crude extract was subjected to a silica gel CC and was separated by a linear gradient of petroleum ether (60–90 °C):EtOAc (50:0, 50:1, 20:1, 10:1, 5:1, 2:1, 1:1, and 0:1) to yield eight fractions (fr. I–fr. VIII). HPLC/MS analyses showed those BNPs were distributed in two fractions: fr. VI–VII. Fractions VI–VII were dissolved in methanol and chromatographed over semi-preparative HPLC (Sunfire, Prep C18 OBD, 10 mm × 250 mm, 5 µm, 7.5 mL/min) with a gradient solvent system from 20% to 60% CH3CN over 30 min, respectively. Eleven BNPs were purified and obtained by repeated semi-preparative HPLC, guided by the HPLC/MS information.
4.5. Bioassays
A previously described bioassay test was used to evaluate the cytotoxicities of those BNPs against 10 human cancer cell lines (K562, A549, Du145, H1975, MCF-7, Huh-7, HL7702, HL60, HeLa, and Molt-4) [18]. The isolated compounds were tested for COX-2 inhibitory activity using the COX (ovine) inhibitor screening kit according to the manufacturer’s instructions, as previously described [20]. Celecoxib (Sigma, St. Louis, MO, USA) was used as the positive control with an IC50 value of 11.3 nM.
Acknowledgments
This work was supported by grants from the Pearl River S&T Nova Program of Guangzhou (201610010017), the National Natural Science Foundation of China (41376162, 21202080, 41406187), Funds for the Development of Science and Technology of Guangdong Province (2016A020222009), the Hubei Agricultural Sciences and Technology Innovation Center (2012-620-008-002), the Guangzhou Scientific Research Project (2014J4100212), and the Special Support Project for Outstanding Young Scholars of Guangdong Province (Zhou X.).
Abbreviations
The following abbreviations are used in this manuscript:
| BNPs | bis-naphtho-γ-pyrones |
| COX | cyclooxygenase |
| CC | column chromatography |
| NSAIDs | nonsteroidal anti-inflammatory drugs |
| SEM | scanning electron microscopy |
| HPLC/MS | high performance liquid chromatography/mass spectrometer |
| NMR | nuclear magnetic resonance |
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/21/7/941/s1.
Author Contributions
W. F. and J. W. contributed to fermentation, analysis and isolation of the compounds, X. L. contributed to the isolation and identification of the fungus, Y. L. and H. T. contributed to identification of the compounds and analyzed the data, X. Z. conceived and designed the experiments, and wrote the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds 1–11 are available from the authors.
References
- 1.Flewelling A.J., Currie J., Gray C.A., Johnson J.A. Endophytes from marine macroalgae: Promising sources of novel natural products. Curr. Sci. India. 2015;109:88–111. [Google Scholar]
- 2.Nielsen K.F., Mogensen J.M., Johansen M., Larsen T.O., Frisvad J.C. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Anal. Bioanal. Chem. 2009;395:1225–1242. doi: 10.1007/s00216-009-3081-5. [DOI] [PubMed] [Google Scholar]
- 3.Choque E., El Rayess Y., Raynal J., Mathieu F. Fungal naphtho-gamma-pyronessecondary metabolites of industrial interest. Appl. Microbiol. Biot. 2015;99:1081–1096. doi: 10.1007/s00253-014-6295-1. [DOI] [PubMed] [Google Scholar]
- 4.Lu S.Q., Tian J., Sun W.B., Meng J.J., Wang X.H., Fu X.X., Wang A.L., Lai D.W., Liu Y., Zhou L.G. Bis-naphtho-gamma-pyrones from fungi and their bioactivities. Molecules. 2014;19:7169–7188. doi: 10.3390/molecules19067169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blobaum A.L., Uddin M.J., Felts A.S., Crews B.C., Rouzer C.A., Marnett L.J. The 2’-trifluoromethyl analogue of indomethacin is a potent and selective COX-2 inhibitor. ACS Med. Chem. Lett. 2013;4:486–490. doi: 10.1021/ml400066a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onions A.H.S. Aspergillus niger. CMI Descr. Pathog. Fungi Bact. 1966;94:1–2. [Google Scholar]
- 7.Campos F.R., Barison A., Daolio C., Ferreira A.G., Rodrigues-Fo E. Spectral assignments and reference data—Complete H-1 and C-13 NMR assignments of aurasperone A and fonsecinone A, two bis-naphthopyrones produced by Aspergillus aculeatus. Magn. Reson. Chem. 2005;43:962–965. doi: 10.1002/mrc.1654. [DOI] [PubMed] [Google Scholar]
- 8.Zhan J.X., Gunaherath G.M.K.B., Wijeratne E.M.K., Gunatilaka A.A.L. Asperpyrone D and other metabolites of the plant-associated fungal strain Aspergillus tubingensis. Phytochemistry. 2007;68:368–372. doi: 10.1016/j.phytochem.2006.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Li X.M., Wang B.G. Nigerasperones A-C, new monomeric and dimeric naphtho-gamma-pyrones from a marine alga-derived endophytic fungus Aspergillus niger EN-13. J. Antibiot. 2007;60:204–210. doi: 10.1038/ja.2007.24. [DOI] [PubMed] [Google Scholar]
- 10.Siriwardane A.M.D.A., Kumar N.S., Jayasinghe L., Fujimoto Y. Chemical investigation of metabolites produced by an endophytic Aspergillus sp. isolated from Limonia acidissima. Nat. Prod. Res. 2015;29:1384–1387. doi: 10.1080/14786419.2015.1025230. [DOI] [PubMed] [Google Scholar]
- 11.Bouras N., Mathieu F., Coppel Y., Lebrihi A. Aurasperone F—A new member of the naphtho-gamma-pyrone class isolated from a cultured microfungus, Aspergillus niger C-433. Nat. Prod. Res. 2005;19:653–659. doi: 10.1080/14786410412331286955. [DOI] [PubMed] [Google Scholar]
- 12.Priestap H.A. New naphthopyrones from Aspergillus fonsecaeus. Tetrahedron. 1984;40:3617–3624. doi: 10.1016/S0040-4020(01)88792-5. [DOI] [Google Scholar]
- 13.Akiyama K., Teraguchi S., Hamasaki Y., Mori M., Tatsumi K., Ohnishi K., Hayashi H. New dimeric naphthopyrones from Aspergillus niger. J. Nat. Prod. 2003;66:136–139. doi: 10.1021/np020174p. [DOI] [PubMed] [Google Scholar]
- 14.Li X.B., Xie F., Liu S.S., Li Y., Zhou J.C., Liu Y.Q., Yuan H.Q., Lou H.X. Naphtho-gamma-pyrones from endophyte Aspergillus niger occurring in the liverwort Heteroscyphus tener (STEPH.) SCHIFFN. Chem. Biodivers. 2013;10:1193–1201. doi: 10.1002/cbdv.201300042. [DOI] [PubMed] [Google Scholar]
- 15.Kjer J., Debbab A., Aly A.H., Proksch P. Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products. Nat. Protoc. 2010;5:479–490. doi: 10.1038/nprot.2009.233. [DOI] [PubMed] [Google Scholar]
- 16.Kawato M., Shinobu R. On Streptomyces herbaricolor sp. nov., supplement: A single technique for microscopical observation. Mem. Osaka Unit Lib. Arts Educ. B Nat. Sci. 1959;8:114–119. [Google Scholar]
- 17.Zhou Z.H., Liu Z.H., Qian Y.D., Kim S.B., Goodfellow M. Saccharopolyspora spinosporotrichia sp. nov., a novel actinomycete from soil. Int. J. Syst. Bacteriol. 1998;48:53–58. doi: 10.1099/00207713-48-1-53. [DOI] [PubMed] [Google Scholar]
- 18.Fang W., Lin X., Zhou X., Wan J., Lu X., Yang B., Ai W., Lin J., Zhang T., Tu Z., Liu Y. Cytotoxic and antiviral nitrobenzoylses quiterpenoids from the marine-derived fungus Aspergillus ochraceus Jcma1F17. MedChemComm. 2014;5:701–705. doi: 10.1039/c3md00371j. [DOI] [Google Scholar]
- 19.Zhou X., Fang W., Tan S., Lin X., Xun T., Yang B., Liu S., Liu Y. Aspernigrins with anti-HIV-1 activities from the marine-derived fungus Aspergillus niger SCSIO Jcsw6F30. Bioorg. Med. Chem. Lett. 2016;26:361–365. doi: 10.1016/j.bmcl.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 20.Ai W., Lin X.P., Tu Z., Tian X.P., Lu X., Mangaladoss F., Zhong Z.L., Liu Y. Axinelline A, a new COX-2 inhibitor from Streptomyces axinellae SCSIO02208. Nat. Prod. Res. 2014;28:1219–1224. doi: 10.1080/14786419.2014.891204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.