Abstract
Glucosinolates are the biochemical compounds that provide defense to plants against pathogens and herbivores. In this study, the relative expression level of 48 glucosinolate biosynthesis genes was explored in four morphologically-different cabbage inbred lines by qPCR analysis. The content of aliphatic and indolic glucosinolate molecules present in those cabbage lines was also estimated by HPLC analysis. The possible association between glucosinolate accumulation and related gene expression level was explored by principal component analysis (PCA). The genotype-dependent variation in the relative expression level of different aliphatic and indolic glucosinolate biosynthesis genes is the novel result of this study. A total of eight different types of glucosinolates, including five aliphatic and three indolic glucosinolates, was detected in four cabbage lines. Three inbred lines BN3383, BN4059 and BN4072 had no glucoraphanin, sinigrin and gluconapin detected, but the inbred line BN3273 had these three aliphatic glucosinolate compounds. PCA revealed that a higher expression level of ST5b genes and lower expression of GSL-OH was associated with the accumulation of these three aliphatic glucosinolate compounds. PCA further revealed that comparatively higher accumulation of neoglucobrassicin in the inbred line, BN4072, was associated with a high level of expression of MYB34 (Bol017062) and CYP81F1 genes. The Dof1 and IQD1 genes probably trans-activated the genes related to biosynthesis of glucoerucin and methoxyglucobrassicin for their comparatively higher accumulation in the BN4059 and BN4072 lines compared to the other two lines, BN3273 and BN3383. A comparatively higher progoitrin level in BN3273 was probably associated with the higher expression level of the GSL-OH gene. The cabbage inbred line BN3383 accounted for the significantly higher relative expression level for the 12 genes out of 48, but this line had comparatively lower total glucosinolates detected compared to the other three cabbage lines. The reason for the genotypic variation in gene expression and glucosinolate accumulation is a subject of further investigation.
Keywords: glucosinolates, biosynthetic genes, expression analysis, Brassica oleracea var. capitata, genotypic variations
1. Introduction
Cabbage, one the most important vegetable crops throughout the world, is a member of the vegetable species Brassica oleracea that contains a wide variety of glucosinolates [1,2,3,4,5]. The subspecies B. oleracea var. capitata produced 12 different kinds of glucosinolates in its edible organ, including eight types of aliphatic, three types of indolic and one type of aromatic glucosinolates in the Korean cabbage varieties [5]. Cabbage is an important ingredient in many important Korean and Chinese dishes. This subspecies was found to produce glucoerucin, which was absent in the edible organs of kale, kohlrabi and cauliflower [5]. However, the number of glucosinolates present in cabbage was found to be variable in different studies [1,2,4,5]. This is probably because the content and type of glucosinolates in cabbage subspecies varies between growing season [2,6], anthocyanin content and leaf age [6] and also between tissue types: roots, shoots [4,7,8], etc. The content and varieties of glucosinolates also widely varies in the edible organ between different cultivars of this subspecies [1,2,9] probably because of the variation in tissue types. Despite a large number of studies reporting that glucosinolate content and types had intra-subspecific variation in cabbage varieties, the associated gene involved in such variation has not been well elucidated.
The biosynthetic pathways of glucosinolates and the break-down of these biomolecules follow species level variation, especially since these pathways are quite different in the vegetable species B. oleracea compared to those in Arabidopsis and B. rapa. B. oleracea has greater diversity compared to B. rapa and B. napus [10]. B. oleracea has about 84 glucosinolate biosynthesis genes and 22 catabolism-related genes [5,10]. A recent study explored two available web sources, Bolbase and EnesmblPlants, to compile the coding DNA sequences of glucosinolate biosynthesis-related genes and then catalogued reverse-transcript PCR-based semi-quantitative expression profiling of 84 genes in the edible organs of four B. oleracea subspecies, including cabbage leaf samples [5]. The authors reported that the biosynthesis of 12 diversified types of glucosinolates was not dependent on the expression of all 84 genes in the cabbage subspecies. Rather, the expression of one of a few genes (probably the key regulatory genes) in each step of glucosinolate biosynthesis might result in the accumulation of a particular glucosinolate in the edible organs of different B. oleracea subspecies [5]. Glucosinolate biosynthesis generally follows a characteristic three-step process. The process commences with the addition and elongation of side chains composed of methylene groups to the amino acid molecules. The second and third steps are the core structure formation and modification in the side chains of the elongated amino acid chain, respectively. The modification of side chains is accomplished by oxidation, hydroxylation, methoxylation, sulfation and glycosylation, processes that involve different genes [11,12,13]. The initial amino acid precursors of glucosinolate biosynthesis are generally methionine (or alanine, leucine, isoleucine and valine) for aliphatic glucosinolates and tryptophan and phenylalanine (or tyrosine), respectively, for indolic and aromatic glucosinolates (Figure 1; [13,14]).
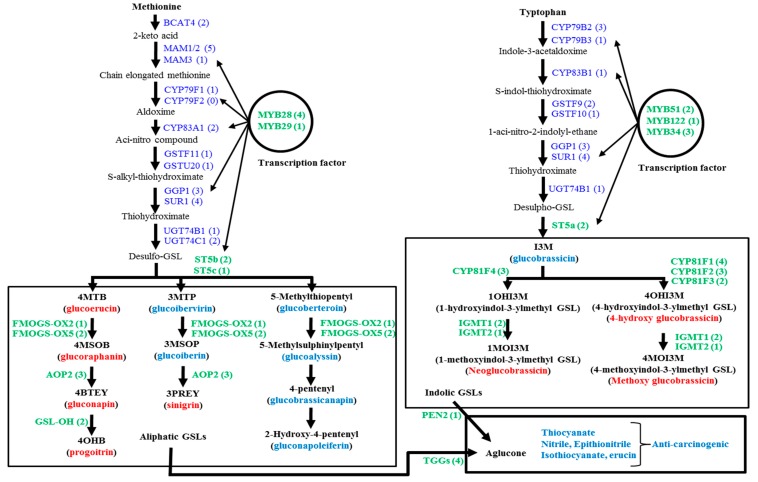
Figure 1.
Metabolic pathways involved in the biosynthesis of aliphatic and indolic glucosinolates (GSL) after Liu et al. [10], along with the related genes of B. oleracea. The GSL molecules measured by HPLC are denoted as red bold (see Table S1 for their chemical structure names). The relative expression level of green bold genes was measured by qPCR. Numbers in parentheses denote the number of genes involved in this process (Adopted from Yi et al. [5]).
In Arabidopsis, the amino acid side chain elongation begins by the action of methylthioalkylmalate synthase (MAM) in association with acid: sodium symporter family protein 5 (BASS5) and branched-chain aminotransferase (BCAT) at the beginning of glucosinolate biosynthesis [15,16,17,18]. The next step is the formation of the core structure, which is a five-step process, including: (i) aldoxime formation by cytochromes P450 of the CYP79 and CYP83 families; (ii) aldoxime oxidation to nitrile oxides or aci-nitro compounds by cytochromes P450 of the CYP83 family; (iii) thiohydroximic acid formation by C-S cleavage; (iv) desulfoglucosinolate formation by S-glucosyltransferase; and (v) the formation of glucosinolates by sulfotransferase [19,20]. The desulfoglucosinolates undergoes secondary modifications to produce particular glucosinolate biomolecules that involve several gene loci, for example: GS-OX, GS-AOP, GS-OH are involved in aliphatic glucosinolate biosynthesis, and CYP81, IGM are involved in indolic glucosinolate biosynthesis (Figure 1). MYB-transcription factor-related genes trans-activate the functions of several genes involved in side chain elongation and core-structure formation (Figure 1; [21,22,23,24,25,26,27]). A few other transcription factor-related genes other than MYB, such as TFL2, IQD1 and Dof1.1, also trans-activate both aliphatic and indolic glucosinolate biosynthesis [28,29,30,31]. In cabbage subspecies, expression profiling of none of those genes was studied to elucidate any intra-subspecies variation in gene expression. The present study therefore was conducted to explore the quantitative expression profiling of glucosinolate biosynthesis in four different cabbage inbred lines with variation in morphological appearance. The glucosinolates present in inbred lines were detected, and their contents were also quantified. An association between the glucosinolate biosynthesis-related gene and the glucosinolate content was explored.
2. Results and Discussion
2.1. Genotypic Variation in the Relative Expression Profiling of Glucosinolate Biosynthesis Genes
This study investigated the relative expression level of a total of 48 genes related to glucosinolate biosynthesis in B. oleracea var. capitata in four inbred lines, BN3273, BN3383, BN4059 and BN4072 (Figure 2) with contrasting morphological variations (Figure 2). The external morphological variations include variation in leaf shape and appearance (Figure 2). Moreover, none of those four inbred lines had any common parents according to Asia Seed Company (Seoul, Korea). The color of the leaves of BN3273 and BN3383 was pale green, and that of BN4059 and BN4072 was green (Figure 2). While calculating the relative expression level of different genes, the inbred line BN3273 was considered as the control. The inbred line BN3383 accounted for a remarkably higher level of relative gene expression for the MYB29 transcription factor-related gene associated with aliphatic glucosinolate biosynthesis compared to the other three lines (Figure 3, Table S2). MYB28 accession Bol036286 recorded the lowest level of gene expression for the same line compared to the other three lines (Figure 3, Table S2). The inbred line BN4072 accounted for a comparatively lower expression level of MYB28 accessions Bol007795 and Bol017019 and MYB29 accession Bol008849 compared to BN4059 (Figure 3, Table S2).
Figure 2.
Genotypic variation in the leaf morphology of middle-aged leaves of four B. oleracea capitata inbred lines. Each column represents the whole plant, the upper- and lower-surface of an individual leaf, respectively. The leaves marked with orange-boxes were sampled. The inbred line BN3273 was the calibrator in the relative expression analysis.
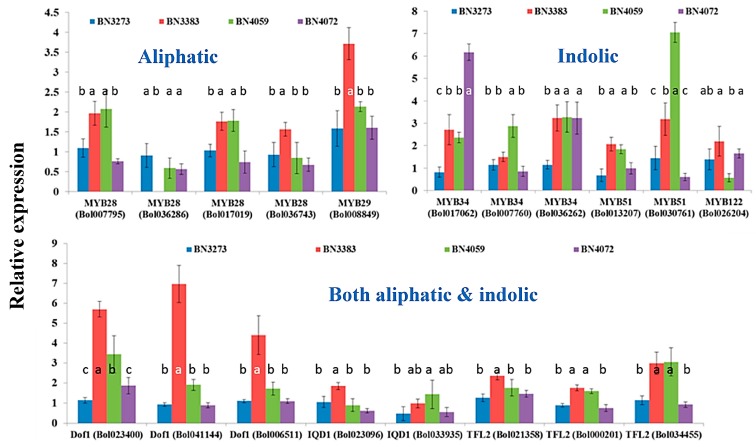
Figure 3.
Relative expression level of transcription factor-related genes in four cabbage inbred lines involved in aliphatic, indolic and both aliphatic and indolic glucosinolate biosynthesis in B. oleracea. Vertical bars indicate the standard deviation of the means of nine observations. Different letters (a, b, c) indicate statistically significant difference between inbred lines.
The inbred line BN4072 accounted for, remarkably, more than a six-fold, higher level of gene expression for the indolic transcription factor-related gene MYB34 accession Bol017062 compared to the control line BN3273 (Figure 3, Table S2). Another line BN4059 accounted for a seven-fold higher expression level compared to the control line BN3273 (Figure 3, Table S2). The same line had about a three-fold higher level of expression for the MYB34 (Bol007760) gene (Figure 3, Table S2). The inbred line BN3383 had a comparatively higher level of expression for three Dof1 genes associated with both aliphatic and indolic glucosinolate biosynthesis compared to the three other lines (Figure 3, Table S2). Another transcription factor-related gene associated with both aliphatic and indolic glucosinolate biosynthesis, TFL2 accession Bol034445, was highly expressed in both BN3383 and BN4059 lines compared to the other two cabbage inbred lines (Figure 3, Table S2).
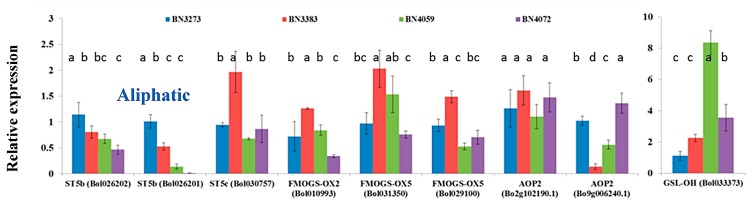
The inbred line BN3383 accounted for a high level of gene expression up to two-fold for the following aliphatic glucosinolate biosynthesis genes: ST5c accession Bol030757, FMOGS-OX2 accession Bol010993, FMOGS-OX5 accessions Bol031350 and Bol029100; compared to the control line BN3273 (Figure 4, Table S2). Inbred lines BN4059 and BN4072 accounted for a strikingly lower expression level of ST5b accession Bol026201 compared to the other two lines, BN3383 and BN3273 (Figure 4, Table S2). The gene AOP2 (Bol9g006240.1) had more than a seven-fold lower level of relative gene expression in the inbred line BN3383 compared to the control line BN3273 (Figure 4, Table S2). Another aliphatic transcription factor-related gene GSL-OH (Bol033373) had more than an eight-fold higher expression level in the inbred line BN4059 (Figure 4, Table S2).
Figure 4.
Relative expression level of aliphatic glucosinolate biosynthesis-related genes in four cabbage inbred lines of B. oleracea. Vertical bars indicate the standard deviation of the means of nine observations. Different letters (a, b, c) indicate statistically significant difference between inbred lines.
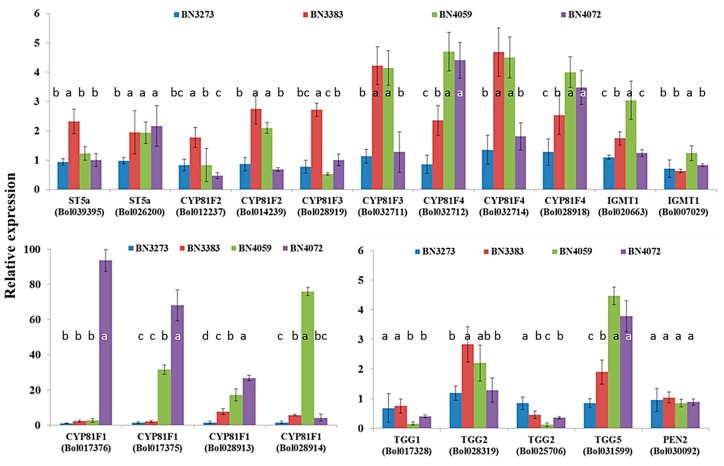
Four indolic biosynthesis-related gene accessions, namely ST5a (Bol039395), CYP81F2 (Bol012237), CYP81F2 (Bol014239) and CYP81F3 (Bol028919), accounted for approximately a two-fold higher level of gene expression for the cabbage line BN3383 compared to the control line BN3273 (Figure 5, Table S2). The inbred line BN4059 coupled with BN3383 accounted for approximately a four-fold higher expression level of the CYP81F3 (Bol028919) and CYP81F4 (Bol032714) genes compared to the two other cabbage lines (Figure 5, Table S2). The relative expression level of CYP81F4 (Bol032712) and CYP81F4 (Bol028918) was significantly higher in the BN4059 and BN4072 lines compared to the other two inbred lines (Figure 5, Table S2). The line BN4072 accounted for more than a 90-fold and a 60-fold higher level of gene expression of CYP81F1 accessions Bol017376 and Bol017375, respectively, compared to the control line BN3273 (Figure 5, Table S2). Another cabbage line, BN4059, accounted for the highest expression level of CYP81F1 (Bol028914) gene, about 80-fold compared to the three other lines (Figure 5). The relative expression level of CYP81F1 accessions Bol017376 and Bol0128913 was comparatively higher in cabbage lines BN4059 and BN4072 compared to BN3273 and BN3383 (Figure 5, Table S2).
Figure 5.
Relative expression level of indolic glucosinolate biosynthesis- and break-down-related genes in four cabbage inbred lines of B. oleracea. Vertical bars indicate the standard deviation of the means of nine observations. Different letters (a, b, c) indicate statistically significant difference between inbred lines.
The glucosinolate break-down-related gene TGG5 (Bol031599) was expressed highly in the inbred lines BN4059 and BN4072 compared to BN3273 and BN3383 (Figure 5, Table S2). The level of expression of two other break-down-related genes, TGG1 (Bol017328) and TGG2 (Bol025706) in the BN4059 and BN4072 lines, was remarkably low compared to the BN3273 and BN3383 lines (Figure 5, Table S2).
2.2. Genotypic Variation in Glucosinolate Contents
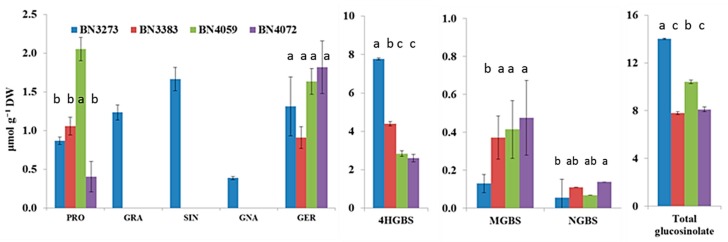
HPLC analysis detected a total of eight types of glucosinolate molecules in four cabbage inbred lines (Figure 6). Those were progoitrin, glucoraphanin, sinigrin, gluconapin, glucoerucin, 4-hydroxy-glucobrassicin (4HGBS), methoxy-glucobrassicin (MGBS) and neoglucobrassicin (NGBS). However, three aliphatic glucosinolates, glucoraphanin, sinigrin and gluconapin, were detected only in the cabbage line BN3273 (Figure 6). The same line also had comparatively higher amounts of 4HGBS and total glucosinolate, but lower MGBS compared to the other three lines (Figure 6, Table S3). Only two other aliphatic glucosinolates, progoitrin and glucoerucin, were detected in all four lines (Figure 6). 4HGBS, MGBS and NGBS are the three indolic glucosinolates that were detected in all four inbred lines (Figure 6). Unlike the relative expression level of a notable number of genes, the cabbage inbred line BN3383 did not have a higher amount of glucosinolates compared to the other three lines (Figure 6). The inbred line BN4059 had the highest content of progoitrin (Figure 6, Table S3), whereas BN4072 had the highest content of NGBS (Figure 6).
Figure 6.
Contents of aliphatic and indolic glucosinolates produced in four inbred lines of cabbage. Vertical bars indicate the standard error of the means of nine observations. The response factors for glucosinolate quantification were obtained from European Economic Community [32], International Standards Organization [33] and American Oil Chemists Society [34]. Different letters (a, b, c) indicate statistically significant difference between inbred lines. PRO, progoitrin; GRA, glucoraphanin; SIN, sinigrin; GNA, gluconapin; GER, glucoerucin; 4HGBS, 4-hydroxy-glucobrassicin; MGBS, methoxy-glucobrassicin and NGBS, neoglucobrassicin.
2.3. Association between Glucosinolate Content and Gene Expression
2.3.1. Association between Aliphatic Glucosinolate Content and Transcription Factor-Related Genes
The association between glucosinolate content and the relative gene expression level was explored through principal component analysis (PCA). In principal component 1 (PC1), the aliphatic transcription factor-related genes and glucosinolate content accounted for a negative contrast (Table 1). The coefficient for MYB genes was positive (except Bol036286), and those for glucoraphanin, sinigrin, gluconapin and glucoerucin content were negative (Table 1). This contrast is largely explained by the significant genotypic variation between the BN3383, BN4059, BN4072 lines and the BN3273 line, as only the BN3273 line contained glucoraphanin, sinigrin and gluconapin (Table 1). PC2 accounted for 23.3% of the data variation and estimated a larger contrast between BN4059, BN4072 and BN3273, BN3383 for a higher content of glucoraphanin, sinigrin and gluconapin, a lower content of glucoerucin and vice versa in association with the expression level of MYB genes (Table 1). The PC3 explaining 15.3% of the data variation accounted for a positive association between higher progoitrin content in line BN4059 and a higher expression level of MYB28 accession Bol007795 (Table 1).
Table 1.
Principal component analysis for aliphatic glucosinolate contents and the relative expression level of transcription factor-related genes in four cabbage lines of B. oleracea. PC, principal component; p, statistical significance; SD, standard deviation.
| Variable | PC1 | PC2 | PC3 |
|---|---|---|---|
| Progoitrin | 0.18 | −0.16 | 0.69 |
| Glucoraphanin | −0.37 | −0.38 | −0.01 |
| Sinigrin | −0.37 | −0.38 | −0.01 |
| Gluconapin | −0.33 | −0.33 | −0.02 |
| Glucoerucin | −0.11 | 0.40 | 0.37 |
| MYB28 (Bol007795) | 0.34 | −0.29 | 0.36 |
| MYB28 (Bol036286) | −0.36 | −0.11 | 0.26 |
| MYB28 (Bol017019) | 0.33 | −0.31 | 0.19 |
| MYB28 (Bol036743) | 0.22 | −0.43 | −0.27 |
| MYB29 (Bol008849) | 0.37 | −0.20 | −0.27 |
| %variation explained | 47.2 | 23.3 | 15.3 |
| p (genotype) | <0.001 | <0.001 | 0.001 |
| Genotype | Mean PC Scores (±SD) | ||
| BN3273 | −2.92 ± 0.43 | −1.31 ± 1.32 | −0.03 ± 0.31 |
| BN3383 | 2.55 ± 0.16 | −0.85 ± 0.27 | −0.99 ± 0.49 |
| BN4059 | 1.24 ± 0.29 | 0.06 ± 0.52 | 1.66 ± 0.90 |
| BN4072 | −0.87 ± 0.16 | 2.01 ± 0.66 | −0.69 ± 0.91 |
2.3.2. Association between Indolic Glucosinolate Content and Transcription Factor-Related Genes
PC1 obtained from a PCA for the indolic glucosinolate content and transcription factor-related genes accounted for 34.3% of the data variation, but the coefficients of this PC were dominated by the higher expression of MYB34 (Bol007760) and MYB51 (Bol030761) genes in BN4059 and BN3383 inbred lines compared to the other two genotypes (Table 2). PC2 accounted for a positive association between NGBS content and the relative expression level of the MYB34 (Bol017062) gene (Table 2). This contrast is largely explained by the higher NGBS content and simultaneously higher expression level of MYB34 (Bol017062) gene in the BN4059 line compared to BN3273 (Table 2). PC2 in Table S4 indicated a notable contrast between the BN4059, BN4072 lines and the BN3273 and BN3383 lines for contrasting higher and lower content of glucoerucin and MGBS, respectively (Figure 6). This variation was largely explained by the contrasting expression level of the Dof1 (Bol041144), Dof1 (Bol006511) and IQD1 (Bol023096) genes (Table S4). Part of this variation is also explained by the absence of glucoraphanin, sinigrin and gluconapin in the BN4059 and BN4072 lines (Table S4).
Table 2.
Principal component analysis for indolic glucosinolate contents and the relative expression level of transcription factor-related genes in four cabbage lines of B. oleracea. PC, principal component; p, statistical significance; SD, standard deviation.
| Variable | PC1 | PC2 |
|---|---|---|
| 4HGBS | −0.24 | −0.29 |
| MGBS | 0.13 | 0.26 |
| NGBS | −0.11 | 0.46 |
| MYB34 (Bol017062) | −0.06 | 0.53 |
| MYB34 (Bol007760) | 0.52 | −0.15 |
| MYB34 (Bol036262) | 0.31 | 0.43 |
| MYB51 (Bol013207) | 0.41 | 0.19 |
| MYB51 (Bol030761) | 0.54 | −0.14 |
| MYB122 (Bol015939) | −0.28 | 0.28 |
| %variation explained | 34.3 | 31.0 |
| p (genotype) | <0.001 | <0.001 |
| Genotype | Mean PC Scores (±SD) | |
| BN3273 | −1.71 ± 0.23 | −2.17 ± 0.38 |
| BN3383 | 0.25 ± 0.20 | 0.86 ± 0.51 |
| BN4059 | 2.64 ± 0.44 | −0.63 ± 0.23 |
| BN4072 | −1.17 ± 0.17 | 1.94 ± 0.81 |
2.3.3. Association between Aliphatic Glucosinolate Content and Biosynthesis-Related Genes
PC1 accounted for a positive association among the higher level of expression of ST5b genes and the lower level of expression of GSL-OH (Bol033373) in BN3273 and their association with the content of glucoraphanin, sinigrin and gluconapin (Table 3). PC2, which explained 31.8% of the data variation, was largely influenced by the higher level of expression of the ST5c and FMOGS-OX genes in BN3383 (Table 3). PC3 accounted for 17.1% of the data variation, indicating that higher progoitrin accumulation in the line BN4059 might be associated with the higher expression level of GSL-OH (Bol033373) and the lower expression level of the AOP2 (Bo2g102190) gene compared to the BN4072 line (Table 3, Figure 4 and Figure 6).
Table 3.
Principal component analysis for aliphatic glucosinolate contents and the relative expression level of the biosynthesis-related genes in four cabbage lines of B. oleracea. PC, principal component; p, statistical significance; SD, standard deviation.
| Variable | PC1 | PC2 | PC3 |
|---|---|---|---|
| Progoitrin | −0.16 | 0.09 | 0.55 |
| Glucoraphanin | 0.42 | −0.08 | 0.17 |
| Sinigrin | 0.42 | −0.07 | 0.17 |
| Gluconapin | 0.42 | −0.09 | 0.14 |
| Glucoerucin | −0.07 | −0.24 | 0.05 |
| ST5b (Bol026201) | 0.34 | 0.09 | 0.12 |
| ST5b (Bol026202) | 0.41 | 0.15 | 0.12 |
| ST5c (Bol030757) | 0.03 | 0.41 | −0.21 |
| FMOGS-OX2 (Bol010993) | 0.01 | 0.44 | 0.18 |
| FMOGS-OX5 (Bol029100) | −0.13 | 0.40 | 0.18 |
| FMOGS-OX5 (Bol031350) | 0.13 | 0.40 | −0.21 |
| AOP2 (Bo2g102190) | 0.01 | 0.10 | −0.52 |
| AOP2 (Bo9g006240) | 0.09 | −0.42 | −0.19 |
| GSL-OH (Bol033373) | −0.34 | −0.09 | 0.35 |
| %variation explained | 36.3 | 31.8 | 17.1 |
| p (genotype) | <0.001 | <0.001 | <0.001 |
| Genotype | Mean PC Scores (±SD) | ||
| BN3273 | 3.58 ± 0.41 | −0.69 ± 0.61 | 0.57 ± 0.74 |
| BN3383 | −0.32 ± 0.35 | 3.29 ± 0.55 | −0.80 ± 0.53 |
| BN4059 | −2.09 ± 0.38 | −0.46 ± 0.18 | 1.88 ± 0.75 |
| BN4072 | −1.17 ± 0.28 | −2.13 ± 0.18 | −1.65 ± 0.95 |
2.3.4. Association between Indolic Glucosinolate Content and Biosynthesis-Related Genes
PC1 in another PCA between indolic glucosinolate content explained 35.1% of the data variation (Table S5). This PC highlighted a contrast in the expression level of most of the indolic biosynthesis-related genes in BN3273 (generally having a lower expression level) compared to the other three lines (Figure 5, Table S5). BN3273 had a comparatively lower expression level for all indolic biosynthesis-related genes compared to the other three lines (Figure 5). The variation explained by PC2 was dominated by the higher expression level of CYP81F1 accessions Bol017376, Bol017375 and Bol028913 and the lower expression level of CYP81F2, CYP81F2 and ST5a (Bol026200) in BN4072 compared to the other two lines, BN3273 and BN4072 (Figure 5, Table S5). The variation in PC2 is largely accounted for by the strikingly higher expression of level of CYP81F1 genes in the inbred line BN4072 (Table S5). PC3, explaining 16.6% of the data variation, is dominated by the higher NGBS content in BN4072 and BN3383 compared to the other two lines (Table S5).
2.4. Discussion
The present study investigated the relative gene expression level of 48 genes from the aliphatic and indolic metabolic pathways of glucosinolate biosynthesis and also estimated the content of glucosinolate in four inbred lines of cabbage with contrasting morphological variation (Figure 2). Only one out of four genotypes, BN3273, had three aliphatic glucosinolates detected, glucoraphanin, sinigrin and gluconapin. The cabbage inbred lines BN3383, BN4059 and BN4072 had none of those three aliphatic glucosinolates. This genotype-specific variation was associated with the expression level of some key genes. PCA analysis between the expression level of aliphatic glucosinolate contents and the relative expression level of aliphatic glucosinolate biosynthesis-related genes indicated that the existence of glucoraphanin, sinigrin and gluconapin might be associated with the comparatively higher level of expression of ST5b genes and the lower expression level of GSL-OH genes in BN3273 (Table 3, Figure 4). Thus, in the inbred lines BN3383, BN4059 and BN4072, the non-existence of glucoraphanin, sinigrin and gluconapin might be associated with the lower level of expression of the ST5b genes (Table 3, Figure 4). The non-existence of those three aliphatic glucosinolates in the BN4059 and BN4072 lines might also be associated with the level of transactivation of ST5b genes by the following transcription factor-related genes: MYB28 (Bol007795), MYB28 (Bol017019), MYB28 (Bol036743) and MYB29 (Bol008849) (Table 1). Such transactivation of ST5b genes by the MYB transcription factors has been discussed by Variyar et al. [35].
Two separate PCs involving indolic transcription factor-related and biosynthesis genes (PC2 in Table 2 and Table S5, respectively) indicated that a notably higher expression level of the MYB34 (Bol017062) gene (Figure 3) is associated with trans-activation of the expression of the CYP81F1 (Bol017375, Bol017376, Bol028913) genes (Figure 5) in the inbred line BN4072 for a comparatively higher accumulation of MGBS and NGBS in that particular line (Figure 6). On the other hand, the strikingly higher level of expression of CYP81F1 (Bol028914) and two IGMT1 genes was associated with comparatively lower content of NGBS in another line, BN4059 (Table S5).
The HPLC analysis in this study only detected eight types of glucosinolates; those included no aromatic glucosinolate compound (Figure 6). In our previous study, we detected a total of 12 glucosinolates, including glucoiberverin (GIV), glucoiberin (GIB), glucobrassicanapin (GBN), glucoraphanin (GRE), glucobrassicin (GBS) and gluconasturtiin (GST, an aromatic glucosinolate), in cabbage subspecies [5]. None of those six glucosinolates were detected in this study. However, in this study, we detected 4HGBS, which is a very close intermediate glucosinolate compound of GBS. The reason behind this variation might be related to the genetic variations of different cabbage genotypes, environmental variation or may be partly related to the HPLC procedure.
A high level of gene expression of the TGG2 (Bol028319) break-down-related gene in the cabbage inbred line BN3383 suggested that break-down of glucosinolate molecules is also high in that inbred line compared to the other three lines (Figure 5). In contrast, the lowest level of expression of TGG1 (Bol017328) and TGG2 (Bol025706) in inbred lines BN4059 and BN4072 suggested a lower level of break-down of glucosinolate molecules compared to the two other inbred lines, although a contrasting expression level was observed for the TGG5 gene (Figure 5). The lower level of expression of break-down-related genes might be associated with the lower level of the defense capacity of the BN4059 and BN4072 lines against herbivory and microbial attack.
One of the notable observations in this study is that out of a the total of 48 genes, 12 genes accounted for a significantly higher level of gene expression in a particular line, BN3383, compared to the other three cabbage lines (Figure 2, Figure 3, Figure 4 and Figure 5). However, none of the eight detected glucosinolate molecules were higher in content in BN3383 compared to the other three lines (Figure 6). The reason behind such variation is a subject of further investigation. However, a lower level of glucosinolate in that particular line might be associated with enhanced glucosinolate break-down through the elevated expression of TGG2 genes (Figure 5). Moreover, sequence variants are another possible explanation, and intuitively, the other includes various protein levels due to post-transcriptional modifications and various levels of enzymatic activities regulated at the post-translational level, which would be specific for the line BN3383.
The general role of the constitutive glucosinolate molecules is providing defense to plants from insects and microbes by their anti-oxidative properties [36,37,38,39]. A notable number of previous studies confirmed that the content of glucosinolates varies between genotypes of cabbage [1,2,3,4]. However, none of those previous studies explored the genetic background of such variation. The reason behind the higher or lower level of gene expression in different cabbage genotypes and associated variation in glucosinolate accumulation is a subject of further variation. Intuitively, there might have been variation in the functional component within the gene sequence, including the presence of the InDel (insertion or deletion) in the base sequence, single nucleotide polymorphism, etc., and also there may be variation in enzymatic activities regulated at the post-translational level.
3. Experimental Section
3.1. Plant Materials and Growth Conditions
A total of four inbred lines with contrasting morphological variation from the cabbage (B. oleracea var. capitata) subspecies were selected for this study (Figure 2). Seeds were obtained from Asia Seed Co., Ltd. (Seoul, Korea). The seeds were sown and seedlings were raised in a garden soil mixture in a culture room. Seedlings were transferred at four weeks of age to a glasshouse. The plants were allowed to grow for another three months before collecting leaf samples for estimating glucosinolates and also for quantifying the relative expression of glucosinolate biosynthetic genes. Leaf samples used for both real-time PCR and metabolite analysis were collected from the middle-aged leaves (see Figure 2).
3.2. Primer Design for Glucosinolate Biosynthesis Genes
As the objective of this study was to investigate the genotypic variation in the expression level of glucosinolate biosynthetic genes related to identified aliphatic and indolic glucosinolates (Figure 6), therefore, a total of 48 genes related to transcription and biosynthesis of glucosinolate were selected from the two respective biosynthetic pathways for relative expression analysis (Figure 1, Table 4). These 48 genes included all transcription factor-related genes associated with side chain elongation and core structure formation and biosynthetic genes that convert crude desulfoglucosinolates to a particular aliphatic or indolic glucosinolate. Among them, five and six genes respectively were aliphatic and indolic transcription factor-related and 10 and 15 genes were aliphatic and indolic glucosinolate biosynthesis related genes, respectively. The other eight genes from the Dof, IQD and TFL subfamilies were also transcription factor related to both pathways. In addition, these 48 genes also included five glucosinolate break-down-related genes. To design primers for each gene, Primer3 web-based software, Primer3web version 4.0.0 (http://primer3.ut.ee/, Table 4), was used. The primer specificity of the genes was initially checked by using the primer-blast option in NCBI (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and also by using the similarity search option in the Bolbase database (http://www.ocri-genomics.org/bolbase/blast/blast.html). As a further confirmatory analysis, the sequences of all highly homologous genes and the designed primers were aligned using ClustalW software [40], and both forward and reverse sequences of the primers of the target gene were obtained from the dissimilar regions of the homologues. To test the efficiency of the primers, the pooled cDNA of equal concentrations, 300 ng·µL−1, of four cabbage inbred lines was serially diluted by 10-fold, and 1.0 µL of the 100, 10−1, 10−2, 10−3, 10−4 and 10−5 diluted cDNA was used as the template in each reaction with forward and reverse primers. The qRT-PCR reaction was performed for 40 cycles along with the melting curve in triplicate along with no template control. A semi-log plot for Ct versus fold dilution was drawn to find out the slope. The efficiency of the primer was calculated by using the following formula, e = 10^(−1/slope). A primer with −3.321928 slopes was found 100% efficient. Primers with an 85%–100% efficiency level were chosen for expression analysis, otherwise redesigned to obtain the efficiency within the expected range.
Table 4.
Primer sequences of 48 glucosinolate biosynthetic-related genes used for the relative expression analysis of four cabbage inbred lines with morphological variation through qPCR.
| Gene Name | Accession Number | cDNA Size (bp) | Forward Primer Sequence | Reverse Primer Sequence | Product Size (bp) |
|---|---|---|---|---|---|
| Transcription factor-related genes (19 genes) | |||||
| Aliphatic | |||||
| MYB28 | Bol007795 | 558 | CCACACCAGTTCAGAGAGGT | GGGAAATGGATCGAAGTCAGC | 221 |
| Bol036286 | 615 | GAAGGTAGCTTGAATGCTAATAC | ATTCATGTAGTGCTCCTCATTC | 249 | |
| Bol017019 | 426 | GTTGCGGCTAAGGTCACTTCT | CAGAAGTAGCGTTGATCTCATGC | 223 | |
| Bol036743 | 426 | CTTGGGCGCTGCTACATTAC | ATCGTTCTCCTCGTTGTGGT | 241 | |
| MYB29 | Bol008849 | 513 | CGCCCAAGACTTCTGAGTT | TGATATTGCCCATGGAAGCTG | 234 |
| Indolic | |||||
| MYB34 | Bol007760 | 843 | TGAAGGAGGATGGCGTACTC | CAGTTCGTCCCGCCAAATTA | 203 |
| Bol017062 | 951 | AAGGTGGATGGCGTACTCTC | TGTGAGTGGTTGGATCGACA | 279 | |
| Bol036262 | 294 | CCCCGAGTTCTTTAGCAACC | TCCAAGTCCAGATCGTCTTCT | 198 | |
| MYB51 | Bol013207 | 1002 | CCAGAGATTCCAGAGAAGC | CAAGTCACACTGCTACTACTAC | 233 |
| Bol030761 | 990 | CAGACAACTATATCGAGTAACG | TATCATTAACGGTCATCTGG | 274 | |
| MYB122 | Bol026204 | 981 | GACCATTCCGAGACATTGCC | GCATCGTGGATCATGTGGAG | 284 |
| Both aliphatic and indolic | |||||
| Dof1.1 | Bol023400 | 1005 | GACGAAACATAGCAGCTCCG | ACCGGGTTGTTCTTCCATCT | 227 |
| Dof1.1 | Bol041144 | TTGGTCACAGCCTACGAACT | ACTTGGTGATGAGGGAGTCG | 260 | |
| Dof1.1 | Bol006511 | ACCAACTTCCACCTTTCCCA | AAGCTGCTCTCATTCTCGGT | 163 | |
| IQD1 | Bol023096 | TGTCCTCGATGCAACTCCAT | ACCGAGAATGAGAGCAGCTT | 287 | |
| IQD1 | Bol033935 | AAGCTGCTCTCATTCTCGGT | CCTCTTTGGTTGCCTTGGTC | 195 | |
| TFL2 | Bol021358 | 1146 | ACGATGCTGCTGAGAAGTCT | CCTGGTCCCCTTAACTCGTT | 199 |
| TFL2 | Bol000201 | AGCGGGAAAAGAAGTGGAGA | GCCTTTCCTCTTTCCCTCCT | 166 | |
| TFL2 | Bol034455 | AAGAGGGAGAAGCAGCCATT | AACCTCCACTGGCTTGATGA | 239 | |
| Aliphatic biosynthesis-related genes (9 genes) | |||||
| ST5b | Bol026201 | 1035 | CCGAGCCGTCAGAATTCAAG | GCTATGGCGAAAGTGAGAGC | 247 |
| Bol026202 | 1035 | AAGCCTTGACTTTCGCCATC | ACTTCACAACTGAGTCCGGT | 204 | |
| ST5c | Bol030757 | 1014 | CCACGCCCAAAACTTCTTCA | TGAGTGGAGAAGAGCGTGTT | 246 |
| FMOGS-OX2 | Bol010993 | 1386 | GAGAAGGTATCCGAGCCACA | GTCCACTGCAAACAACGACT | 200 |
| FMOGS-OX5 | Bol029100 | 1347 | CTTGCTCCAACGCTTTCCTT | CCTCAGCTCTCCAGTGTTCA | 280 |
| Bol031350 | 1380 | GACACTACACAGAGCCTCGT | CCCCGGGAAGCTTCTCATAT | 234 | |
| AOP2 | Bo2g102190 | GGAACGTGTCTCCAAAACCC | TAGCACCATCACCAGCATCA | 354 | |
| Bo9g006240 | CCAGGAAGTGAGAAGTGGGT | TAGCACCATCACCAGCATCA | 517 | ||
| GSL-OH | Bol033373 | 243 | GATTGTGCAAAAGGCTTGT | AGAGCATTAGGATTAGGAGGA | 188 |
| Indolic biosynthesis-related genes (15 genes) | |||||
| ST5a | Bol026200 | 1017 | GTCCGGTTGCAAGATGGTTT | CCTCTCCGGGTTCTCTTTGT | 214 |
| Bol039395 | 1014 | TGCCGTTTGTGAAGAGGTTG | CCCAATCTCCAACCTTCCCT | 210 | |
| CYP81F4 | Bol032712 | 1506 | CGGTGGAGGAGAAGGAGAAA | CTGACACATGGCTCGTAACG | 226 |
| Bol032714 | 960 | ACCCTGGTGAATACTTGCCA | GAAACACACTGAAGCAGAAC | 239 | |
| Bol028918 | 1503 | GTTTGCGGCATCAGAGACAT | GAATAGTCCACGCGTTCACC | 299 | |
| CYP81F1 | Bol017375 | 369 | AAGCAGAGCGGTTCAAGAAG | GCGTGACCATTGTGTTACCA | 204 |
| Bol017376 | 246 | CCGTCTCCTTCAACGGTTCT | CGACGTATTTACCGGTGAGC | 170 | |
| Bol028913 | 1500 | GAGACCTCCGCAGTAACCTT | GTCCTCCGTCGGTCTTCTAG | 222 | |
| Bol028914 | 1497 | CTTTCCAACTGACGGCCAAA | CGTTAGGTCCGAGAAAAGCG | 257 | |
| CYP81F2 | Bol012237 | 933 | GCAGCCGTGACACTAGAATG | TCCGCCAATCTTGAGGTCTT | 231 |
| Bol014239 | 1482 | TTGTACCGCGTTCTCCTTCT | GACACCATCCTCTGACCCAA | 238 | |
| CYP81F3 | Bol028919 | 1500 | TAACAGCGGAGGAGAAGACG | CACCTTCTAACTGGGCCTGA | 260 |
| Bol032711 | 1492 | CCGTCTCACCAACTTCCTCT | CTTCTCAAAGCTCCCTCCCA | 292 | |
| IGMT1 | Bol007029 | 1119 | GTGTTCCTCTCACCTTCCGA | GTGTTGAGGAAGACGCTGTC | 260 |
| Bol020663 | 342 | AGATGCCATGATCTTGAAACGT | CCAGCAATGATAAGCCTGACA | 298 | |
| Glucosinolate break-down-related genes (5 genes) | |||||
| TGG1 | Bol017328 | 822 | TCTTAACGTGTGGGATGGCT | CCTCCTTTGTTCACTCCCCT | 210 |
| TGG2 | Bol028319 | 1179 | TCGTCTCAACAGTAGCAGCT | AGTAGCGTTGAGTTCGTCCA | 220 |
| Bol025706 | 663 | GGTGAGTAGGGGAGTGAACC | TTCCTCGGTGAAGTTGGGAA | 244 | |
| TGG5 | Bol031599 | 1326 | CCAGATCACAGTTCCGGAGA | ACTATACGCCGGCTCAAGAA | 293 |
| PEN2 | Bol030092 | 1299 | GCATCATCATCCAACAGCGT | ACGCCTTGATCAGTTCTCCA | 207 |
| House-keeping genes | |||||
| actin1 | AF044573 | TTCTCTCTTCCACACGCCAT | CTTGTCCTGCGGGTAATTCG | 235 | |
| cctin2 | JQ435879 | GTCGCTATTCAAGCTGTTCTCT | GAGAGCTTCTCCTTGATGTCTC | 251 | |
| actin3 | XM_013753106 | ATCACACTTTCTACAATGAGC | TCGTAGATTGGCACAGTGTGAG | 241 | |
3.3. cDNA Synthesis and Real-Time Quantitative PCR Analysis
An RNeasy mini kit (Catalogue No. 74106, Qiagen, Valencia, CA, USA) was used to extract the total RNA of the leaf samples. cDNA was synthesized from the total RNA of each sample using a PrimeScript-based kit (Takara Bio, Inc., Shiga, Japan). iTaq™ SYBR® Green Super-mix was used with ROX (ROX is a positive reference dye) (Foster City, CA, USA) for real-time PCR. The reaction volume was 20 µL, where 1 µL cDNA of each sample was used. The concentration of cDNA of all genotypes was diluted from 60–70 ng·µL−1. The targeted DNA segment was amplified by denaturation at 95 °C for 10 min, followed by 40 cycles of amplification with denaturation at 94 °C for 20 s, annealing at 58 °C for 20 s and a final incubation and signal acquisition at 72 °C for 25 s. For some genes, the annealing temperature was increased from 58 °C–63 °C to improve the melting peaks. The fluorescence was recorded for each sample, while Light Cycler 96 software was used for quantification (Cq) analysis (Roche, Germany). There were three biological replicates for each line, and each biological replicate was repeated three times. The relative expression level was calculated by the comparative 2−ΔΔCt method [41]. Three different actin genes, actin1, actin2 and actin3, GenBank Accession Nos. AF044573 [42], JQ435879 [43] and XM_013753106 [44], respectively, which were expressed in all inbred lines, were the reference (Table 4 and Table S6, Figure S1). cDNA of four cabbage lines was diluted to 60, 120 and 180 ng·µL−1 concentrations to test the expression efficiency of the three reference genes. Reference genes were found quite stable under three different cDNA concentrations (Figure S2). The expression stability of all genes, including three reference genes, was calculated using NormFinder software (see the Supplementary excel data file) [45]. All genes accounted for the low intra-group (within genotype) variation, but inter-group variation was high for some notable genes that accounted for significant genotypic variation, for example CYP81F1, ST5b (Bol026201) and MYB28 (Bol036286), suggesting that the expression of the genes was valid. The average Cq value of these three actin genes was used for calculating the relative expression of each glucosinolate biosynthesis-related gene. While calculating the relative expression level of the each glucosinolate biosynthesis gene, the cabbage inbred line BN3273 was considered as the calibrator. The relative expression of the other three lines for each glucosinolate biosynthesis-related gene was then accounted for comparing to the expression level of BN3273.
3.4. Desulfoglucosinolate Extraction for HPLC Analysis
The modified HPLC protocol of Choi et al. [6] was used to extract desulfoglucosinolates from the leaf samples. About 10 g of frozen tissue were ground after treatment with 70% ethyl alcohol. The ground samples were incubated initially at 70 °C for 10 min followed by incubation at room temperature for 1 h. Then, the samples were centrifuged at 10,000× g for 8 min at 4 °C to precipitate the tissues and proteins. The supernatant was collected to conduct anion-exchange chromatography. This centrifugation process was repeated twice, and the supernatants from three steps were pooled in a 5-mL tube. The pooled supernatants contained crude glucosinolates. The crude glucosinolate solution was then mixed with 0.5 mL 50 mM barium acetate and 0.5 mL 50 mM lead acetate. The solution was then passed through a desulfation process. In this process, the solution was centrifuged once more at 2000× g for 10 min before loading into a pre-equilibrated column. In this column, the crude glucosinolate sample was rinsed with distilled water several times. The desulfation commenced after the addition of 250 μL aryl sulfatase and continued for 16 h. The desulfated solution was eluted with 1 mL distilled water. To remove any impurities in the desulfated glucosinolates, the eluted solution was centrifuged at 20,000× g for 4 min at 4 °C and filtered through a PTFE filter (13 mm, 0.2 μm, Advantec, Pleasanton, CA, USA). The desulfoglucosinolate samples were then analyzed in a Waters 2695 HPLC system (Waters, Milford, MA, USA) equipped with a C18 column (Zorbax Eclipse XBD C18, 4.6 mm × 150 mm, Agilent Technologies, Palo Alto, CA, USA). The mobile phase solvents used in the HPLC system were water and acetonitrile. The flow of desulfoglucosinolates into the system was 0.4 mL min−1 at room temperature. A PDA 996 UV-visible detector (Waters) was used to detect the desulfoglucosinolates at a wavelength of 229 nm. The detected glucosinolates were quantified by comparison with a standard curve prepared of commercial sinigrin. Mass spectrometry analysis (HPLC/MS, Agilent 1200 series, Agilent Technologies) facilitated the identification of individual glucosinolate molecules (HPLC/MS, Agilent 1200 series, Agilent Technologies).
3.5. Statistical Analysis
Analysis of variance (ANOVA) for the relative expression level of each gene and glucosinolate content against four different genotypes was analyzed via the one-way command using MINITAB 17 statistical software (Minitab Inc., 1829 Pine Hall Rd, State College, PA, USA). Tukey’s pairwise comparison was conducted for mean separation. A PCA was conducted taking either aliphatic or indolic glucosinolate content with the corresponding relative expression level of biosynthesis genes as a set of variables. PC scores obtained under four cabbage lines were also analyzed using one-way ANOVA.
4. Conclusions
The present study investigated the relative expression level of 48 genes related to glucosinolate biosynthesis and also estimated glucosinolate content in four cabbage inbred lines with contrasting morphological variation. The expression level of the majority of genes and glucosinolate content varied significantly in a genotype-dependent manner. A total of eight glucosinolates were detected in four inbred lines with significant variation in their content among them. Only one inbred line BN3273 had glucoraphanin, sinigrin and gluconapin detected. The biosynthesis and accumulation of these three aliphatic glucosinolates were associated with the variable expression level of ST5b and GSL-OH genes that were trans-activated by the MYB28 and MYB29 genes. For the indolic glucosinolates, higher accumulation of NGBS in the line BN4072 was associated with a higher level of expression of the MYB34 and CYP81F1 genes. The relative expression level of ST5b (Bol026201), TGG1 (Bol017328) and TGG2 (Bol025706) was consistently lower in the BN4059 and BN4072 lines compared to the other two inbred lines. A total of 12 genes were highly expressed in the inbred line BN3383, but that particular line had the least content of total glucosinolates measured. The possible reason for such genotypic variation is a matter of further investigation. However, the candidate genes with a contrasting level of relative expression might be cloned and sequenced to find out any possible existence of single nucleotide polymorphism, and any variation in enzymatic activities regulated at the post-translational level might be explored.
Acknowledgments
We thank the Asia Seed Co., Ltd., Republic of Korea, for providing B. oleracea seeds. This study was supported by the Golden Seed Project (Center for Horticultural Seed Development, No. 213003-04-4-SB110) of the Ministry of Agriculture, Food and Rural affairs in the Republic of Korea (MAFRA).
Abbreviations
- 4HGBS
4-hydroxyglucobrassicin
- MGBS
methoxyglucobrassicin
- NGBS
neoglucobrassicin
Supplementary Materials
Supplementary materials can be accessed at: http://www.mdpi.com/1420-3049/21/6/787/s1.
Author Contributions
I.-S.N., J.-I.P. and A.H.K.R. conceived of and designed the study. A.H.K.R. managed the experimental plants, collected samples, prepared cDNA, the qPCR analysis, prepared samples for HPLC and wrote the manuscript. H.R.K. conducted the HPLC analysis. R.L., G.-E.Y. and K.Y. assisted with the laboratory works.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Seeds of the inbred lines are available from the authors.
References
- 1.Van Etten C.H., Daxenbichler M.E., Williams P.H., Kwolek W.F. Glucosinolates and derived products in cruciferous vegetables. Analysis of the edible part from twenty-two varieties of cabbage. J. Agric. Food Chem. 1976;24:452–455. doi: 10.1021/jf60205a049. [DOI] [PubMed] [Google Scholar]
- 2.Cartea M.E., Velasco P., Obregón S., Padilla G., de Haro A. Seasonal variation in glucosinolate content in Brassica oleracea crops grown in northwestern Spain. Phytochemistry. 2008;69:403–410. doi: 10.1016/j.phytochem.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 3.Kusznierewicz B., Bartoszek A., Wolska L., Drzewiecki J., Gorinstein S., Namieśnik J. Partial characterization of white cabbages (Brassica oleracea var. capitata f. alba) from different regions by glucosinolates, bioactive compounds, total antioxidant activities and proteins. LWT–Food Sci. Technol. 2008;41:1–9. [Google Scholar]
- 4.Kabouw P., Biere A., van der Putten W.H., van Dam N.M. Intra-specific differences in root and shoot glucosinolate profiles among white cabbage (Brassica oleracea var. capitata) cultivars. J. Agric. Food Chem. 2009;58:411–417. doi: 10.1021/jf902835k. [DOI] [PubMed] [Google Scholar]
- 5.Yi G.E., Robin A.H.K., Yang K., Park J.I., Kang J.G., Yang T.J., Nou I.S. Identification and Expression Analysis of Glucosinolate Biosynthetic Genes and Estimation of Glucosinolate Contents in Edible Organs of Brassica oleracea Subspecies. Molecules. 2015;20:13089–13111. doi: 10.3390/molecules200713089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi S.H., Park S., Lim Y.P., Kim S.J., Park J.T., An G. Metabolite profiles of glucosinolates in cabbage varieties (Brassica oleracea var. capitata) by season, color, and tissue position. Hortic. Environ. Biotechnol. 2014;55:237–247. [Google Scholar]
- 7.Sang J.P., Minchinton I.R., Johnstone P.K., Truscott R.J.W. Glucosinolate profiles in the seed, root and leaf tissue of cabbage, mustard, rapeseed, radish and swede. Can. J. Plant Sci. 1984;64:77–93. doi: 10.4141/cjps84-011. [DOI] [Google Scholar]
- 8.Bellostas N., Kachlicki P., Sørensen J.C., Sørensen H. Glucosinolate profiling of seeds and sprouts of B. oleracea varieties used for food. Sci. Hort. 2007;114:234–242. doi: 10.1016/j.scienta.2007.06.015. [DOI] [Google Scholar]
- 9.VanEtten C.H., Daxenbichler M.E., Tookey H.L., Kwolek W.F., Williams P.H., Yoder O.C. Glucosinolates: Potential toxicants in cabbage cultivars. J. Am. Soc. Hort. Sci. 1980;105:710–714. [Google Scholar]
- 10.Liu S., Liu Y., Yang X., Tong C., Edwards D., Parkin I.A., Waminal N.E. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5 doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sørensen H. Canola and Rapeseed. Springer; New York, NY, USA: 1990. Glucosinolates: Structure-properties-function; pp. 149–172. [Google Scholar]
- 12.Mikkelsen M.D., Petersen B.L., Olsen C.E., Halkier B.A. Biosynthesis and metabolic engineering of glucosinolates. Amino Acids. 2002;22:279–295. doi: 10.1007/s007260200014. [DOI] [PubMed] [Google Scholar]
- 13.Sønderby I.E., Geu-Flores F., Halkier B.A. Biosynthesis of glucosinolates-gene discovery and beyond. Trends Plant Sci. 2010;15:283–290. doi: 10.1016/j.tplants.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 14.Grubb C.D., Abel S. Glucosinolate metabolism and its control. Trends Plant Sci. 2006;11:89–100. doi: 10.1016/j.tplants.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 15.Kroymann J., Textor S., Tokuhisa J.G., Falk K.L., Bartram S., Gershenzon J., Mitchell-Olds T. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol. 2001;127:1077–1088. doi: 10.1104/pp.010416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Textor S., de Kraker J.W., Hause B., Gershenzon J., Tokuhisa J.G. MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 2007;144:60–71. doi: 10.1104/pp.106.091579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gigolashvili T., Yatusevich R., Rollwitz I., Humphry M., Gershenzon J., Flügge U.I. The plastidic bile acid transporter 5 is required for the biosynthesis of methionine-derived glucosinolates in Arabidopsis thaliana. Plant Cell. 2009;21:1813–1829. doi: 10.1105/tpc.109.066399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawada Y., Toyooka K., Kuwahara A., Sakata A., Nagano M., Saito K., Hirai M.Y. Arabidopsis bile acid: Sodium symporter family protein 5 is involved in methionine-derived glucosinolate biosynthesis. Plant Cell Physiol. 2009;50:1579–1586. doi: 10.1093/pcp/pcp110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wittstock U., Halkier B.A. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002;7:263–270. doi: 10.1016/S1360-1385(02)02273-2. [DOI] [PubMed] [Google Scholar]
- 20.Brader G., Mikkelsen M.D., Halkier B.A., Tapio Palva E. Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 2006;46:758–767. doi: 10.1111/j.1365-313X.2006.02743.x. [DOI] [PubMed] [Google Scholar]
- 21.Celenza J.L., Quiel J.A., Smolen G.A., Merrikh H., Silvestro A.R., Normanly J., Bender J. The Arabidopsis ATR1 Myb transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 2005;137:253–262. doi: 10.1104/pp.104.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy M., Wang Q., Kaspi R., Parrella M.P., Abel S. Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 2005;43:79–96. doi: 10.1111/j.1365-313X.2005.02435.x. [DOI] [PubMed] [Google Scholar]
- 23.Skirycz A., Reichelt M., Burow M., Birkemeyer C., Rolcik J., Kopka J., Witt I. DOF transcription factor AtDof1. 1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. Plant J. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- 24.Gigolashvili T., Berger B., Mock H.P., Müller C., Weisshaar B., Flügge U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- 25.Gigolashvili T., Yatusevich R., Berger B., Müller C., Flügge U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;51:247–261. doi: 10.1111/j.1365-313X.2007.03133.x. [DOI] [PubMed] [Google Scholar]
- 26.Sønderby I.E., Hansen B.G., Bjarnholt N., Ticconi C., Halkier B.A., Kliebenstein D.J. A systems biology approach identifies a R2R3 MYB gene subfamily with distinct and overlapping functions in regulation of aliphatic glucosinolates. PLoS ONE. 2007;2:e1322. doi: 10.1371/journal.pone.0001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gigolashvili T., Engqvist M., Yatusevich R., Müller C., Flügge U.I. HAG2/MYB76 and HAG3/MYB29 exert a specific and coordinated control on the regulation of aliphatic glucosinolate biosynthesis in Arabidopsis thaliana. New Phytol. 2008;177:627–642. doi: 10.1111/j.1469-8137.2007.02295.x. [DOI] [PubMed] [Google Scholar]
- 28.Kliebenstein D., Pedersen D., Barker B., Mitchell-Olds T. Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics. 2002;161:325–332. doi: 10.1093/genetics/161.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mewis I., Appel H.M., Hom A., Raina R., Schultz J.C. Major signaling pathways modulate Arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 2005;138:1149–1162. doi: 10.1104/pp.104.053389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikkelsen M.D., Petersen B.L., Glawischnig E., Jensen A.B., Andreasson E. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 2003;131:298–308. doi: 10.1104/pp.011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brader G., Tas E., Palva E.T. Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 2001;126:849–860. doi: 10.1104/pp.126.2.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.European Economic Community (EEC) European Economic Community, Commission Regulation, EEC No 1864/90. Oilseeds—Determination of glucosinolates—High performance liquid chromatography. Off. J. Eur. Community L. 1990;170:27–34. [Google Scholar]
- 33.International Standards Organization (ISO) ISO 9167-1 (E). Rapeseed: Determination of Glucosinolates Content. ISO; Geneva, Switzerland: 1992. Part 1: Method using high performance liquid chromatography; pp. 1–9. [Google Scholar]
- 34.American Oil Chemists Society (AOCS) Official Methods and Recommended Practices of the American Oil Chemists Society. 5th ed. AOCS; Champaign, IL, USA: 1998. [Google Scholar]
- 35.Variyar P.S., Banerjee A., Akkarakaran J.J., Suprasanna P. Emerging Technologies and Management of Crop Stress Tolerance. Academic Press; San Diego, CA, USA: 2014. Role of glucosinolates in plant stress tolerance; pp. 271–291. [Google Scholar]
- 36.Hansen B.G., Kerwin R.E., Ober J.A., Lambrix V.M., Mitchell-Olds T., Gershenzon J., Kliebenstein D.J. A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol. 2008;148:2096–2108. doi: 10.1104/pp.108.129981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlaeppi K., Bodenhausen N., Buchala A., Mauch F., Reymond P. The glutathione-deficient mutant pad2-1 accumulates lower amounts of glucosinolates and is more susceptible to the insect herbivore Spodoptera littoralis. Plant J. 2008;55:774–786. doi: 10.1111/j.1365-313X.2008.03545.x. [DOI] [PubMed] [Google Scholar]
- 38.Neilson E.H., Goodger J.Q., Woodrow I.E., Møller B.L. Plant chemical defense: At what cost? Trends Plant Sci. 2013;18:250–258. doi: 10.1016/j.tplants.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 39.Vicas S.I., Teusdea A.C., Carbunar M., Socaci S.A., Socaciu C. Glucosinolates profile and antioxidant capacity of Romanian Brassica vegetables obtained by organic and conventional agricultural practices. Plant Foods Human Nutr. 2013;68:313–321. doi: 10.1007/s11130-013-0367-8. [DOI] [PubMed] [Google Scholar]
- 40.Multiple Sequence Alignment by ClustalW. [(accessed on 15 April 2016)]. Available online: http://www.genome.jp/tools/clustalw/
- 41.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Zhang B., Hu Z., Zhang Y., Li Y., Zhou S., Chen G. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica Oleracea var. acephala f. tricolor) Plant Cell Rep. 2012;31:281–289. doi: 10.1007/s00299-011-1162-3. [DOI] [PubMed] [Google Scholar]
- 43.Nawaz I., Iqbal M., Hakvoort H.W., Bliek M., de Boer B., Schat H. Expression levels and promoter activities of candidate salt tolerance genes in halophytic and glycophytic Brassicaceae. Environ. Exp. Bot. 2014;99:59–66. doi: 10.1016/j.envexpbot.2013.10.006. [DOI] [Google Scholar]
- 44.Lee J., Yang K., Lee M., Kim S., Kim J., Lim S., Jang Y.S. Differentiated cuticular wax content and expression patterns of cuticular wax biosynthetic genes in bloomed and bloomless broccoli (Brassica oleracea var. italica) Process Biochem. 2015;50:456–462. doi: 10.1016/j.procbio.2014.12.012. [DOI] [Google Scholar]
- 45.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.