Abstract
The aim of the work was to determine the interactions of a set of anti-cancer compounds with bovine serum albumin (BSA) using a ProteOn XPR36 array biosensor and molecular docking studies. The results revealed that a total of six anti-cancer compounds: gallic acid, doxorubicin, acteoside, salvianolic acid B, echinacoside, and vincristine were able to reversibly bind to the immobilized BSA. The sensorgrams of these six compounds were globally fit to a Langmuir 1:1 interaction model for binding kinetics analysis. There were significant differences in their affinity for BSA, with doxorubicin, the weakest binding compound having 1000-fold less affinity than salvianolic acid B, the strongest binding compound. However, compounds with a similar KD often exhibited markedly different kinetics due to the differences in ka and kd. Molecular docking experiments demonstrated that acteoside was partially located within sub-domain IIA of BSA, whereas gallic acid bound to BSA deep within its sub-domain IIIA. In addition, the interactions between these compounds and BSA were dominated by hydrophobic forces and hydrogen bonds. Understanding the detailed information of these anti-cancer compounds can provide important insights into optimizing the interactions and activity of potential compounds during drug development.
Keywords: surface plasmon resonance, compounds/BSA interaction, kinetics, molecular docking
1. Introduction
Numerous small-molecule compounds are capable of binding various target proteins in order to exhibit their antitumor activity. Serum albumin (SA), exemplified by bovine serum albumin (BSA) and human serum albumin (HSA), is the most abundant transport protein in plasma, playing important roles in the absorption, metabolism, pharmacological, and toxicological effects of compounds [1,2]. In order to effectively determine the overall pharmacokinetic profile of small-molecule compounds, it is important to investigate the affinity of compounds on SA. The use of BSA is, thus, considered as a model protein for studying compound-protein interactions due to its low cost, availability, and similarity to HSA [3,4]. Currently, various techniques have been used to study the binding affinity of compounds on proteins in vitro, including equilibrium dialysis, ultrafiltration, spectroscopy, capillary electrophoresis, nuclear magnetic resonance (NMR), surface plasmon resonance (SPR) technology, and molecular docking [5,6,7,8,9,10,11,12,13].
Recently, SPR technology has been developed to detect and analyze changes in the refractive index of buffers, where various molecules interact with the targets immobilized on the surface [14,15]. A ProteOn XPR36 parallel array biosensor based on SPR technology allows for real-time, quantitative detection of biomolecular interactions without the need for labeling and displays increased sensitivity, higher throughput. Therefore, this instrument provides enhanced support for small-molecule compound discovery and development, while being less expensive and time-consuming [16,17,18]. Molecular docking is another important technique which can provide better understanding of the various compound interactions with BSA by identifying the possible binding sites and primary forces involved [11,19,20].
In this study, we utilized the ProteOn XPR36 system to detect the interactions between BSA and a set of anti-cancer compounds including fluorouracil, hydroxytyrosol, gallic acid, matrine, salidroside, curcumin, oxaliplatin, paeoniflorin, doxorubicin, acteoside, ginsenoside Rh1, salvianolic acid B, echinacoside, and vincristine [21,22,23,24,25,26,27,28,29,30]. In addition, we performed molecular docking studies to evaluate the possible binding sites, interaction forces as well as residues of subdomain interaction of acteoside and gallic acid with BSA, respectively. The study can provide important insights into optimizing the interactions and activity of potential compounds during compound development.
2. Results
2.1. Sensorgrams of Anti-Cancer Compounds
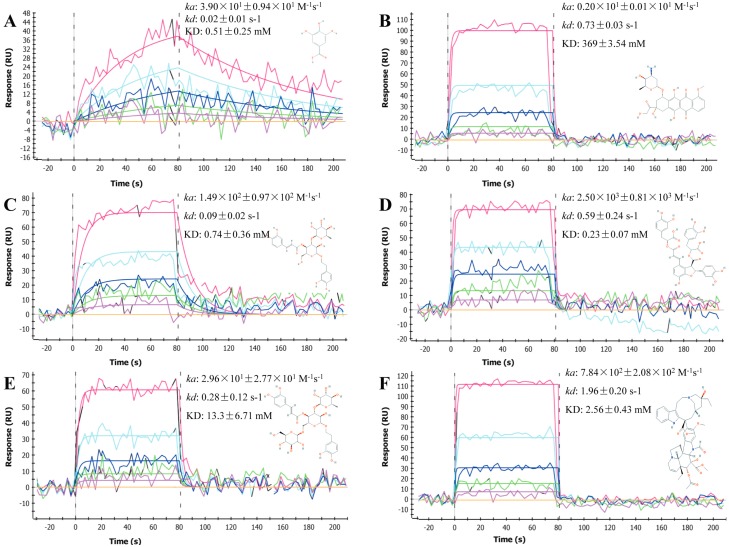
BSA was immobilized onto the GLH sensor surface via amine coupling, which resulted in final immobilization levels of approximately 11,000 RU (resonance units). Next, the immobilized BSA was interacted with various anti-cancer compounds, with large variations in molecular masses from fluorouracil (130.08 Da) to vincristine (824.96 Da) (Table 1) and different gradient concentrations. All resulting sensorgrams were collected and analyzed using the ProteOn manager software (Figure 1 and Figure S1). We determined that a total of six anti-cancer compounds: gallic acid, doxorubicin, acteoside, salvianolic acid B, echinacoside and vincristine were able to reversibly bind to the immobilized BSA (Figure 1). Interestingly, other than gallic acid, the association phases of these compounds all reached the steady state within a few seconds, while their dissociation phase signals rapidly returned to baseline almost immediately following injection. The sensorgrams of these six compounds were globally fit to a Langmuir 1:1 interaction model (for overlays of modeled and experimental data, see Figure 1), which demonstrated that it was possible to use a simple model to analyze the various number of interactions recorded on the biosensor.
Table 1.
Compounds used in analysis.
| Sample | Compound | Molecular Mass (Da) |
|---|---|---|
| A | Fluorouracil | 130.08 |
| B | Hydroxytyrosol | 138.16 |
| C | Gallic acid | 170.12 |
| D | Matrine | 248.37 |
| E | Salidroside | 300.31 |
| F | Curcumin | 368.39 |
| G | Oxaliplatin | 397.29 |
| H | Paeoniflorin | 480.45 |
| I | Doxorubicin | 543.52 |
| J | Acteoside | 624.59 |
| K | Ginsenoside Rh1 | 638.87 |
| L | Salvianolic acid B | 718.62 |
| M | Echinacoside | 786.72 |
| N | Vincristine | 824.96 |
Figure 1.
Sensorgrams for the interactions of BSA with the six test compounds; the structure of each compound was shown in the insets. Overlays of the models represented a global fit of the experimental data to a Langmuir 1:1 interaction model. Sensorgrams were representative of three independent experiments. (A) gallic acid; (B) doxorubicin; (C) acteoside; (D) salvianolic acid B; (E) echinacoside; and (F) vincristine.
2.2. Determination and Comparison of Kinetic Binding Constants
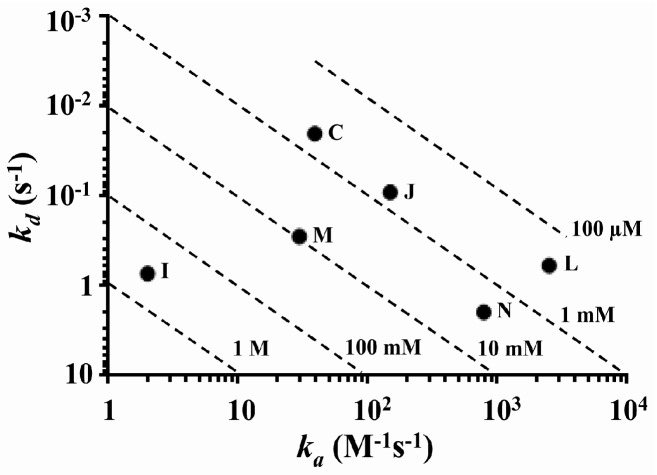
Determination of kinetic binding constants for all six compounds was provided in Figure 1, and the comparisons between their association and dissociation rates were plotted (Figure 2). Association rate was indicated by the x-axis, where 1 had the slowest and 104 had the fastest association rate. Dissociation rate was indicated by the y-axis, where 10−3 had the slowest and 10 had the fastest dissociation rate. Diagonal dashed lines represented the equilibrium dissociation constant (KD), from 1 M to 100 µM. We determined that the difference in affinity between doxorubicin (I), the weakest binding compound to salvianolic acid B (L), the strongest binding compound was greater than 1000-fold. Vincristine (N) had by far the fastest dissociation rate; salvianolic acid B (L), doxorubicin (I) and echinacoside (M) had intermediate dissociation rates, whereas acteoside (J) and gallic acid (C) had the slowest dissociation rates. Compounds with similar KD often displayed observable differences in their kinetics, as indicated by their ka and kd. For instance, acteoside (J) and gallic acid (C) bound to BSA with approximately the same KD; however, these two compounds exhibited markedly different kinetics, where acteoside had significantly faster ka and kd compared to gallic acid.
Figure 2.
Kinetic profile plot of the six test compounds; Compounds: (C) gallic acid; (I) doxorubicin; (J) acteoside; (L) salvianolic acid B; (M) echinacoside; and (N) vincristine. The diagonal dashed lines represented the equilibrium dissociation constant (KD).
2.3. Molecular Docking Studies
We demonstrated above that six anti-cancer compounds were able to reversibly bind to the immobilized BSA. In order to evaluate the possibility that additional compounds could reversibly bind to BSA, molecular docking was carried out using LibDock protocol, with Discovery Studio 2.5 software. We selected acteoside and gallic acid as reference compounds in order to identify the possible binding sites on BSA, and to better understand the interaction mechanisms. And the results of other compounds were provided in Figures S2–S5. BSA is a globular protein which contains 583 amino acids and consists of three homologous domains (I, II, and III): I (residues 1–183), II (184–376), III (377–583), each containing two sub-domains (A and B). Two hydrophobic cavities are present in sub-domain IIA and IIIA, which form the site of binding for various compounds [31,32].
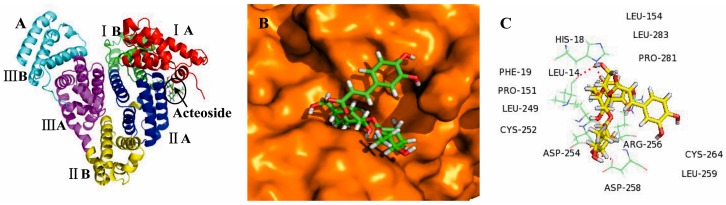
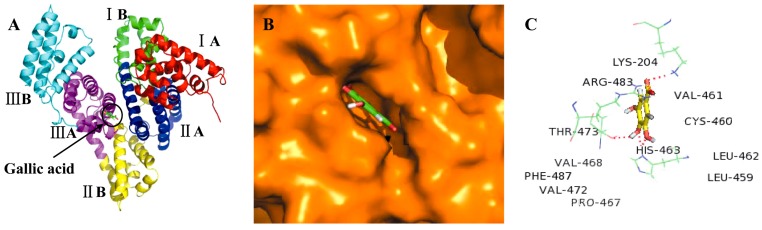
We showed that the best energy conformation model of acteoside was partially located within sub-domain IIA of BSA (Figure 3A,B). Notably, acteoside was surrounded by various hydrophobic residues including Leu-154, Leu-249, Leu-259, Leu-283, Pro-281, Pro-151, Cys-252, Cys-264, and Phe-19. The molecular docking results also indicated that there were hydrogen bonding interactions of acteoside with Arg-256, Asp-254, Asp-258, Leu-14, and His-18 residues (Figure 3C). In contrast, gallic acid bound deep within the pocket at sub-domain IIIA of BSA, in the minimum energy conformation (Figure 4A,B). Gallic acid was also adjacent to many hydrophobic residues including Val-461, Val-468, Val-472, Leu-459, Leu-462, Cys-460, Phe-487, and Pro-467. In addition, there were four hydrogen bonding interactions of gallic acid with Lys-204, His-463, Thr-473, and Arg-483 (Figure 4C). Therefore, we can conclude that acteoside and gallic acid could both bind to BSA, and the interactions between them were dominated by hydrophobic forces and hydrogen bonds. These molecular docking results were consistent with data obtained from SPR technology and, therefore, can provide a solid structural basis for understanding ligand–protein binding interactions.
Figure 3.
(A) The docking conformation of acteoside-BSA complex with the lowest energy conformation; the BSA and acteoside were represented in the cartoon as indicated; (B) Molecular docking model of acteoside partially located within sub-domain IIA of BSA; BSA and acteoside were represented by the orange sphere model and green stick model, respectively; (C) The surrounding hydrophobic amino acid residues within 6 Å and hydrogen bond interactions between acteoside and BSA; hydrogen bonds, amino acids, and acteoside were represented by red dashed lines, green lines, and yellow stick model, respectively.
Figure 4.
(A) The docking conformation of gallic acid-BSA complex with the lowest energy conformation; the BSA and gallic acid were represented in the cartoon as indicated; (B) Molecular docking model of gallic acid deeply inserted into the sub-domain IIIA of BSA; BSA and gallic acid were represented by the orange sphere model and green stick model, respectively; (C) The surrounding hydrophobic amino acid residues within 6 Å and hydrogen bond interactions between gallic acid and BSA; hydrogen bonds, amino acids and gallic acid were represented by red dashed lines, green lines, and yellow stick model, respectively.
3. Discussion
SPR technology is a useful analytical technology for detecting interactions between various biomolecules, via relaying changes in the refractive index at the surface of a gold chip. Recently, SPR technology has been used commercially through advanced systems such as Biacore S51 and ProteOn XPR36, which are capable of detecting minute changes in the mass of analytes binding to the chip at a temporal resolution of only 0.1 s [33,34]. In this study, we used the ProteOn XPR36 system to determine the interactions of a set of anti-cancer compounds with BSA. It is commonly accepted that the value of KD is positively correlated to temperature [16]. Therefore, variations in temperature which can result in deviations of KD should be avoided via simultaneous injection of analytes at various concentrations. In this regard, the ProteOn XPR36 system can accurately measure concentration-dependent binding data of six different concentrations of analytes simultaneously flowing over the immobilized targets. Perhaps the most important advantage was that this system eliminated the need to regenerate the surface, and allowed the simple fit of all test compounds with the reaction surfaces into a 1:1 interaction model. A possible limitation was that it was difficult to determine the KD of compounds with multiple binding sites.
Generally, the weak interactions of compounds with serum albumin lead to a short lifetime or the poor distribution of compounds. In contrast, the strong binding displays the reverse effect [20]. The weak/strong affinity of compound-BSA is preliminarily evaluated by KD that lower of KD value indicates higher affinity. However, compounds with a similar KD often exhibit markedly different kinetics due to the differences in ka and kd, which are also key factors from a pharmacological point of view. As the potential anti-cancer agent, the kinetic binding constants of compounds should be comprehensively considered. Therefore, the selection of anti-cancer compounds is depended on not only the relatively low of KD value, but also weak kd and strong ka, which acteoside and gallic acid exhibited this feature among these compounds.
However, there were a few inconsistencies between previous research and our results on the interactions of BSA with the tested anti-cancer compounds [12,35,36,37,38]. In general, they use UV-VIS spectroscopy, fluorescence spectroscopy, Fourier transform infrared (FTIR), circular dichrosim (CD), and mass spectrometry to detect the interactions of compounds with BSA. The principle of each methodology is different. The determination of interactions between compounds with BSA relies on changes in the mass-to-charge ratio by mass spectrometry. Meanwhile, variations in light or band absorption, as well as shifting of the maximum peak position or maximum emission wavelength of compound-BSA were used to reflect the interactions of compounds with BSA via spectroscopic methodologies. For instance, Cheng reported that salidroside could bind to BSA via spectroscopic investigation, whereas our study identified no such binding using SPR technology [12]. Cheng had studied the interaction of salidroside with BSA under physiological conditions, whereas in our study the determination of BSA binding affinity was carried out in PBS-T buffer (10 mM Na-phosphate, 150 mM NaCl, and 0.005% Tween 20, pH 7.4). Therefore, a possible reason was the use of different methodologies and experimental conditions which resulted in the inconsistency. Further studies carried out under various conditions are required to confirm the actual binding affinity of salidroside to BSA.
Molecular docking studies were performed to determine the possible binding sites between the test compounds with BSA, and to provide a molecular explanation for these binding interactions. Using molecular modeling, we determined that the interactions between the tested anti-cancer compounds and BSA were dominated by hydrophobic forces and hydrogen bonds. In addition, we found that the test compounds bound to sub-domain IIA and sub-domain IIIA of BSA, and these sites have been recognized to play important roles in absorption, metabolism and transportation of various compounds. Taken together, studying the interactions between anti-cancer compounds with BSA via SPR technology and molecular docking techniques could provide significant contributions to the improved understanding and future pharmaceutical applications of these compounds.
4. Materials and Methods
4.1. Materials
All experiments were performed using ProteOn XPR36 instruments from Bio-Rad. A GLH Sensor chip, 50% glycerol, sodium acetate, N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide (EDC), sulfo-N-hydroxysuccinimide (sulfo-NHS) and ethanolamine-HCl were purchased from Bio-Rad (Hercules, CA, USA). Buffer reagents and BSA were purchased from Sigma-Aldrich (St. Louis, MO, USA). Fluorouracil, hydroxytyrosol, gallic acid, matrine, salidroside, curcumin, oxaliplatin, paeoniflorin, doxorubicin, acteoside, ginsenoside Rh1, salvianolic acid B, echinacoside, and vincristine were all purchased from the National Institutes for Food and Compound Control (Beijing, China).
4.2. Initialization of GLH Sensor Chip
The GLH sensor chip, consisting of glass prisms coated with gold and an alginate-based layer, was an ideal choice for examining protein-small molecule interactions. The chip was initialized with 50% glycerol for several minutes prior to use.
4.3. Preconditioning of GLH Sensor Chip
Following initialization, the GLH sensor chip was washed with 0.5% SDS, 50 mM NaOH and 100 mM HCl, respectively, for 1 min each, at a flow rate of 100 µL/min. PBS-T buffer (10 mM sodium phosphate, 150 mM NaCl, and 0.005% Tween 20, pH 7.4) was then injected in three pulses for baseline stabilization.
4.4. Immobilization of BSA
One of the six channels was activated with a mixture of EDC (40 mM) and sulfo-NHS (10 mM) in the horizontal direction for 3 min, at a flow rate of 30 µL/min. BSA (2 mg/mL) was diluted to 300 nM in 10 mM sodium acetate (pH 4.0) and immobilized on the channel for 5 min, at a flow rate of 30 µL/min. Next, the channel was blocked by injection of 1 M ethanolamine-HCl for 5 min. The final immobilization level was approximately 11,000 RU. Baseline equilibration was performed by multiple washes with running buffer (PBS-T) across the chip surface for 1 h.
4.5. Injection of Analytes
The highest concentrations of fluorouracil, hydroxytyrosol, gallic acid, matrine, salidroside, curcumin, oxaliplatin, paeoniflorin, doxorubicin, acteoside, ginsenoside Rh1, salvianolic acid B, echinacoside, and vincristine were 1920, 1500, 500, 200, 1000, 200, 251.7, 200, 36.8, 200, 100, 200, 800, and 400 µM, respectively. Each compound was tested at six concentrations using a two-fold dilution series. Gradient concentrations of each compound were simultaneously injected over the BSA-immobilized surface at a flow rate of 30 µL/min in the vertical direction, where the association and dissociation times were 80 s and 60 s, respectively. All interaction analyses were performed at 25 °C.
4.6. Data Processing and Analysis
All sensorgrams for protein binding were collected, processed, and analyzed using the ProteOn Manager software. We utilized two types of reference sources, the standard blank buffer reference, and interspot reference, which was the average response before and after each interaction spot to correct for nonspecific binding. The target protein was washed off in the dissociation phase, which caused baseline drift, although real-time injection of a blank reference can completely correct this baseline drift by subtracting artifacts in real time. The dual references were both subtracted from the interaction spot data to correct for any bulk shifts due to baseline drift and nonspecific binding. Finally, the data for each compound collected over the same target protein were globally fit to a Langmuir 1:1 binding model with a global ka and kd, where ka is the association rate constant and kd is the dissociation rate constant for the protein-small molecule binding. The ratio of (kd/ka) indicated the value for the equilibrium dissociation constant (KD).
4.7. Molecular Docking
The structures of acteoside and gallic acid were obtained from the PubChem Compound Database for the molecular docking study. The MMFF force field was utilized for the energy minimization of compounds using Discovery Studio 2.5 software. The crystal structure of BSA (PDB ID: 4F5S) was obtained from RCSB Protein Data Bank. Binding interactions of test compounds with BSA were simulated by molecular docking using LibDock protocol with Discovery Studio 2.5 software. BSA was prepared by the removal of all water molecules and heteroatoms, and addition of hydrogen atoms. According to the scoring function, the compound–BSA complex conformation with the lowest binding energy was used for subsequent analysis. Molecular docking results were illustrated using the PyMol molecular graphic program.
Acknowledgments
This study was sponsored by Fujian Province Natural Science Foundation (2015J01687) and the Youth Science Foundation of Fujian Provincial Health Department (2014-2-29).
Abbreviations
| SPR | surface plasmon resonance |
| ka | association rate constant |
| kd | dissociation rate constant |
| KD | equilibrium dissociation constant |
| SA | Serum albumin |
| HSA | human serum albumin |
| BSA | bovine serum albumin |
| EDC | N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide |
| sulfo-NHS | sulfo-N-hydroxysuccinimide |
| RU | Resonance Units |
Supplementary Materials
The following are available online at http://www.mdpi.com/1420-3049/21/12/1706/s1.
Author Contributions
Ling Zhang conceived and designed the experiments, analyzed the data and wrote the manuscript; Ling Zhang, Qiao-Yan Cai and Yi Fang performed the ProteOn experiments; Zhi-Xiong Cai, and Chun-Song Zheng carried out the molecular docking; Li-Li Wang, Shan Lin and Da-Xin Chen contributed reagents; Jun Peng revised the manuscript critically for important intellectual content, provided scientific advice and final approval of the version to be published.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds fluorouracil, hydroxytyrosol, gallic acid, matrine, salidroside, curcumin, oxaliplatin, paeoniflorin, doxorubicin, acteoside, ginsenoside Rh1, salvianolic acid B, echinacoside, and vincristine are available from the authors.
References
- 1.Qin P., Liu R., Pan X., Fang X., Mou Y. Impact of carbon chain length on binding of perfluoroalkyl acids to bovine serum albumin determined by spectroscopic methods. J. Agric. Food Chem. 2010;58:5561–5567. doi: 10.1021/jf100412q. [DOI] [PubMed] [Google Scholar]
- 2.Sugio S., Kashima A., Mochizuki S., Noda M., Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 3.Han X.-L., Tian F.-F., Ge Y.-S., Jiang F.-L., Lai L., Li D.W., Yu Q.L., Wang J., Lin C., Liu Y. Spectroscopic, structural and thermodynamic properties of chlorpyrifos bound to serum albumin: A comparative study between BSA and HSA. J. Photochem. Photobiol. B Biol. 2012;109:1–11. doi: 10.1016/j.jphotobiol.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Atanu Singha R., Debi Ranjan T., Angshuman C., Swagata D. A spectroscopic study of the interaction of the antioxidant naringin with bovine serum albumin. J. Biophys. Chem. 2010;1:141–152. [Google Scholar]
- 5.Liang H., Xin B., Wang X., Yuan Y., Zhou Y., Shen P. Equilibrium dialysis study on the interaction between Cu (II) and HSA or BSA. Sci. Bull. 1998;43:404–409. doi: 10.1007/BF02883720. [DOI] [Google Scholar]
- 6.Wang Z., Zhang Z., Fu Z., Xiong Y., Zhang X. A flow-injection ultrafiltration sampling chemiluminescence system for on-line determination of drug–protein interaction. Anal. Bioanal. Chem. 2003;377:660–665. doi: 10.1007/s00216-003-2125-5. [DOI] [PubMed] [Google Scholar]
- 7.Shao L.W., Dong C.Q., Huang X.Y., Ren J.C. Using capillary electrophoresis mobility shift assay to study the interaction of CdTe quantum dots with bovine serum albumin. Chin. Chem. Lett. 2008;19:707–710. doi: 10.1016/j.cclet.2008.04.012. [DOI] [Google Scholar]
- 8.Van der Vlies C. Binding of a diphenhydramine analogue to BSA: An NMR investigation. Biochem. Pharmacol. 1970;19:859–864. doi: 10.1016/0006-2952(70)90248-0. [DOI] [PubMed] [Google Scholar]
- 9.Mir M.U.H., Maurya J.K., Ali S., Ubaid-ullah S., Khan A.B., Patel R. Molecular interaction of cationic gemini surfactant with bovine serum albumin: A spectroscopic and molecular docking study. Process Biochem. 2014;49:623–630. doi: 10.1016/j.procbio.2014.01.020. [DOI] [Google Scholar]
- 10.Zhang X., Li L., Xu Z., Liang Z., Su J., Huang J., Li B. Investigation of the interaction of naringin palmitate with bovine serum albumin: Spectroscopic analysis and molecular docking. PLoS ONE. 2013;8:E59106. doi: 10.1371/journal.pone.0059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skrt M., Benedik E., Podlipnik Č., Ulrih N.P. Interactions of different polyphenols with bovine serum albumin using fluorescence quenching and molecular docking. Food Chem. 2012;135:2418–2424. doi: 10.1016/j.foodchem.2012.06.114. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Z., Zhang Y. Spectroscopic investigation on the interaction of salidroside with bovine serum albumin. J. Mol. Struct. 2008;889:20–27. doi: 10.1016/j.molstruc.2008.01.013. [DOI] [Google Scholar]
- 13.Gelamo E., Silva C., Imasato H., Tabak M. Interaction of bovine (BSA) and human (HSA) serum albumins with ionic surfactants: Spectroscopy and modelling. Biochim. Biophys. Acta. 2002;1594:84–99. doi: 10.1016/S0167-4838(01)00287-4. [DOI] [PubMed] [Google Scholar]
- 14.Myszka D.G. Analysis of small-molecule interactions using Biacore S51 technology. Anal. Biochem. 2004;329:316–323. doi: 10.1016/j.ab.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 15.Long S., Myszka D. In: Affinity-Based Optical Biosensors. Handbook of Affinity Chromatography. Hage D.S., Cazes J., editors. CRC Press; Boca Raton, FL, USA: 2005. pp. 685–698. [Google Scholar]
- 16.Bravman T., Bronner V., Lavie K., Notcovich A., Papalia G.A., Myszka D.G. Exploring “one-shot” kinetics and small molecule analysis using the ProteOn XPR36 array biosensor. Anal. Biochem. 2006;358:281–288. doi: 10.1016/j.ab.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Rich R.L., Myszka D.G. Survey of the year 2004 commercial optical biosensor literature. J. Mol. Recognit. 2005;18:431–478. doi: 10.1002/jmr.753. [DOI] [PubMed] [Google Scholar]
- 18.Reichmann D., Rahat O., Albeck S., Meged R., Dym O., Schreiber G. The modular architecture of protein–protein binding interfaces. Proc. Natl. Acad. Sci. USA. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ao J., Gao L., Yuan T., Jiang G. Interaction mechanisms between organic UV filters and bovine serum albumin as determined by comprehensive spectroscopy exploration and molecular docking. Chemosphere. 2015;119:590–600. doi: 10.1016/j.chemosphere.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 20.Shi J.-H., Wang J., Zhu Y.-Y., Chen J. Characterization of interaction between isoliquiritigenin and bovine serum albumin: Spectroscopic and molecular docking methods. J. Lumin. 2014;145:643–650. doi: 10.1016/j.jlumin.2013.08.042. [DOI] [Google Scholar]
- 21.Hu X., Lin S., Yu D., Qiu S., Zhang X., Mei R. A preliminary study: The anti-proliferation effect of salidroside on different human cancer cell lines. Cell Biol. Toxicol. 2010;26:499–507. doi: 10.1007/s10565-010-9159-1. [DOI] [PubMed] [Google Scholar]
- 22.Sun L., Luo C., Liu J. Hydroxytyrosol induces apoptosis in human colon cancer cells through ROS generation. Food Funct. 2014;5:1909–1914. doi: 10.1039/C4FO00187G. [DOI] [PubMed] [Google Scholar]
- 23.Li Y., Zhang J., Ma H., Chen X., Liu T., Jiao Z., He W., Wang F., Liu X., Zeng X. Protective role of autophagy in matrine‑induced gastric cancer cell death. Int. J. Oncol. 2013;42:1417–1426. doi: 10.3892/ijo.2013.1817. [DOI] [PubMed] [Google Scholar]
- 24.Villegas I., Sánchez-Fidalgo S., Alarcón de la Lastra C. New mechanisms and therapeutic potential of curcumin for colorectal cancer. Mol. Nutr. Food Res. 2008;52:1040–1061. doi: 10.1002/mnfr.200700280. [DOI] [PubMed] [Google Scholar]
- 25.Lee K.-W., Kim H.J., Lee Y.S., Park H.-J., Choi J.-W., Ha J., Kyung-Tae L. Acteoside inhibits human promyelocytic HL-60 leukemia cell proliferation via inducing cell cycle arrest at G0/G1 phase and differentiation into monocyte. Carcinogenesis. 2007;28:1928–1936. doi: 10.1093/carcin/bgm126. [DOI] [PubMed] [Google Scholar]
- 26.Fang S., Zhu W., Zhang Y., Shu Y., Liu P. Paeoniflorin modulates multicompound resistance of a human gastric cancer cell line via the inhibition of NF-κB activation. Mol. Med. Rep. 2012;5:351–356. doi: 10.3892/mmr.2011.652. [DOI] [PubMed] [Google Scholar]
- 27.Lee Y., Jin Y., Lim W., Ji S., Choi S., Jang S., Lee S. A ginsenoside-Rh1, a component of ginseng saponin, activates estrogen receptor in human breast carcinoma MCF-7 cells. J. Steroid Biochem. Mol. Biol. 2003;84:463–468. doi: 10.1016/S0960-0760(03)00067-0. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y., Guo Y., Gu X. Salvianolic Acid B, a potential chemopreventive agent, for head and neck squamous cell cancer. J. Oncol. 2011;2010 doi: 10.1155/2011/534548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You B.R., Park W.H. Gallic acid-induced lung cancer cell death is related to glutathione depletion as well as reactive oxygen species increase. Toxicol. In Vitro. 2010;24:1356–1362. doi: 10.1016/j.tiv.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Dong L., Yu D., Wu N., Wang H., Niu J., Wang Y., Zou Z. Echinacoside Induces Apoptosis in Human SW480 Colorectal Cancer Cells by Induction of Oxidative DNA Damages. Int. J. Mol. Sci. 2015;16:14655–14668. doi: 10.3390/ijms160714655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bujacz A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012;68:1278–1289. doi: 10.1107/S0907444912027047. [DOI] [PubMed] [Google Scholar]
- 32.Paul B.K., Samanta A., Guchhait N. Exploring hydrophobic subdomain IIA of the protein bovine serum albumin in the native, intermediate, unfolded, and refolded states by a small fluorescence molecular reporter. J. Phys. Chem. B. 2010;114:6183–6196. doi: 10.1021/jp100004t. [DOI] [PubMed] [Google Scholar]
- 33.Frostell-Karlsson Å., Remaeus A., Roos H., Andersson K., Borg P., Hämäläinen M., Karlsson R. Biosensor analysis of the interaction between immobilized human serum albumin and drug compounds for prediction of human serum albumin binding levels. J. Med. Chem. 2000;43:1986–1992. doi: 10.1021/jm991174y. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen B., Tanious F.A., Wilson W.D. Biosensor-surface plasmon resonance: Quantitative analysis of small molecule–nucleic acid interactions. Methods. 2007;42:150–161. doi: 10.1016/j.ymeth.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Abdi K., Nafisi S., Manouchehri F., Bonsaii M., Khalaj A. Interaction of 5-Fluorouracil and its derivatives with bovine serum albumin. J. Photochem. Photobiol. B Biol. 2012;107:20–26. doi: 10.1016/j.jphotobiol.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Sun X., Xu X., Liu M., Li L., Sun D. Study on the interaction of matrine with bovine serum albumin. J. Solut. Chem. 2010;39:77–85. doi: 10.1007/s10953-009-9487-z. [DOI] [Google Scholar]
- 37.Barik A., Mishra B., Kunwar A., Priyadarsini K.I. Interaction of curcumin with human serum albumin: Thermodynamic properties, fluorescence energy transfer and denaturation effects. Chem. Phys. Lett. 2007;436:239–243. doi: 10.1016/j.cplett.2007.01.006. [DOI] [Google Scholar]
- 38.Qiu J., Shao S., You J.-Z. Studies on the Interaction between Oxaliplatin and Bovine Serum Albumin by Fluorescence Spectrometry. J. Instrum. Anal. 2007;26:51–54. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.