Abstract
The aim of the present study was to establish a new method based on Similarity Analysis (SA), Cluster Analysis (CA) and Principal Component Analysis (PCA) to determine the quality of different samples of Poria cocos (Schw.) Wolf obtained from Yunnan, Hubei, Guizhou, Fujian, Henan, Guangxi, Anhui and Sichuan in China. For this purpose 15 samples from the different habitats were analyzed by HPLC-PAD and HPLC-MSn. Twenty-three compounds were detected by HPLC-MSn, of which twenty compounds were tentatively identified by comparing their retention times and mass spectrometry data with that of reference compounds and reviewing the literature. The characteristic fragmentations were summarized. 3-epi-Dehydrotumulosic acid (F13), 3-oxo-16α,25-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid (F4), 3-oxo-6,16α-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid (F7) and dehydropachymic acid (F15) were deemed to be suitable marker compounds to distinguish between samples of different quality according to CA and PCA. This study provides helpful chemical information for further anti-tumor activity and active mechanism research on P. cocos. The results proved that fingerprint combined with a chemometric approach is a simple, rapid and effective method for the quality discrimination of P. cocos.
Keywords: triterpene acids, fingerprints, cluster analysis, principal component analysis
1. Introduction
Poria cocos (Schw.) Wolf is a saprophytic fungus that grows on diverse species of Pinus. Its sclerotium, called Fu-Ling or hoelen, is used in traditional Chinese and Japanese medicine for its diuretic, sedative, and tonic effects. Poria cocos (Schw.) Wolf is widely used in Yunnan, Guizhou, Hubei, Anhui, Fujian, Sichuan and Guangxi provinces in China, Modern medical research has indicated that P. cocos had comprehensive biological activities, such as antitumor [1,2,3,4,5,6,7,8,9,10,11], anti-inflammatory [12,13,14,15,16,17,18], immune-modulating [19,20], liver protecting [21,22] and so on, but particularly antitumor activity. Poria cocos (Schw.) Wolf contains a variety of triterpene acids found to be the bioactive components [2,3,4,5,6,7,8], for example pachymic acid, tumulosic acid, polyporenic acid C, dehydroeburicoic acid, dehydropachymic acid and so on. The type and content of triterpene acids reflect the quality of P. cocos so triterpene acids could be used as marker components to evaluate the quality of P. cocos.
The therapeutic effects of traditional Chinese medicines (TCMs) are based on the complex interactions of complicated chemical constituents as a whole system, so methods are needed in order to control the quality of the complex system. In this case, HPLC fingerprints of key components provide a new approach for quality control of traditional Chinese medicines. There are many studies about fingerprints analysis combined with chemometrics for the quality control of traditional Chinese medicines and to find the bioactive components [23,24,25,26,27].
Some studies on the fingerprints of Poria cocos (Schw.) Wolf have been reported [28,29,30,31], but in those reports only a few compounds were identified by HPLC-MSn and the characteristic fragmentations were not summarized. No marker compounds were found from cluster analysis (CA) and principal component analysis (PCA).
In the present study, nineteen common peaks and four other peaks which have not been detected using HPLC were identified by high–resolution liquid mass spectrometry. To the best of our knowledge, this is the first time that so many compounds were identified and their characteristic fragmentations summarized. We also found for the first time that 3-epi-dehydrotumulosic acid (F13), 3-oxo-16α,25-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid (F4), 3-oxo-6,16α-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid (F7) and dehydropachymic acid (F15) might be suitable marker compounds to distinguish between P. cocos samples with different quality according to CA and PCA. This study provides helpful chemical information for further anti-tumor activity and active mechanism research on P. cocos. The method developed in our study also provides a scientific foundation for the origin discrimination and quality control of P. cocos.
2. Results and Discussion
2.1. Validation of the Method
The relative retention time, relative peak area and similarities were used to evaluate the quality of the fingerprints. Dehydrotumulosic acid (peak 8) which is a large single peak in the middle of the chromatogram, was assigned as the reference peak to calculate relative retention times and relative peak areas.
The precision was determined by replicate injection with the same sample solution six consecutive times. The RSDs of relative retention time and relative peak area of the common peaks were all below 0.87% and 1.47%, respectively; the similarities of different chromatograms were all above 0.995.
The repeatability was evaluated by the analysis of six prepared samples. The RSDs of relative retention time and relative retention time of the common peaks were all below 1.59% and 1.97%, respectively; the similarities of different chromatograms were all above 0.995.
Stability testing was performed with one sample over 24 h. The RSDs of relative retention time and relative retention time of the common peaks were all below 0.96% and 1.98%; the similarities of different chromatograms were all 1.000. All these results indicated that the samples remained stable during the testing period and the conditions for the fingerprint analysis were satisfactory.
2.2. Similarity Analysis (SA)
The chromatographic profile must be representative of all the samples and have the features of integrity and fuzziness. By analyzing the mutual pattern of chromatograms, the identification and authentication of the samples can be conducted well even if the amounts of some chemical constituents are different from the others.
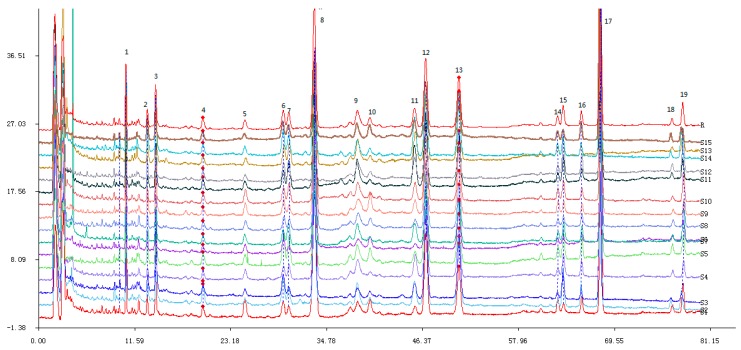
Fifteen batches of samples from different habitats were determined and the chromatograms analyzed by SES to generate a common pattern R (Figure 1). SES for Chromatographic Fingerprint was performed to calculate the similarities of different chromatograms compared to the common pattern. The results are shown in Table 1.
Figure 1.
Overlaid HPLC chromatograms of samples from No. S1–S15. The common pattern (marked R) was obtained by using Similarity Evaluation System (SES) for Chromatographic Fingerprint of TCM.
Table 1.
The results of similarities of the chromatograms from different origins.
| NO. | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 | S15 | R |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | 1.000 | 0.848 | 0.860 | 0.897 | 0.927 | 0.800 | 0.717 | 0.962 | 0.944 | 0.953 | 0.806 | 0.828 | 0.819 | 0.843 | 0.804 | 0.935 |
| S2 | 0.848 | 1.000 | 0.944 | 0.803 | 0.953 | 0.874 | 0.884 | 0.911 | 0.941 | 0.930 | 0.831 | 0.923 | 0.863 | 0.982 | 0.934 | 0.973 |
| S3 | 0.860 | 0.944 | 1.000 | 0.730 | 0.912 | 0.844 | 0.841 | 0.914 | 0.911 | 0.940 | 0.707 | 0.920 | 0.916 | 0.914 | 0.797 | 0.943 |
| S4 | 0.897 | 0.803 | 0.730 | 1.000 | 0.921 | 0.660 | 0.641 | 0.879 | 0.866 | 0.886 | 0.933 | 0.767 | 0.723 | 0.799 | 0.852 | 0.880 |
| S5 | 0.927 | 0.953 | 0.912 | 0.921 | 1.000 | 0.821 | 0.815 | 0.952 | 0.962 | 0.967 | 0.902 | 0.921 | 0.869 | 0.942 | 0.931 | 0.985 |
| S6 | 0.800 | 0.874 | 0.844 | 0.660 | 0.821 | 1.000 | 0.902 | 0.876 | 0.922 | 0.832 | 0.607 | 0.812 | 0.649 | 0.856 | 0.818 | 0.885 |
| S7 | 0.717 | 0.884 | 0.841 | 0.641 | 0.815 | 0.902 | 1.000 | 0.804 | 0.865 | 0.811 | 0.632 | 0.843 | 0.721 | 0.870 | 0.827 | 0.872 |
| S8 | 0.962 | 0.911 | 0.914 | 0.879 | 0.952 | 0.876 | 0.804 | 1.000 | 0.974 | 0.984 | 0.814 | 0.905 | 0.813 | 0.896 | 0.853 | 0.971 |
| S9 | 0.944 | 0.941 | 0.911 | 0.866 | 0.962 | 0.922 | 0.865 | 0.974 | 1.000 | 0.959 | 0.820 | 0.911 | 0.809 | 0.933 | 0.918 | 0.985 |
| S10 | 0.953 | 0.930 | 0.940 | 0.886 | 0.967 | 0.832 | 0.811 | 0.984 | 0.959 | 1.000 | 0.835 | 0.937 | 0.891 | 0.913 | 0.858 | 0.980 |
| S11 | 0.806 | 0.831 | 0.707 | 0.933 | 0.902 | 0.607 | 0.632 | 0.814 | 0.820 | 0.835 | 1.000 | 0.772 | 0.716 | 0.856 | 0.900 | 0.860 |
| S12 | 0.828 | 0.923 | 0.920 | 0.767 | 0.921 | 0.812 | 0.843 | 0.905 | 0.911 | 0.937 | 0.772 | 1.000 | 0.875 | 0.910 | 0.854 | 0.940 |
| S13 | 0.819 | 0.863 | 0.916 | 0.723 | 0.869 | 0.649 | 0.721 | 0.813 | 0.809 | 0.891 | 0.716 | 0.875 | 1.000 | 0.853 | 0.737 | 0.875 |
| S14 | 0.843 | 0.982 | 0.914 | 0.799 | 0.942 | 0.856 | 0.870 | 0.896 | 0.933 | 0.913 | 0.856 | 0.910 | 0.853 | 1.000 | 0.945 | 0.965 |
| S15 | 0.804 | 0.934 | 0.797 | 0.852 | 0.931 | 0.818 | 0.827 | 0.853 | 0.918 | 0.858 | 0.900 | 0.854 | 0.737 | 0.945 | 1.000 | 0.927 |
| R | 0.935 | 0.973 | 0.943 | 0.880 | 0.985 | 0.885 | 0.872 | 0.971 | 0.985 | 0.980 | 0.860 | 0.940 | 0.875 | 0.965 | 0.927 | 1.000 |
The conclusion can be drawn from the results that the similarities of different chromatograms compared to the common pattern are all above 0.900 except for samples S4 (0.880), S6 (0.885), S7 (0.872), S11 (0.860) and S13 (0.875), which indicates that the chemical constituents of different samples are not influenced highly by sources. The common pattern is a very positive identification for the samples of P. cocos.
2.3. Identification of the Compounds Present
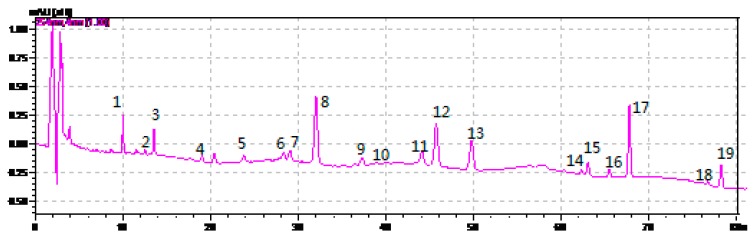
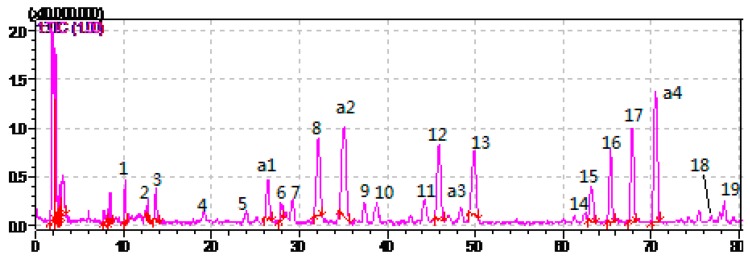
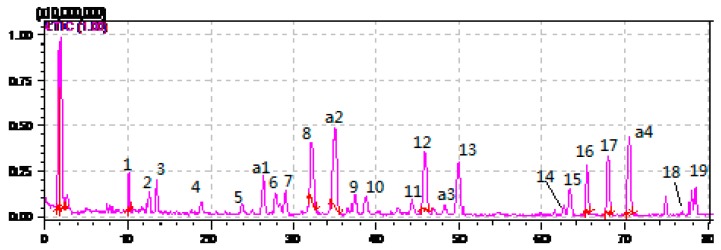
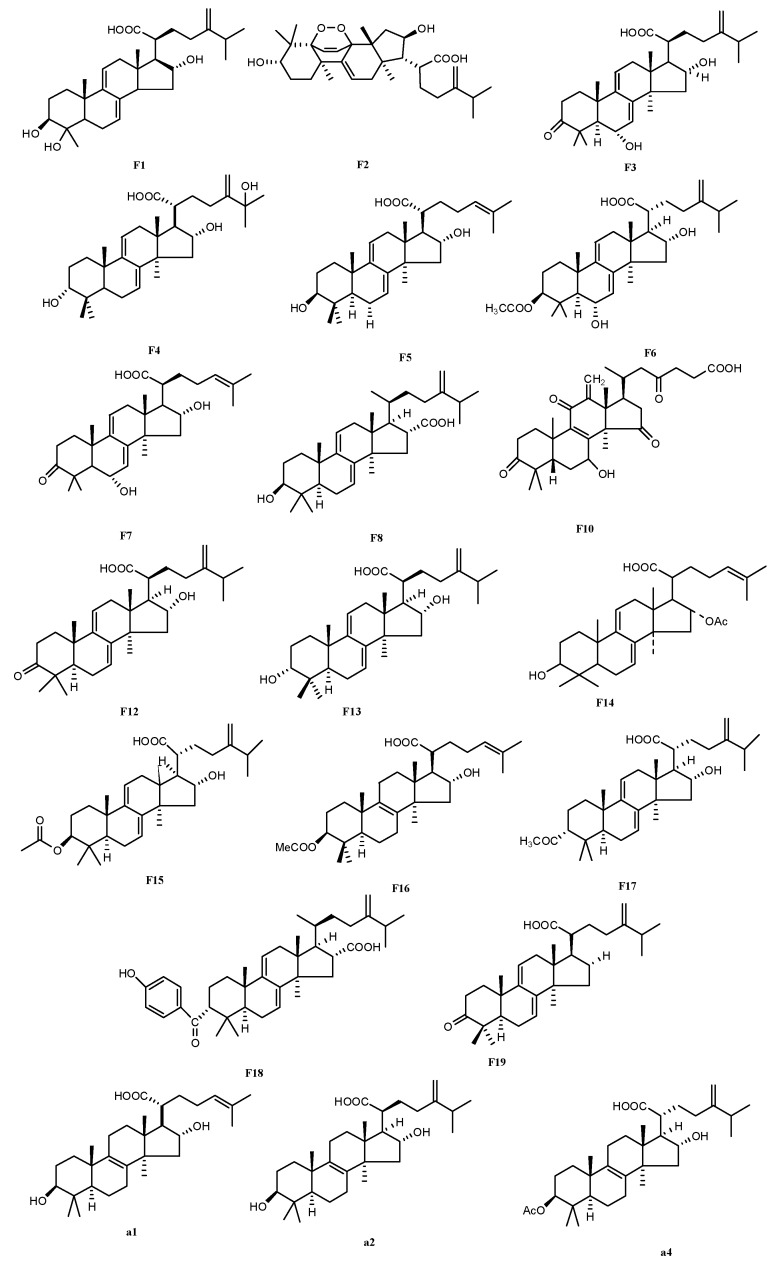
HPLC-ESI-MSn method was employed to identify the components in P. cocos (Figure 2, Figure 3 and Figure 4) Molecular weight and fragmentation information (Table 2) were obtained. The possible structures of these 19 common peaks and four other peaks a1, a2, a3 and a4 were deduced as it shown in Figure 5. Under the optimized MS conditions, positive mode and negative mode were used to identify the peaks.
Figure 2.
HPLC chromatograms of P. cocos.
Figure 3.
Positive mode of the HPLC-MSn chromatograms of P. cocos.
Figure 4.
Negative mode of the HPLC-MSn chromatograms of P. cocos.
Table 2.
The HPLC-MSn data and compound names of the 20 peaks.
| Peak No. | tR (min) | [M − H]− [M + H]+ |
Negative Mode | Positive Mode | Identification |
|---|---|---|---|---|---|
| F1 | 10.228 | 499.3346 501.3562 |
MS1: 499.3346 [M – H]− MS2: 499.3346→481.3221 [M – 18(H2O) − H]−, 467.3075 [M − 32(2CH4) − H]−, 437.2931 [M − 62(CH4 + HCOOH) − H]−, 419.2964 [M − 80(CH4 + HCOOH + H2O) − H]−, 325.2526 [M − 174(H2O + side chainon D ring) − H]− MS3: 419.2964→403.2698 [M − 80(H2O + CH4 + HCOOH) − 16(CH4) − H]− |
MS1: 501.3562 [M + H]+, 483.3447 [M − 18(H2O) + H]+, 465.3384 [M − 36(2H2O) + H]+
MS2: 501.3562→483.3460 [M − 18(H2O) + H]+, 465.3330 [M − 36(2H2O) + H]+ MS3: 483.3460→465.3324 [M − 18(H2O) − 18(H2O) + H]+, 447.3232 [M − 18(H2O) − 36(2H2O) + H]+, 419.3319 [M − 18(H2O) − 64(H2O + HCOOH) + H]+, 327.2338 [M − 18(H2O) − 156(side chain on D ring) + H]+, 309.2208 [M − 18(H2O) − 174(H2O + side chain on D ring) + H]+, 465.3324→447.3118 [M − 36(2H2O) − 18(H2O) + H]+, 309.2139 [M − 36(2H2O) − 156(side chain on D ring) + H]+, 291.2027 [M − 36(2H2O) − 174(H2O + side chain on D ring) + H]+ |
29-Hydroxy-dehydrotumulosic acid [32] |
| F2 | 12.637 | 513.3213 515.3352 |
MS1: 513.3213 [M − H]−
MS2: 513.3213→481.3303 [M − 32(2CH4) − H]− MS3: 481.3303→466.3146 [M − 32(2CH4) − 15(CH3) − H]− |
MS1: 515.3352 [M + H]+, 497.3238 [M − 18(H2O) + H]+ MS2: 515.3352→497.3235 [M − 18(H2O) +H]+, 461.3047 [M − 54(3H2O) + H]+, 433.3021 [M − 82(2H2O + HCOOH) + H]+, 341.2130 [M − 174(H2O + side chain on D ring) + H]+ MS3: 497.3238→341.2081 [M − 18(H2O) − 156(side chain on D ring) + H]+, 323.1985 [M − 18(H2O) − 174(H2O + side chain on D ring) + H]+ |
5α, 8α-Peroxy-dehydrotumulosic acid [5] |
| F3 | 13.665 | 497.3254 499.3394 |
MS1: 497.3254 [M − H]−
MS2: 497.3254→479.3096 [M − 18(H2O) − H]−, 435.3142 [M − 64(H2O + HCOOH) − H]−, 419.2947 [M − 78(2CH4 + HCOOH) − H]−, 401.2782 [M − 96(2CH4 + H2O + HCOOH) − H]− MS3: 419.2947→403.2698 [M − 78(2CH4 + HCOOH) − 16(CH4) − H]− |
MS1: 499.3494 [M + H]+, 481.3307 [M − 18(H2O) + H]+, 463.3196 [M − 36(2H2O) + H]+, 346.3312 [M − 153(CH3 + RDA fragmentation of B ring) + H]+
MS2: 499.3394→481.3306 [M − 18(H2O) + H]+, 463.3205 [M − 36(2H2O) + H]+ MS3: 481.3306→463.3220 [M − 18(H2O) − 18(H2O) + H]+, 445.3217 [M − 18(H2O) − 36(2H2O) + H]+, 417.3181 [M − 18(H2O) − 64(H2O + HCOOH) + H]+, 325.2182 [M − 18(H2O) − 156(side chain on D ring) + H]+ or [M − 18(H2O) − 156(H2O + RDA fragmentation of B ring) + H]+, 307.2059 [M − 18(H2O) − 174(H2O + side chain on D ring) + H]+, 463.3205→445.3125 [M − 36(2H2O) − 18(H2O) + H]+, 417.3209 [M − 36(2H2O) − 46(HCOOH) + H]+, 307.1993 [M − 36(2H2O) − 156(side chain on D ring) + H]+ |
6α-Hydroxy-polyporenic acid C [33] |
| F4 | 19.150 | 497.3287 499.3436 |
MS1: 497.3287 [M − H]−
MS2: 497.3287→467.3141 [M − 30(2CH3) − H]− MS3: 467.3141→421.3067 [M − 30(2CH3) − 46 (HCOOH) − H]−, 389.2857 [M − 30(2CH3) − 78 (2CH4 + HCOOH) − H]− |
MS1: 499.3436 [M + H]+, 521.3314 [M + Na]+, 481.3343 [M − 18(H2O) + H]+
MS2: 499.3436→481.3343 [M − 18(H2O) + H]+, 469.3290 [M − 30(2CH3) + H]+, 451.3264 [M − 48(2CH3 + H2O) + H]+, 330.6835 [M − 68(2CH3 + RDA fragmentation of B ring) + H]+, 327.1625 [M − 172(side chain on D ring) + H]+, 325.2160 [M − 174(2H2O + RDA fragmentation of B ring) + H]+, 297.1598 [M − 202(2CH3 + side chain on D ring) + H]+, 279.1605 [M − 220(2H2O + HCOOH + RDA fragmentation of B ring) + H]+ MS3: 481.3433→463.3205 [M − 18(H2O) − 18(H2O) + H] +, 451.3202 [M − 18(H2O) − 30(2CH3) + H]+, 325.2160 [M − 18(H2O) − 156(H2O + RDA fragmentation of B ring) + H]+, 295.2101 [M − 18(H2O) − 186(2CH3 + H2O + RDA fragmentation of B ring) + H]+, 451.3264→295.2045 [M − 48(2CH3 + H2O) − 156(H2O + RDA fragmentation of B ring) + H]+ |
3-oxo-16α,25-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid |
| F5 | 23.845 | 469.3329 471.3508 |
MS1: 469.3329 [M − H]−
MS2: 469.3329→425.3429 [M − 44(CO2) − H]− MS3: 425.3429→409.3112 [M − 44(CO2) − 16(CH4) − H]− |
MS1: 471.3508 [M + H]+, 509.3063 [M + K]+, 493.3397 [M + Na]+, 453.3371 [M − 18(H2O) + H]+, 435.3267 [M − 36(2H2O) + H]+ MS2: 471.3508→453.3350 [M − 18(H2O) + H]+ MS3: 453.3350→435.3268 [M − 18(H2O) − 18(H2O) + H]+, 311.2349 [M − 18(H2O) − 142(side chain on D ring) + H]+, 293.2289 [M − 18(H2O) − 160(H2O + side chain on D ring) + H]+, 311.2349→293.2229 [M − 18(H2O) − 142(side chain on D ring) − 18(H2O) + H]+, 278.2023 [M − 18(H2O) − 142(side chain on D ring) − 33(H2O + CH3) + H]+ |
3β,16α-Dihydroxy-lanosta-7,9(11),24-trien-21-oic acid [34] |
| F6 | 28.422 | 541.3569 543.3700 |
MS1: 541.3569 [M − H]−, 481.3308 [M − 60(CH3COOH) − H]−, 384.9361 [M − 156(side chain on D ring) − H]− MS2: 541.3569→481.3293 [M − 60(CH3COOH) − H]− |
MS1: 543.3700 [M + H]+, 525.3596 [M − 18(H2O) + H]+, 507.3346 [M − 36(2H2O) + H]+, 465.3378 [M − 78(H2O + CH3COOH) + H]+, 447.3277 [M − 96(2H2O + CH3COOH) + H]+, 361.6931 [M − 182(RDA fragmentation of B ring) + H]+ MS2: 543.3700→525.3521 [M − 18(H2O) + H]+, 507.3462 [M − 36(2H2O) + H]+, 465.3363 [M − 78(H2O + CH3COOH) + H]+, 447.3277 [M − 96(2H2O + CH3COOH) + H]+, 369.2441 [M − 174(H2O + side chain on D ring) + H]+, 361.6931 [M − 182(RDA fragmentation of B ring) + H]+, 291.2117 [M − 18(H2O) − 234(H2O + side chain on D ring + CH3COOH) + H]+ MS3: 465.3378→447.3228 [M − 78(H2O + CH3COOH) − 18(H2O) + H]+, 429.3027 [M − 78(H2O + CH3COOH) − 36(2H2O) + H]+, 419.3290 [M − 78(H2O + CH3COOH) − 46(HCOOH) + H]+, 309.2194 [M − 78(H2O + CH3COOH) − 156(side chain on D ring) + H]+, 291.2111 [M − 78(H2O + CH3COOH) − 174(H2O + side chain on D ring) + H]+, 447.3277→291.2103 [M − 96(2H2O+CH3COOH) − 156(side chain on D ring) + H]+ |
6α-Hydroxy-dehydropachymic acid [34] |
| F7 | 29.043 | 483.2973 485.3333 |
MS1: 483.2973 [M − H]−
MS2: 483.2973→465.2966 [M − 18(H2O) − H]− |
MS1: 485.3332 [M + H]+, 507.3094 [M + Na]+, 467.3190 [M − 18(H2O) + H]+, 449.3071 [M − 36(2H2O) + H]+, 328.9961 [M − 156(H2O + RDA fragmentation of B ring) + H]+, 326.0151 [M − 158(CH4 + side chain on D ring) + H]+, 311.1511 [M − 174(2CH4 + side chain on D ring) + H]+ or [M − 174(2H2O + RDA fragmentation of B ring) + H]+ MS2: 485.3333→467.3242 [M − 18(H2O) + H] +, 449.3070 [M − 36(2H2O) + H]+, 325.2146 [M − 160(H2O + side chain on D ring) + H]+ MS3: 467.3242→449.3048 [M − 18(H2O) − 18(H2O) + H]+, 431.2976 [M − 18(H2O) − 36(2H2O) + H]+, 325.2218 [M − 18(H2O) − 142(side chain on D ring) + H]+, 449.3070→ 431.3046 [M − 36(2H2O) − 18(H2O) + H]+, 307.2038 [M − 36(H2O) − 142(side chain on D ring) + H]+ |
3-oxo-6,16α-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid [35] |
| F8 | 32.068 | 483.3425 485.3602 |
MS1: 483.3425 [M − H]−, 295.2370 [M − 188(2CH4 + side chain on D ring) − H]−
MS2: 483.3425→465.2955 [M − 18(H2O) − H]−, 437.3440 [M − 46(HCOOH) − H]−, 311.1998 [M − 172(CH4+side chain on D ring) − H]−, 295.2036 [M − 188(2CH4 + side chain on D ring) − H]− |
MS1: 485.3602 [M + H]+, 523.3198 [M + K]+, 507.3451 [M + Na]+, 467.3518 [M − 18(H2O) + H]+, 449.3387 [M − 36(2H2O) + H]+, 439.3618 [M − 46(HCOOH) + H]+, 311.1665 [M − 174(H2O + side chain on D ring) + H]+ MS2: 485.3602→467.3512 [M − 18(H2O) + H]+, 449.3496 [M − 36(2H2O) + H]+, 311.2428 [M − 174(H2O + side chain on D ring) + H]+ MS3: 467.3512→449.3409 [M − 18(H2O) − 18(H2O) + H]+, 431.2786 [M − 18(H2O) − 36(2H2O) + H]+, 327.2293 [M − 18(H2O) − 140(RDA fragmentation of B ring) + H]+, 311.2351 [M − 18(H2O) − 156(side chain on D ring) + H]+, 293.2248 [M − 18(H2O) − 174(H2O + side chain on D ring) + H]+, 311.2428→293.2308 [M − 174(H2O + side chain on D ring) − 18(H2O) + H]+, 281.6503 [M − 174(H2O + side chain on D ring) − 30(2CH3) + H]+ |
Dehydrotumulosic acid [32] |
| F9 | 37.338 | 497.3263 499.3444 |
MS1: 497.3263 [M − H]−, 479.3193 [M − 18(H2O) − H]−, 452.9247 [M − 45(3CH3) − H]−, 248.9602 [M − 248 (4CH4 + HCOOH + RDA fragmentation of B ring) − H]− MS2: 497.3263→479.3161 [M − 18(H2O) − H]−, 452.9247 [M − 45(3CH3) − H]−, 249.9602 [M − 248(4CH4 + HCOOH + RDA fragmentation of B ring) − H]− |
MS1: 521.3305 [M + Na]+, 499.3444 [M + H]+, 481.3334 [M − 18(H2O) + H]+, 463.3219 [M − 36(2H2O) + H]+, 405.2614 [M − 93(2CH3 + H2O + HCOOH) + H]+, 310.1678 [M − 189(CH3 + H2O + side chain on D ring) + H]+, 279.1589 [M − 220(2H2O + HCOOH + RDA fragmentation of B ring) + H]+ MS2: 499.3444→481.3324 [M − 18(H2O) + H]+, 463.3205 [M − 36(2H2O) + H]+, 325.2130 [M − 174(H2O + side chain on D ring) + H]+ or [M − 174(2H2O + RDA fragmentation of B ring) + H]+ MS3: 481.3324→463.3215 [M − 18(H2O) − 18(H2O) + H]+, 445.3115 [M − 18(H2O) − 36(2H2O) + H]+, 325.2132 [M − 18(H2O) − 156(side chain on D ring) + H]+ or [M − 174(2H2O + RDA fragmentation of B ring) + H]+, 463.3205→445.3046 [M − 36(2H2O) − 18(H2O) + H]+, 417.3143 [M − 36(2H2O) − 46(HCOOH) + H]+, 307.2058 [M − 36(2H2O) − 156(side chain on D ring) + H]+ |
Unknown |
| F10 | 38.513 | 485.3269 487.3491 |
MS1: 485.3269 [M − H]−, 469.3311 [M − 16(CH4) − H]−, 248.9582 [M − 236(CH4 + 2H2O + HCOOH + RDA fragmentation of B ring) − H]− MS2: 485.3269→441.3391 [M − 44(CO2) − H]−, 423.3255 [M − 62(CH4 + HCOOH) − H]−, 248.9582 [M − 236(CH4 + 2H2O + HCOOH + RDA fragmentation of B ring − H)]− |
MS1: 509.3283 [M + Na]+, 487.3491 [M + H]+, 469.3318 [M − 18(H2O) + H]+, 451.3180 [M − 36(2H2O) + H]+, 433.3214 [M − 54(3H2O) + H]+, 405.2659 [M − 82(2H2O + HCOOH) + H]+, 348.9844 [M − 138(RDA fragmentation of B ring) + H]+, 327.0051 [M − 18(H2O) − 142(side chain on D ring) + H]+, 313.1531 [M − 174(2H2O + RDA fragmentation of B ring) + H]+, 312.1531 [M − 18(H2O) − 15(CH3) − 142(side chain on D ring) + H]+ MS2: 487.3491→469.3290 [M − 18(H2O) + H]+, 451.3169 [M − 36(2H2O) + H]+ |
3-oxo-6,16α-Dihydroxytra-metenolic acid [36] |
| F11 | 44.363 | −−−−− | −−−−−− | −−−−−− | |
| F12 | 45.727 | 481.3333 483.3448 |
MS1: 481.3333 [M − H]−
MS2: 481.3333→437.3338 [M − 44(CO2) − H]−, 435.3259 [M − 46(HCOOH) − H]−, 403.2999 [M − 78(2CH4 + HCOOH) − H]−, 311.2015 [M − 170(2CH4 + RDA fragmentation of B ring) − H]− MS3: 311.2015→293.2008 [M − 170(2CH4 + RDA fragmentation of B ring) − 18(H2O) − H]− |
MS1: 483.3463 [M + H]+, 505.3322 [M + Na]+, 465.3360 [M − 18(H2O) + H]+, 437.3412 [M − 46(HCOOH) + H]+, 327.0080 [M − 156(side chain on D ring) + H]+ or [M − 156(H2O + RDA fragmentation of B ring) + H]+ MS2: 483.3448→465.3357 [M − 18(H2O) + H]+, 447.2191 [M − 36(2H2O) + H]+, 309.2130 [M − 174(H2O + side chain on D ring) + H]+ MS3: 465.3357→447.3255 [M − 18(H2O) − 18(H2O) + H]+, 419.3318 [M − 18(H2O) − 46(HCOOH) + H]+, 309.2194 [M − 18(H2O) − 156(side chain on D ring) + H]+ |
Polyporenic acid C [32] |
| F13 | 49.785 | 483.3478 485.3613 |
MS1: 483.3425 [M − H]−, 295.2370 [M − 188(2CH4 + side chain on D ring) − H]−
MS2: 483.3425→465.2955 [M − 18(H2O) − H]−, 437.3440 [M − 46(HCOOH) − H]−, 311.1998 [M − 172(CH4 + side chain on D ring) − H]−, 295.2036 [M − 188(2CH4 + side chain on D ring) − H]− |
MS1: 485.3602 [M + H]+, 523.3198 [M + K]+, 507.3451 [M + Na]+, 467.3518 [M − 18(H2O) + H]+, 449.3387 [M − 36(2H2O) + H]+, 439.3618 [M − 46(HCOOH) + H]+, 311.1665 [M − 174(H2O+side chain on D ring) + H]+ MS2: 485.3602→467.3512 [M − 18(H2O) + H]+, 449.3496 [M − 36(2H2O) + H]+, 311.2428 [M − 174(H2O + side chain on D ring) + H]+ MS3: 467.3512→449.3409 [M − 18(H2O) − 18(H2O) + H]+, 431.2786 [M − 18(H2O) − 36(2H2O) + H]+, 327.2293 [M − 18(H2O) − 140(RDA fragmentation of B ring) + H]+, 311.2351 [M − 18(H2O) − 156(side chain on D ring) + H]+, 293.2248 [M − 18(H2O) − 174(H2O + side chain on D ring) + H]+, 311.2428→293.2308 [M − 174(H2O + side chain on D ring) − 18(H2O) + H]+, 281.6503 [M − 174(H2O + side chain on D ring) − 30(2CH3) + H]+ |
3-epi-Dehydrotumulosic acid [36] |
| F14 | 62.492 | 511.3436 513.3544 |
MS1: 511.3433 [M − H]−
MS2: 511.3436→467.3499 [M − 44(CO2) − H]−, 451.3122 [M − 60(CH3COOH) − H]−, 355.2211 [M − 156(2H2O + CO2 + CH4 + CH3COOH) − H]− MS3: 467.3499→451.3222 [M − 44(CO2) − 16(CH4) − H]− |
MS1: 513.3544 [M + H]+, 495.3478 [M − 18(H2O) + H]+, 477.3357 [M − 36(2H2O) + H]+, 435.3200 [M − 18(H2O) − 60(CH3COOH) + H]+, 337.6933 [M − 176(2H2O + RDA fragmentation of B ring) + H]+
MS2: 513.3544→495.3446 [M − 18(H2O) + H]+, 477.3298 [M − 36(2H2O) + H]+, 435.3185 [M − 78(H2O + CH3COOH) + H]+, 353.2502 [M − 160(H2O + side chain on D ring) + H]+ MS3: 495.3446→435.3266 [M − 18(H2O) − 60(CH3COOH) + H]+, 353.2445 [M − 18(H2O) − 142(side chain on D ring) + H]+, 293.2276 [M − 18(H2O) − 202(CH3COOH + side chain on D ring) + H]+, 435.3185→293.2244 [M − 78(H2O + CH3COOH) − 142(side chain on D ring) + H]+ |
3β-Hydroxy-16α-acetoxylanosta-7,9(11),24-trien-21-oic acid [36] |
| F15 | 63.130 | 525.3603 527.3735 |
MS1: 525.3581 [M − H]−
MS2: 525.3603→509.3196 [M − 16(CH4) − H]−, 465.3379 [M − 60(CH3COOH) − H]−, 447.3200 [M − 78(H2O + CH3COOH) − H]−, 432.3020 [M − 93(CH3 + H2O + CH3COOH) − H]− |
MS1: 527.3735 [M + H]+, 549.3522 [M + Na]+, 509.3624 [M − 18(H2O) + H]+, 481.3769 [M − 46(HCOOH) + H]+, 467.3539 [M − 60(CH3COOH) + H]+, 449.3400 [M − 78(H2O + CH3COOH) + H]+ MS2: 527.3735→509.3624 [M − 18(H2O) + H]+, 449.3465 [M − 78(H2O + CH3COOH) + H]+ MS3: 509.3624→491.3414 [M − 18(H2O) − 18(H2O) + H]+, 449.3399 [M − 18(H2O) − 60(CH3COOH) + H]+, 353.2453 [M − 18(H2O) − 156(side chain on D ring) + H]+, 293.2240 [M − 18 (H2O) − 216(CH3COOH + side chain on D ring) + H]+, 449.3465→293.2249 [M − 78(H2O + CH3COOH) − 156(side chain on D ring) + H]+ |
Dehydropachymic acid [32] |
| F16 | 65.458 | 513.3579 515.3762 |
MS1: 513.3580 [M − H]−, 487.3071 [M − 36(2H2O) − H]−
MS2: 513.3579→467.3514 [M − 46(HCOOH) − H]−, 453.3324 [M − 60(CH3COOH) − H]−, 451.3184 [M − 46(HCOOH) − 16(CH4) − H]− MS3: 451.3184→391.3126 [M − 46(HCOOH) − 16(CH4) − 60(CH3COOH) − H]− |
MS1: 515.3761 [M + H]+, 497.3646 [M − 18(H2O) + H]+, 479.3530 [M − 36(2H2O) + H]+, 471.3044 [M − 44(CO2) + H]+, 455.3505 [M − 60(CH3COOH) + H], 437.3442 [M − 18(H2O) − 60(CH3COOH) + H]+, 419.3207 [M − 60(CH3COOH) − 36(2H2O) + H]+
MS2: 515.3762→497.3629 [M − 18(H2O) + H]+, 479.3437 [M − 36(2H2O) + H]+, 437.3399 [M − 78(H2O + CH3COOH) + H]+ MS3: 497.3626→437.3391 [M − 18(H2O) − 60(CH3COOH) + H]+, 419.3360 [M − 18(H2O) − 78(H2O + CH3COOH) + H]+, 355.2645 [M − 18(H2O) − 142(side chain on D ring) + H]+, 295.2417 [M − 18(H2O) − 202(CH3COOH + side chain on D ring) + H]+, 437.3391→419.3295 [M − 78(H2O + CH3COOH) − 18(H2O) + H]+, 391.3359 [M − 78(H2O + CH3COOH) − 46(HCOOH) + H], 295.2419 [M − 78(H2O + CH3COOH) − 142(side chain on D ring) + H]+ |
3-O-Acetyl-16α-hydroxydehydrotra-metenolic acid [37] |
| F17 | 67.750 | 525.3584 527.3719 |
MS1: 525.3581 [M − H]−
MS2: 525.3603→509.3196 [M − H − 16(CH4)]−, 465.3379 [M − 60(CH3COOH) − H]−, 447.3200 [M − H − 78(H2O + CH3COOH)]−, 432.3020 [M − 93(CH3 + H2O + CH3COOH) − H]− |
MS1: 527.3735 [M + H]+, 549.3522 [M + Na]+, 509.3624 [M − 18(H2O) + H]+, 481.3769 [M − 46(HCOOH) + H]+, 467.3539 [M − 60(CH3COOH) + H]+, 449.3400 [M − 78(H2O + CH3COOH) + H]+ MS2: 527.3735→509.3624 [M − 18(H2O) + H]+, 449.3465 [M − 78(H2O + CH3COOH) + H]+ MS3: 509.3624→491.3414 [M − 18(H2O) − 18(H2O) + H]+, 449.3399 [M − 18(H2O) − 60(CH3COOH) + H]+, 353.2453 [M − 18(H2O) − 156(side chain on D ring) + H]+, 293.2240 [M − 18(H2O) − 216(CH3COOH + side chain on D ring) + H]+, 449.3465→293.2249 [M − 78(H2O + CH3COOH) − 156(side chain on D ring) + H]+ |
3-epi-Dehydro-pachymic acid [37] |
| F18 | 76.858 | 587.3756 589.3883 |
MS1: 587.3756 [M − H]−
MS2: 587.3756→465.3296 [M − 122(HCO−Ar−OH) − H]− |
MS1: 589.3883 [M + H]+, 611.3583 [M + Na]+, 571.3853 [M − 18(H2O) + H]+, 449.3413 [M − 18(H2O) − 122(HCO−Ar−OH) + H]+, 430.9047 [M − 36(2H2O) − 122(HCO−Ar−OH) + H]+, 406.2653 [M − 15(CH3) − 46(HCOOH) − 122(HCO−Ar−OH) + H]+
MS2: 589.3883→571.3813 [M − 18(H2O) + H]+ MS3: 571.3813→449.3361 [M − 18(H2O) − 122(HCO−Ar−OH) + H]+, 415.2603 [M − 18(H2O) − 156(side chain on D ring) + H], 403.0557 [M − 18(H2O) − 168(HCOOH+HCO−Ar−OH) + H]+, 293.2261 [M − 18(H2O) − 278(HCO−Ar−OH+ side chain on D ring) + H]+, 449.3413→419.0319 [M − 18(H2O) − 122(HCO−Ar−OH) − 30(2CH3) + H]+, 293.2260 [M − 18(H2O) − 122(HCO−Ar−OH) − 156(side chain on D ring) + H]+ |
3β-p-Hydroxybenzoyl-dehydrotumulosic acid [4] |
| F19 | 78.300 | 467.3152 469.3617 |
MS1: 467.3152 [M − H]−, 439.3557 [M − 28(CO) − H]−
MS2: 467.3152→451.3368 [M − 16(CH4) − H]−, 421.3552 [M − 46(HCOOH) − H]−, 292.9842 [M − 174(H2O + side chain on D ring) − H]− |
MS1: 469.3617 [M + H]+, 451.3574 [M − 18(H2O) + H]+
MS2: 469.3617→451.3574 [M − 18(H2O) + H]+, 433.3207 [M − 36(2H2O) + H]+, 423.3302 [M − 46 (HCOOH) + H]+, 328.9961 [M − 140(RDA fragmentation of B ring) + H]+, 313.2356 [M − 156(side chain on D ring) + H]+ MS3: 451.3574→433.3207 [M − 18(H2O) − 18(H2O) + H]+, 311.2370 [M − 18(H2O) − 140(RDA fragmentation of B ring) + H]+, 295.2413 [M − 18(H2O) − 156(side chain on D ring) + H]+ |
Dehydroeburicoic acid [35] |
| a1 | 26.285 | 471.3478 473.3639 |
MS1: 471.3478 [M − H]−
MS2: 471.3478→409.3100 [M − 62(CH4 + HCOOH) − H]− |
MS1: 473.3639 [M + H]+, 495.3469 [M + Na]+, 511.3243 [M + K]+, 457.3665 [M − 16(CH4) + H]+, 455.3527 [M − 18(H2O) + H]+, 437.3413 [M − 36(2H2O) + H]+, 429.2905 [M − 44(CO2) + H]+, 317.6939 [M − 156(CH4 + RDA fragmentation of B ring) + H]+
MS2: 473.3639→455.3515 [M − 18(H2O) + H]+, 437.3438 [M − 36(2H2O) + H]+ MS3: 455.3508→437.3415 [M − 18(H2O) − 18(H2O) + H]+, 313.2512 [M − 18(H2O) − 142(side chain on D ring) + H]+, 295.2432 [M − 18(H2O) − 160(H2O + side chain on D ring) + H]+, 437.3415→419.3394 [M − 36(2H2O) − 18(H2O) + H]+, 295.2422 [M − 36(2H2O) − 142(side chain on D ring) + H]+ |
16α-Hydroxy-trametenolic acid [34] |
| a2 | 35.110 | 485.3641 487.3779 |
MS1: 485.3641 [M − H]−
MS2: 485.3641→437.3421 [M − 48(3CH4) − H]−, 423.3261 [M − 62(CH4 + HCOOH) − H]− MS3: 423.3261→407.3050 [M − 62(CH4 + HCOOH) − 6(CH4) − H]− |
MS1: 487.3779 [M + H]+, 525.3343 [M + K]+, 509.3610 [M + Na]+, 469.3686 [M − 18(H2O) + H]+, 451.3542 [M − 36(2H2O) + H]+ MS2: 487.3779→469.3669 [M − 18(H2O) + H]+, 451.3542 [M − 36(2H2O) + H]+ MS3: 469.3669→451.3509 [M − 18(H2O) − 18(H2O) + H]+, 313.2502 [M − 18(H2O) − 156(side chain on D ring) + H]+, 295.2419 [M − 18(H2O) − 174(H2O+side chain on D ring) + H]+, 451.3542→433.3188 [M − 36(2H2O) − 18(H2O) + H]+, 309.2200 [M − 36(2H2O) − 142(side chain on D ring) + H]+, 295.2425 [M − 36(2H2O) − 156(H2O + side chain on D ring) + H]+ |
Tumulosic acid [32] |
| a3 | 48.572 | 483.3478 485.3613 |
MS1: 483.3478 [M − H]−
MS2: 483.3478→437.3382 [M − 46(HCOOH) − H]−, 421.3146 [M − 62(CH4 + HCOOH) −H]−, 405.3155 [M − 78(2CH4 + HCOOH) − H]−, 389.2812 [M − 94(3CH4 + HCOOH) − H]−, 369.2392 [M − 114(2CH4 + 2H2O + HCOOH) − H]−, 295.1952 [M − 188(2CH4 + side chain on D ring) − H] |
MS1: 485.3613 [M + H]+, 507.3456 [M + Na]+, 467.3515 [M − 18(H2O) + H]+, 449.3399 [M − 36(2H2O) + H]+, 311.1682 [M − 174(H2O + side chain on D ring) + H]+ or [M − 174(2H2O+RDA fragmentation of B ring) + H]+, 301.1401 [M − 184(HCOOH+RDA fragmentation of B ring) + H]+ MS2: 485.3613→467.3506 [M − 18(H2O) + H]+, 449.3390 [M − 36(2H2O) + H]+, 311.2428 [M − 174(H2O + side chain on D ring) + H]+ MS3: 467.3506→449.3399 [M − 18(H2O) − 18(H2O) + H]+, 431.3310 [M − 18(H2O) − 36(2H2O) + H]+, 421.3452 [M − 18(H2O) − 46(HCOOH) + H]+, 311.2353 [M − 18(H2O) − 156(side chain on D ring) + H]+, 293.2247 [M − 18(H2O) − 174(H2O + side chain on D ring) + H]+, 449.3390→431.3378 [M − 36(2H2O) − 18(H2O) + H]+, 403.3292 [M − 36(2H2O) − 46(HCOOH) + H]+, 293.2258 [M − 36(2H2O) − 156(side chain on D ring) + H]+, 311.2428→293.2250 [M − 174(H2O + side chain on D ring) − 18(H2O) + H]+ |
Unknown |
| a4 | 70.508 | 527.3730 529.3897 |
MS1: 527.3730 [M − H]−
MS2: 527.3730→481.3658 [M − 46(HCOOH) − H]−, 465.3329 [M − 62(CH4 + HCOOH) − H]−, 431.2794 [M − 96(2H2O + CH3COOH) − H]−, 405.3045 [M − 122(CH3COOH + HCOOH + CH4) − H]− MS3: 465.3329→405.3168 [M − 62(CH4 + HCOOH) − 60(CH3COOH) − H]− |
MS1: 567.3456 [M + K]+, 551.3703 [M + Na]+, 529.3897 [M + H]+, 511.3759 [M − 18(H2O) + H]+, 493.3662 [M − 36(2H2O) + H]+, 469.3707 [M − 60(CH3COOH) + H]+, 451.3572 [M − 78(H2O + CH3COOH) + H]+
MS2: 529.3897→511.3764 [M − 18(H2O) + H]+, 451.3559 [M − 78(H2O + CH3COOH) + H]+ MS3: 511.3764→451.3555 [M − 18(H2O) − 60(CH3COOH) + H]+, 433.3480 [M − 18(H2O) − 78(H2O + CH3COOH) + H]+, 355.2589 [M − 18(H2O) − 156(side chain on D ring) + H]+, 295.2407 [M − 18(H2O) − 216(CH3COOH + side chain on D ring) + H]+, 451.3559→433.3485 [M − 78(H2O + CH3COOH) − 18(H2O) + H]+, 405.3540 [M − 78(H2O + CH3COOH) − 46(HCOOH) + H]+, 295.2412 [M − 78(H2O + CH3COOH) − 156(side chain on D ring) + H]+, 295.2407→280.2181 [M − 78(H2O + CH3COOH) − 156(side chain on D ring) − 15(CH3) + H]+ |
Pachymic acid [32] |
Figure 5.
The chemical structures of the identified compounds.
As shown in Table 2, in the positive mode ESI-MS1 spectra, the [M + H]+ and [M − H2O + H]+ ions were observed for all 23 compounds except for compound F11. The [M + Na]+ ions were seen for all the compounds except for compounds F1–F3, F6, F11, F16 and F19. The [M − 2H2O + H]+ ions were seen for all the compounds except for compounds F2, F4, F11, F12, F15, F17, F18 and F19. Compounds F6, F14–F17 and a4 showed the corresponding [M − H2O − CH3COOH + H]+ ions. [M + K]+ ions were observed for compounds F5, F8, F13, a1, a2 and a4. The [M − HCOOH + H]+ ions were found for compounds F8, F12, F13, F15 and F17. [M − 2H2O − RDA fragmentation of B ring + H]+ ions were found for compounds F7, F10, F14 and a3. The [M − CH3COOH + H]+ ions were observed for compounds F15−F17 and a4. Compounds F8, F13 and a3 presented [M − H2O − side chain on D ring + H]+ ions, while F3 and a1 showed [M − CH3 − RDA fragmentation of B ring + H]+ ions. [M − 2H2O − CH3COOH + H]+ ions were found for compounds F6 and F16. [M − RDA fragmentation of B ring + H]+ ions were found for compounds F6 and F10. Compounds F7 and F12 presented [M − H2O − RDA fragmentation of B ring + H]+ ions, while F7 and F10 displayed [M − CH3 − side chain on D ring + H]+ions and F7 also displayed an [M − 2CH3 − side chain on D ring + H]+ ion. The [M − CO2 + H]+ ion was observed compounds a1 and F16.Compound F9 presented [M − CH3 − H2O − side chain on D ring + H]+, [M − 2CH3 − H2O − HCOOH + H]+ and [M − 2H2O − HCOOH − RDA fragmentation of B ring + H]+ ions, while F10 had [M − 3H2O + H]+, [M − 2H2O − HCOOH + H]+ and [M − H2O − CH3 − side chain on D ring + H]+ ones. The [M − side chain on D ring + H]+ ion was seen in the spectrum of compound F12. A [M − HCOOH − RDA fragmentation of B ring + H]+ ion was seen for compound a3. Compound a2, on the other hand, displayed [M − H2O − HCO−Ar−OH + H]+, [M − 2H2O–HCO–Ar–OH + H]+ and [M − CH3 − HCOOH–HCO–Ar–OH + H]+ ions, whereas a1 had an [M − CH4 + H]+ ion.
In the ESI-MS2 spectra, all 23 compounds except for compound F11 displayed the corresponding [M − H2O + H]+ ions.[M − 2H2O + H]+ ions were found for all the compounds except for compounds F2, F4, F5, F11, F15, F17, F18 and a4. Compounds F2, F6–F9, F12–F14 and a3 showed [M − (H2O + side chain on D ring) + H]+ ions and compounds F5, F14−F17 and a4 showed [M − (H2O + CH3COOH) + H]+ ones. [M − side chain on D ring + H]+ ions were seen for compoundsF4, F19, while F6 and F19 had [M − RDA fragmentation of B ring + H]+ ions. The [M − RDA fragmentation of B ring − 2H2O + H]+ ion was observed for F4 and F9. Only F2 had [M − 3H2O + H]+ and [M − 2H2O − HCOOH + H]+ ions. The ions [M − 2CH3 + H]+, [M − 2CH3 − H2O + H]+, [M − 2CH3 − RDA fragmentation of B ring + H]+, [M − RDA fragmentation of B ring − 2H2O − HCOOH + H]+ and [M − side chain on D ring − 2CH3 + H]+ were only found for compound F4, and only compound F5 had a [M − (2H2O + CH3COOH) + H]+ ion. Other ions seen in only one compound were [M − side chain on D ring − 2H2O − CH3COOH + H]+ in compound F6 and [M − HCOOH + H]+ for F19.
In the ESI-MS3 spectra all 23 compounds except for compounds F2, F11 and F18 displayed [MS2 − H2O + H]+ ions, while compounds F1, F3, F6–F9 and a3 also showed a [MS2 − 2H2O + H]+ ion. All 23 compounds except for F4 and F11 had [MS2 − side chain on D ring + H]+ ions. The [MS2 − side chain on D ring − H2O + H]+ ion was observed in the spectra of compounds F1−F3, F5, F6, F8, F13 and a1–a4 and F1, F3, F6–F9 and a3 showed a [MS2 − 2H2O + H]+ ion. [MS2 − HCOOH + H]+ ions were seen for compounds F3, F6, F9, F12, F16, a3 and a4, while F14−F17 and a4 showed both [MS2 − CH3COOH + H]+ and [MS2 − CH3COOH − side chain on D ring + H]+ ions. Compounds F4, F8, F13 and F18 produced [MS2 − 2CH3 + H]+ ions and F8, F13 and F19 had an ion corresponding to a [MS2 − RDA fragmentation of B ring + H]+ species. The [MS2 − CH3COOH − H2O + H]+ ion was noted for compounds F16 and a4. The latter compound also had a [MS2 − CH3 + H]+ ion. [MS2 − HCOOH − H2O + H]+ ions were found for compounds F1 and F3. Compound F5 displayed a [MS2 − CH3 − H2O + H]+ ion while compound F4 showed [MS2 − RDA fragmentation of B ring − H2O + H]+ and [MS2 − RDA fragmentation of B ring − H2O − 2CH3 + H]+ ones and compound F9 showed a [MS2 − RDA fragmentation of B ring − 2H2O + H]+ ion. The [MS2 − HCO–Ar–OH + H]+, [MS2 − HCOOH − (HCO–Ar–OH) + H]+ and [MS2 − side chain on D ring − (HCO–Ar–OH) + H]+ ions were observed in the spectrum of compound F18.
In the negative mode ESI-MS1 spectra, the [M − H]− ions were found for all 23 compounds except for compound F11. Compound F6 had [M − CH3COOH − H]− and [M − side chain on D ring − H]− ion. Compound F8 presented [M − 2CH3 − side chain on D ring − H]−, [M − 4CH3 − HCOOH − RDA fragmentation of B ring − H]−, [M − H2O − H]− and [M − 3CH3 − H]− ions, while compound F10 had [M − CH4 − H]− and [M − CH4 − 2H2O − HCOOH − RDA fragmentation of B ring − H]− ions. A [M − K]− ion was found for compound F12. [M − 2CH4 − side chain on D ring − H]−, [M − 2H2O − H]− and [M − CO − H]− ions were only seen for compounds F13, F16 orF19, respectively.
In the ESI-MS2 spectra, [M − H2O − H]− ions were found for compounds F1, F3, F7–F9 and F13, while [M − CH4 − HCOOH − H]− ions were seen for compounds F1, F10, F16 and a1–a4. Meanwhile, compounds F8, F12, F13, F16, a3 and a4 showed a [M − HCOOH − H]− ion and F6, F14–F17 formed a [M − CH3COOH − H]− ion. The [M − CO2 − H]− ion was seen for compounds F5, F10, F12 and F14 and F15, F17 and F19 displayed a [M − CH4 − H]− ion. The [M − 2CH4 − HCOOH − H]− ion was observed for compounds F3, F12 and a3. The latter compound, F8 and F13 displayed [M − 2CH4 − side chain on D ring − H]− ions. [M − 2CH4 − H]− ions were found for compounds F1 and F2, while F1 and F19 had [M − H2O − side chain on D ring − H]− ions. Compounds F3 and a3 showed a [M − 2CH4 − H2O − HCOOH − H]− ion, F8 and F13 had a [M − CH4 − side chain on D ring − H]− ion and F9 and a2 had [M − 3CH3 − H]− ion. [M − H2O − CH3COOH − H]− and [M − H2O − CH4 − CH3COOH − H]− ions were found for compounds F15 and F17. Compound a3 had [M − 2H2O − 2CH4 − HCOOH − H]− and [M − 3CH4 − HCOOH − H]− ions, whereas a4 showed [M − 2H2O − CH3COOH − H]− and [M − CH4 − HCOOH − CH3COOH − H]− ions and a2 showed a [M − 3CH4 − H]− ion. Compound F10 presented a [M − CH4 − 2H2O − HCOOH − RDA fragmentation of B ring − H]− ion while compound F1 had a [M − CH4 − H2O − HCOOH − H]− one. A [M − H2O − HCOOH − H]− ions was observed for compound F3. Compound F4 showed a [M − 2CH3 − H]− ion. The [M − CH4 − HCOOH − RDA fragmentation of B ring − H]− ion was observed for compound F9 and compound F12 presented a peak for a [M − 2CH4 − RDA fragmentation of B ring − H]− ion. The [M − 2H2O − CH4 − CO2 − CH3COOH − H]− ion was observed in the spectrum of compound F14 and an [M − HCO−Ar−OH −H]− ion was found for compound F18.
In the ESI-MS3 spectra, [MS2 − CH4 − H]− ions were seen for compounds F1, F3, F5, F14 and a2, while compounds F16 and a4 presented [MS2 − CH3COOH − H]− ions and compound 2 showed a [MS2 − CH3 − H]− ion. Compound F4 showed [MS2 − HCOOH − H]− and [MS2 − 2CH4 − HCOOH − H]− ions. Finally, compound F12 showed the corresponding [MS2 − H2O − H]− ion.
2.4. Cluster Analysis (CA)
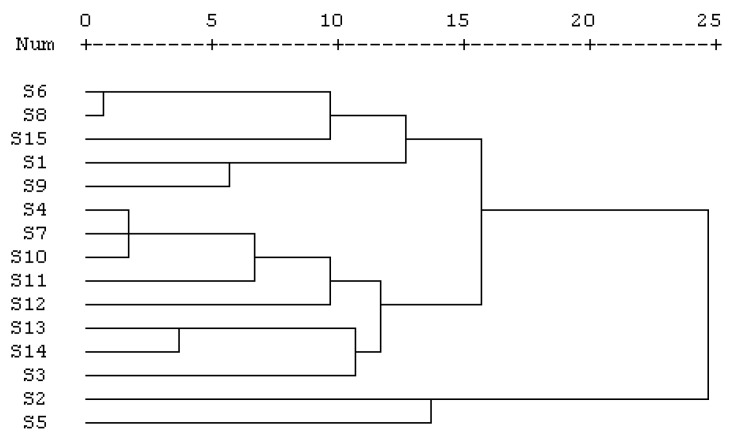
Cluster Analysis is a multivariate analysis technique that is used to sort samples into groups. It is widely applied for fingerprint analysis, because it is a nonparametric data interpretation method and simple to use. CA provides a visual representation of complex data. Average linkage between groups was applied, and Pearson correlation was selected as a measurement. The method can classify different herbs by measuring the peak areas from their corresponding HPLC fingerprints. The common characteristic peaks, which were calculated by the Similarity Evaluation System, were selected for the CA. Cluster analysis of P. cocos samples was performed based on the relative peak areas of all 19 common peaks.
The results of CA are shown in Figure 6, where the quality characteristics are revealed more clearly. The cluster analysis results show that the samples could be divided into three quality clusters. Among them, Cluster I includes the samples S6, S8, S15, S1 and S9, Cluster III includes S2, S5 and the others are in Cluster II. All the compounds in Cluster III had much higher concentrations than the other two clusters.
Figure 6.
Results of cluster analysis of 15 samples.
Cluster I was distinguished as it contains less 3-epi-dehydrotumulosic acid (F13), 3-oxo-16α,25-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid (F4), 3-oxo-6,16α-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid (F7), dehydropachymic acid (F15), Unknown F9, and Unknown F11 than Clusters II and III. The low concentration of these six compounds in Cluster I may be due to the poor herb quality of P. cocos. This indicated that these compounds could be used as marker compounds to distinguish the P. cocos samples with different quality. The results of CA could be validated against each other and provided more references for the quality evaluation of P. cocos.
2.5. Principal Components Analysis (PCA)
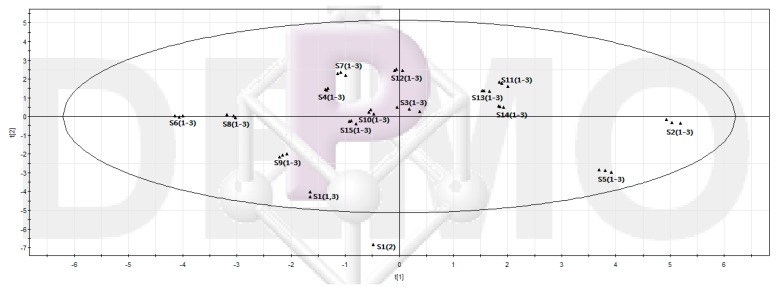
To evaluate the variations in quality of the 15 samples, PCA was carried out with the relative amounts of each identified component. The contents of 19 fingerprint peaks were applied to evaluate the sample variations. Figure 7 shows the score plots obtained by PCA. The first six principal components accounted for 89.329% of the total variance. Examination of the score plots indicates that the main components responsible for the separation were 3-epi-dehydrotumulosic acid (F13), 6α-hydroxyldehydropachymic acid (F6), 24(31)-trien-21-oic acid (F4), 24(31)-trien-21-oic acid (F7), 3-oxo-6,16α-dihydroxylanosta-7, 9 (F15), 29-hydroxydehydrotumulosic acid (F1), dehydropachymic acid (F12), as shown in Table 3. These components were deemed to be the marker compounds of sample variation. This result is in accord with the one obtained from the cluster analysis (CA). The combination of PCA and CA was thus a useful tool for quality control and evaluation of P. cocos.
Figure 7.
PCA scores plots of the sample from different regions with 95% confidence ellipses.
Table 3.
The factor loading matrix.
| Peak No. | Six Principal Components a | |||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| F13 | 0.855 | 0.027 | −0.389 | −0.055 | 0.071 | −0.212 |
| F6 | 0.848 | 0.165 | −0.113 | 0.030 | −0.367 | 0.183 |
| F4 | 0.808 | 0.190 | −0.015 | −0.474 | −0.006 | 0.130 |
| F7 | 0.754 | −0.186 | 0.255 | 0.214 | 0.214 | −0.207 |
| F15 | 0.744 | −0.352 | 0.089 | −0.293 | −0.100 | 0.251 |
| F1 | 0.648 | 0.133 | −0.260 | 0.309 | 0.279 | 0.457 |
| F12 | 0.596 | 0.529 | −0.359 | −0.396 | 0.258 | −0.030 |
| F16 | 0.559 | −0.080 | 0.528 | −0.289 | −0.460 | 0.028 |
| F9 | 0.549 | −0.499 | 0.407 | −0.127 | 0.263 | −0.281 |
| F11 | 0.535 | −0.533 | 0.160 | 0.245 | 0.322 | −0.349 |
| F8 | −0.310 | 0.810 | −0.045 | −0.240 | 0.329 | 0.042 |
| F10 | −0.244 | 0.768 | 0.174 | 0.190 | −0.346 | −0.279 |
| F17 | 0.516 | 0.707 | 0.223 | −0.075 | −0.108 | −0.303 |
| F5 | −0.124 | 0.688 | 0.232 | −0.054 | 0.645 | 0.114 |
| F18 | 0.186 | 0.604 | 0.349 | −0.222 | −0.114 | 0.050 |
| F19 | 0.032 | −0.208 | 0.767 | 0.186 | 0.280 | 0.446 |
| F3 | 0.540 | 0.102 | −0.630 | 0.494 | 0.133 | −0.015 |
| F2 | 0.383 | 0.397 | 0.069 | 0.614 | −0.458 | 0.180 |
| F14 | 0.267 | 0.500 | 0.474 | 0.505 | 0.168 | −0.120 |
Extraction method: principal components. a The six components has extracted.
3. Experimental Section
3.1. Samples and Reagents
Fifteen P. cocos samples were purchased from different regions of China and were authenticated by Professor Chun-Sheng Liu (School of Chinese Materia Medica, Beijing University of Chinese Medicine, Beijing, China). The samples were harvested between July and September. The samples were processed as follows: the sediment was removed after them digging up, and the material was piled to “sweat”, spread out until the surface was dry, then “sweated” again. This was repeated several times until the surface of the samples was wrinkled and the water in the sample was almost dissipated. Samples were then dried in the shade, peeled and cut into cubes. The surface of the blocks is white or faint red in color. Each sample (three replicates) was placed in a dark and dry environment. The regions where the 15 samples were obtained are shown in Table 4. Pachymic acid (Batch number: 130306, purity ≥ 98%) and dehydroeburicoic acid (Batch number: 131027, purity ≥ 98%) were obtained from Chengdu MUST BioTechnology Co., Ltd. (Chengdu, China); HPLC grade acetonitrile and acetic acid were obtained from Fisher (Waltham, MA, USA); distilled water was bought from Watsons (Beijing, China) and was filtered through a 0.45 µm membrane (Dikma, Beijing, China) prior to use. All other reagents were of analytical grade.
Table 4.
The regions of origin of the 15 samples.
| No. | Region | No. | Region |
|---|---|---|---|
| S1 | Yuxi, Yunnan | S9 | Xiangxi, Hunan |
| S2 | Chuxiong, Yunnan | S10 | Xinxiang, Henan |
| S3 | Dali, Yunnan | S11 | Yulin, Guangxi |
| S4 | Lijiang, Yunnan | S12 | Jinzhai, Anhui |
| S5 | Luotian, Hubei | S13 | Chengdu, Sichuan |
| S6 | Shennongjia, Hubei | S14 | Suining, Sichuan |
| S7 | Yundu, Guizhou | S15 | Yuexi, Anhui |
| S8 | Fujian |
3.2. Sample Preparation
3.2.1. Preparation of Reference Substance
Stock solutions of individual reference substance were prepared by dissolving each compound in 50% methanol at a concentration of 212 µg·mL−1 for pachymic acid and 22.9 µg·mL−1 for dehydroeburicoic acid. Both solutions were stored at approximately 4 °C.
3.2.2. Preparation of Sample Solution
Dried powder of P. cocos from different regions (1 g) was accurately weighed out and transferred into a 100 mL conical flask. Methanol (10 mL) was added to the flask and the flask with the methanol and powder was accurately weighed and placed in an ultrasonic extraction device and extracted for 30 min. The flask was weighed again and methanol was added to make up the weight. The solution was filtered through a 0.45 µm membrane filter for fingerprint analysis.
3.3. Apparatus and Parameters
A Waters Alliance HPLC 2695 series instrument (Waters, Manchester, UK) was used to perform the high performance liquid chromatography (HPLC) analysis. Mobile phase: A (acetonitrile); B (H2O:CH3COOH, 100:0.2, v/v). Column: Diamansil™ C18 (250 × 4.6 mm, 5 μm), maintained at 30 °C with flow rate of 1.0 mL·min. The detection wavelength was set at 254 nm for acquiring chromatograms. The injection volume was 20 µL. Gradient elution procedure: 0 min (45% A) → 8 min (55% A) → 22 min (55% A) → 55 min (65% A) → 56 min (70% A) → 80 min (90% A).
The LCMS-IT-TOF instrument (Shimadzu, Kyoto, Japan) was equipped with an ESI source used in positive and negative ionization mode. The interface and MS parameters were as follows: nebulizer pressure, 100 kPa; dry gas, N2 (1.5 L/min); drying gas temperature, 200 °C; spray capillary voltage, 4000 V; scan range, m/z 100–1500.
4. Conclusions
The therapeutic effects of traditional Chinese medicines (TCM) are based on the complex interactions of complicated chemical constituents as a whole system. HPLC and HPLC-MSn fingerprint analysis combined with chemometrics were employed to study the complex P. cocos system. Triterpenoid acids were the most important chemical components in the samples, which had a variety of potential biological activities, according to previous extensive phytochemical and pharmacological studies. The qualitative analysis and quantification of triterpenoid acids can better reflect the therapeutic effects and quality of P. cocos. The chromatographic method is predominantly to control the quality and stability of the complex system. This study provided a systematic method for the quality control of P. cocos by HPLC fingerprinting and the HPLC-MSn evaluation system based on Similarity Analysis (SA), Cluster Analysis (CA) and Principal Component Analysis (PCA). As a result, a common mutual pattern was established by determining and comparing the fingerprints of 15 samples of P. cocos from different regions. Twenty-three compounds were detected by HPLC-MSn, of which twenty were tentatively identified by comparing their retention times, and mass spectrometry data with that of reference compounds and literature data. The characteristic fragmentations were summarized. 3-epi-Dehydrotumulosic acid (F13), 3-oxo-16α,25-dihydroxy-lanosta-7,9(11),24(31)-trien-21-oic acid (F4), 3-oxo-6,16α-dihydroxylanosta-7,9(11),24(31)-trien-21-oic acid (F7) and dehydropachymic acid (F15) were deemed to be the markers to distinguish between P. cocos samples of different quality. The proposed method can be used to improve the quality control of P. cocos, thus ensuring the effectiveness of Poria herbs. There are still three peaks—F9, F11 and a3—which were not identified by HPLC-MSn, of which F9 and F11 were used as marker compounds to distinguish the P. cocos of different quality. These two components require further study.
Acknowledgments
The authors gratefully acknowledge the financial support from the Ministry of Science and Technology support project (No. 2012BAI29B01).
Author Contributions
Conceived and designed the experiments: Lan-Zhen Zhang, Ling-Fang Wu. Performed the experiments: Ling-Fang Wu, Kun-Feng Wang, Xin-Mao, Wen-Jing Chen, Wen-Yi Liang, Shi Li, Qi Qi, Ya-Ping Cui. Analyzed the data: Ling-Fang Wu. Wrote the paper: Ling-Fang Wu, Lan-Zhen Zhang.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds Pachymic acid and Dehydroeburicoic acid are available from the authors.
References
- 1.Zhang M., Chiu L., Cheung P., Ool V. Growth-inhibitory effects of a β-glucan from the mycelium of Poria cocos on human breast carcinoma MCF-7 cells cell-cycle arrest and apoptosis induction. Oncol. Rep. 2006;15:637–643. doi: 10.3892/or.15.3.637. [DOI] [PubMed] [Google Scholar]
- 2.Huang Q.L., Jin Y., Zhang L.N. Structure, molecular size and antitumor activities of polysaccharides from Poria cocos mycelia produced in fermenter. Carbohydr. Polym. 2007;70:324–333. doi: 10.1016/j.carbpol.2007.04.015. [DOI] [Google Scholar]
- 3.Chen Y.Y., Chang H.M. Antiproliferative and differentiating effects of polysaccharide fraction from fu-ling (Poria cocos) on human leukemic U937 and HL-60 cells. Food. Chem. Toxicol. 2004;42:759–7691. doi: 10.1016/j.fct.2004.01.018. [DOI] [PubMed] [Google Scholar]
- 4.Akihisa T., Uchiyama E., Kikuchi T., Tokuda H., Suzuki T., Kimura Y. Anti-tumor-promoting effects of 25-methoxyporicoic acid A and other triterpene acids from Poria cocos. J. Nat. Prod. 2009;72:1786–1792. doi: 10.1021/np9003239. [DOI] [PubMed] [Google Scholar]
- 5.Akihisa T., Nakamura Y., Tokuda H., Uchiyama E., Suzuki T., Kimura Y., Uchikura K., Nishino H. Triterpene acids from Poria cocos and their anti-tumor-promoting effects. J. Nat. Prod. 2007;70:948–953. doi: 10.1021/np0780001. [DOI] [PubMed] [Google Scholar]
- 6.Ukiya M., Akihisa T., Tokuda H., Hirano M., Oshikubo M., Nobukuni Y. Inhibition of tumor-promoting effects by poricoic acids G and H and other lanostane-type triterpenes and cytotoxic activity of poricoic acids A and G from Poria cocos. J. Nat. Prod. 2002;65:462–465. doi: 10.1021/np0103721. [DOI] [PubMed] [Google Scholar]
- 7.Kikuchi T., Uchiyama E., Ukiya M., Tabata K., Kimura Y., Suzuki T., Akihisa T. Cytotoxic and apoptosis-inducing activities of triterpene acids from Poria cocos. J. Nat. Prod. 2011;74:137–144. doi: 10.1021/np100402b. [DOI] [PubMed] [Google Scholar]
- 8.Gapter L., Wang Z., Glinski J., Ng K.Y. Induction of apoptosis in prostate cancer cells by pachymic acid from Poria cocos. Biochem. Biophys. Res. Commun. 2005;332:1153–1161. doi: 10.1016/j.bbrc.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 9.Ling H., Zhang Y.C., Ng K.Y., Chew E.H. Pachymic acid impairs breast cancer cell invasion by suppressing nuclear factor-κB-dependent matrix metalloproteinase-9 expression. Breast Cancer Res. Treat. 2011;126:609–620. doi: 10.1007/s10549-010-0929-5. [DOI] [PubMed] [Google Scholar]
- 10.Ling H., Zhou L., Jia X., Gapter L.A., Agarwal R., Ng K.Y. Polyporenic acid C induces caspase-8-mediated apoptosis in human lung cancer A549 cells. Mol. Carcinogen. 2009;48:498–507. doi: 10.1002/mc.20487. [DOI] [PubMed] [Google Scholar]
- 11.Ke R.D., Lin S.F., Chen Yi., Ji C.R., Shu Q.G. Analysis of chemical composition of polysaccharides from Poria cocos Wolf and its anti-tumor activity by NMR spectroscopy. Carbohydr. Polym. 2010;80:31–34. [Google Scholar]
- 12.Lu M.K., Cheng J.J., Lin C.Y., Chang C.C. Purification, structural elucidation, and anti-inflammatory effect of a wate-soluble 1, 6–branched 1, 3-α-d-galactan from cultured wycelia of Poria cocos. Food. Chem. 2010;2:349–356. doi: 10.1016/j.foodchem.2009.04.126. [DOI] [Google Scholar]
- 13.Deng J.S., Huang S.S., Lin T.H., Lee M.M., Kuo C.C., Sung P.J., Hou W.C., Huang G.J., Kuo Y.H. Analgesic and anti-inflammatory bioactivities of eburicoic acid and dehydroeburicoic acid isolated from Antrodia camphorata on the inflammatory mediator expression in mice. J. Agric. Food. Chem. 2013;61:5064–5071. doi: 10.1021/jf303820k. [DOI] [PubMed] [Google Scholar]
- 14.Jeong J.W., Lee H.H., Han M.H., Kim G.Y., Hong H., Park C., Choi Y.H. Ethanol extract of Poria cocos reduces the production of inflammatory mediators by suppressing the NF-κB signaling pathway in lipopolysaccharide-stimulated RAW 264.7 macrophages. BMC Complement. Altern. Med. 2014;14:101. doi: 10.1186/1472-6882-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.GinerLarza E.M., Manez S., GinerPons R.M., CarmenRecio M., Rios J.L. On the anti-inflammatory and anti-phospholipase A(2) activity of extracts from lanostane-rich species. J. Ethnopharmacol. 2000;73:61–69. doi: 10.1016/S0378-8741(00)00276-2. [DOI] [PubMed] [Google Scholar]
- 16.Yasukawa K., Kaminaga T., Kitanaka S., Tai T., Nunoura Y., Natori S., Takido M. 3β-p-Hydroxybenzoyldehydrotumulosic acid from Poria cocos, and its anti-inflammatory effect. Phytochemistry. 1998;48:1357–1360. doi: 10.1016/S0031-9422(97)01063-7. [DOI] [PubMed] [Google Scholar]
- 17.Lee H.C., Cheng W.Y., Huang B., Hsu Y.H., Huang S.Y. Anti-inflammatory and hypoglycemic efficacy of Poria cocos and Dioscorea opposita in prediabetes mellitus rats. Rsc. Adv. 2014;4:55649–55657. doi: 10.1039/C4RA10539G. [DOI] [Google Scholar]
- 18.Fuchs S.M., Heinemann C., Fluhr J.W., Schliemann-Willers S., Grafe U., Elsner P. Anti-inflammatory efficacy of Poria cocos in SLS induced irritant contact dermatitis and UVB-induced erythema. Skin Res. Technol. 2003;2:178–179. doi: 10.1111/j.0909-752X.2006.00168.x. [DOI] [PubMed] [Google Scholar]
- 19.Spelman K., Burns J.J., Nichols D., Winters N., Ottersberg S., Tenborg M. Modulation of cytokine expression by traditional medicines: A review of herbal immunomodulators. Altern. Med. Rev. 2006;11:128–150. [PubMed] [Google Scholar]
- 20.Ma C.Y., Chang W.C., Chang H.M., Wu J. Immunomodulatory effect of the polysaccharide-rich fraction from sclerotium of medicinal mushroom Poria cocos F.A. Wolf (Aphyllophoromycetideae) on Balb/c Mice. Int. J. Med. Mushrooms. 2010;12:111–121. doi: 10.1615/IntJMedMushr.v12.i2.10. [DOI] [Google Scholar]
- 21.Huang G.J., Deng J.S., Huang S.S., Lee C.Y., Hou W.C., Wang S.Y., Sung P.J., Kuo Y.H. Hepatoprotective effects of eburicoic acid and dehydroeburicoic acid from Antrodia camphorata in a mouse model of acute hepatic injury. Food. Chem. 2013;141:3020–3027. doi: 10.1016/j.foodchem.2013.03.061. [DOI] [PubMed] [Google Scholar]
- 22.Wen Y.Q., Jia B., Peng T. Research on chemical constituents and pharmacological action of polysaccharide in Poria cocos. J. Med. Plants Res. 2014;5:51–54. [Google Scholar]
- 23.Donno D., Beccaro G.L., Carlen C., Ancay A., Cerutti A.K., Mellano M.G., Bounous G. Analytical fingerprint and chemometrics as phytochemical composition control tools in food supplement analysis: characterization of raspberry bud-preparations of different cultivars. J. Sci Food. Agric. 2015 doi: 10.1002/jsfa.7494. [DOI] [PubMed] [Google Scholar]
- 24.Donno D., Boggia R., Zunin P., Cerutti A.K., Guido M., Mellano M.G., Prgomet Z., Beccaro G.L. Phytochemical fingerprint and chemometrics for natural food preparation pattern recognition: An innovative technique in food supplement quality control. J. Food. Sci. Technol. 2015 doi: 10.1007/s13197-015-2115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun H., Chen X., Zhang A., Sakurai T., Jiang J., Wang X. Chromatographic fingerprinting analysis of Zhizhu Wan preparation by high-performance liquid chromatography coupled with photodiode array detector. Pharmacogn. Mag. 2014;10:470–476. doi: 10.4103/0973-1296.141819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yongyu Z., Shujun S., Jianye D., Wenyu W., Huijuan C., Jianbing W., Xiaojun G. Quality Control of Herbal Medicines and Related Areas. InTech; Vienna, Austria: 2011. Quality control method for herbal medicine—Chemical fingerprint analysis; pp. 171–194. [Google Scholar]
- 27.Zhu J.Q., Fan X.H., Cheng Y.Y., Agarwal R., Moore C.M.V., Chen S.T., Tong W.D. Chemometric analysis for identification of botanical raw materials for pharmaceutical use: A case study using Panax notoginseng. PLoS ONE. 2014;9:e87462. doi: 10.1371/journal.pone.0087462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song X., Xie Z.M., Huang D. Study on HPLC Fingerprint of Poria cocos. China Pharm. 2015;15:2109–2111. [Google Scholar]
- 29.Ding G., Wang Z.Z., Zhang C.F., Sheng L.S. Study on HPLC fingerprint of the triterpene acids in Poria cocos. Zhongguo Zhong Yao Za Zhi. 2012;27:756–758. [PubMed] [Google Scholar]
- 30.Li K., Zhang L.Q., Nie J. Study on UPLC-UV-MS fingerprints of different medicinal parts of Poria cocos. Zhong Yao Cai. 2013;36:382–387. [PubMed] [Google Scholar]
- 31.Zhou X., Zhang Y.S., Zhao Y., Gong X.J., Zhao C., Chen H.G. An LC fingerprint study of Poria cocos (Schw.) wolf. Chromatographia. 2009;69:1283–1289. doi: 10.1365/s10337-009-1038-7. [DOI] [Google Scholar]
- 32.Cai T.G., Cai Y. Triterpenes from the fungus Poria cocos and their inhibitory activity on nitric oxide production in mouse macrophages via blockade of activating Protein-1 pathway. Chem Biodivers. 2011;8:2135–2143. doi: 10.1002/cbdv.201100013. [DOI] [PubMed] [Google Scholar]
- 33.Pinhey J.T., Ralph B.J., Simes J.J.H., Wootton M. Extractives of fungi. II. The constituents of Trametes feei. 6α-Hydroxypolyporenic acid C. Aust. J. Chem. 1971;24:609–619. doi: 10.1071/CH9710609. [DOI] [Google Scholar]
- 34.Nukaya H., Yamashiro H., Fuzakawa H. Isolation of inhibitors of TPA-induced mouse ear edema from hoelen, Poria cocos. Chem. Pharm. Bull. 1996;44:847–849. doi: 10.1248/cpb.44.847. [DOI] [PubMed] [Google Scholar]
- 35.Tai T., Shingu T., Kikuchi T., Tezuka Y., Akahori A. Triterpenes from the surface layer of Poria cocos. Phytochemistry. 1995;39:1165–1169. doi: 10.1016/0031-9422(95)00110-S. [DOI] [Google Scholar]
- 36.Wang L.Y., Wan H.J. Studies on the chemical constituents of Fuling (Poria cocos) Chin. Tradit. Herb. Drugs. 1998;3:145–148. [Google Scholar]
- 37.Tai T., Shingu T., Kikuchi T. Isolation of lanostane-type triterpense acids having an acetoxyl group from sclerotia of Poria cocos. Phytochemistry. 1995;40:225–231. doi: 10.1016/0031-9422(95)00182-7. [DOI] [Google Scholar]