Abstract
We describe the infrastructure and functionality for a centralized preclinical and clinical data repository and analytic platform to support importing heterogeneous multi-modal data, automatically and manually linking data across modalities and sites, and searching content. We have developed and applied innovative image and electrophysiology processing methods to identify candidate biomarkers from MRI, EEG, and multi-modal data. Based on heterogeneous biomarkers, we present novel analytic tools designed to study epileptogenesis in animal model and human with the goal of tracking the probability of developing epilepsy over time.
Keywords: biomarkers, EEG, epilepsy, epileptogenesis, informatics, MRI, neuroimaging, TBI
INTRODUCTION
The goal of the Epilepsy Bioinformatics Study for Antiepileptogenic Therapy (EpiBioS4Rx) is to identify relevant biomarkers of epileptogenesis after traumatic brain injury (TBI) and perform rigorous preclinical trials that permit the future design and performance of economically feasible full-scale clinical trials of antiepileptogenic therapies. A fundamental challenge in discovering these biomarkers of epileptogenesis is that this process is multifactorial and crosses multiple modalities. Rather than considering one type of data, we have been collecting and analyzing multi-modal data, including neuroimaging, electrophysiology, and molecular/serological/tissue. Furthermore, to facilitate analysis and collaboration among scientists from various centers around the world, we have created the informatics infrastructure needed for a large dataset of this size. We have also developed innovative analytic tools that are shared with the broader epilepsy research community, including any other interested researchers outside of the epilepsy research community who might find these data useful for their research, so that others may use our tools in addition to their own tools to advance research in this field in general, in addition to identifying biomarkers of epileptogenesis after TBI.
Investigators must have access to a large number of high quality, well-curated data points and study subjects in order for biomarker signals to be detectable above the noise inherent in complex phenomena, such as epileptogenesis, TBI, and conditions of data collection. Additionally, data generating and collecting sites are spread worldwide among different laboratories, clinical sites, heterogeneous data types, and formats, and across multi-center preclinical trials. Before the data can even be analyzed, a central platform is needed to standardize these data and provide tools for searching, viewing, annotating, and analyzing them. By centralizing an enduring data archive, biobank, and analytic tools, researchers may identify and validate biomarkers of epileptogenesis in studies using various types of data. Beyond creating a centralized data repository, we have pioneered innovative standardization/co-registration references, fully supported by novel image and electrophysiology processing methods to extract candidate biomarkers from the diverse data. Not only does a well-curated and standardized multi-modal dataset facilitate the development of models of epileptogenesis, but it also ensures that such models are statistically significant and can be validated.
EEG and MRI Databases
There have been other efforts to create centralized data archives, but it has proven to be especially challenging for human neurophysiological data for many reasons, such as large file sizes, varying formats, privacy constraints, and funding. Two examples of centralized EEG databases that have been developed include Epilepsiae (A. et al., 2009; Ihle et al., 2012; Klatt et al., 2012; Schulze-Bonhage et al., 2010), a European Union-funded project, and IEEG.ORG, an NINDS-funded cloud-based platform (Kini et al., 2016). Epilepsiae stores recordings from 275 individuals with epilepsy, with a total recording time of more than 40,000 hours. Investigators can export the data locally for analysis. IEEG.ORG (Brinkmann et al., 2009; Kini et al., 2016; Wagenaar et al., 2015) hosts academic and clinical datasets of scalp and intracranial EEG, just over 800 of which are shared publicly, from both animal models of epilepsy and patients. This platform uses Amazon cloud services. Access for Epilepsiae is restricted to scientific groups that financially contribute to the maintenance of the database, which has resulted in fewer people using the platform. IEEG.ORG is free and accessible to the epilepsy research community.
The Laboratory of Neuro Imaging (LONI) at the University of Southern California (USC) has experience with many major clinical consortia and big data projects, such as the Biomedical Informatics Resource Network (BIRN)(Astakhov et al., 2005; Helmer et al., 2011), the Alzheimer’s Disease Neuroimaging Initiative (ADNI)(Toga and Crawford, 2010a), the Michael J. Fox Foundation’s Parkinson’s Progressive Markers Initiative (PPMI)(Marek et al., 2011), the Human Connectome Project (HCP)(Marcus et al., 2013; Van Essen et al., 2013), the Big Data to Knowledge (BD2K)(Bourne et al., 2015; Margolis et al., 2014) program, the NIH Autism Centers of Excellence (ACE)(Rakap et al., 2015) program, The Enhancing Neuroimaging Genetics through Meta-Analysis (ENIGMA) Consortium (Thompson et al., 2014), and the Global Alzheimer’s Association Interactive Network (GAAIN)(Toga et al., 2016), among others. The challenges that we have faced in these projects and the knowledge and experience that we have gained from them have informed and guided us to design an optimal platform to focus on clinical and preclinical TBI data in EpiBioS4Rx.
The number of large databases and related neurological disease-focused consortia around the world has grown rapidly in recent years (Lim, 2014), which demonstrates the importance of transparency in large-scale projects and the sharing of data that are collected. The larger datasets from preclinical studies, such as the one generated in EpiBioS4Rx are now emerging. Beyond sharing data, to encourage the most impactful outside collaborations and scientific discoveries, the data must be well organized and annotated (i.e., for EEG). Furthermore, the data sharing platform must be user friendly and straightforward to use. What makes our project unique is the ability to store and share disparate types of data, including imaging, electrophysiology, and clinical data, from both humans and animals, on one platform that includes not only options for data visualization but also a wide variety of analytic tools that are integrated across different programming languages.
LONI INFRASTRUCTURE, DATA STORAGE, AND PROCESSING
The total amount of data that are planned to be collected in EpiBioS4Rx is unprecedented: video-electroencephalography (EEG) from cohorts of animals after TBI (using a fluid percussion model) recorded continuously for six months, in addition to prolonged continuous intensive care unit (ICU) EEG recordings from 300 humans and intermittent sampling of brain images, blood, and tissue data(Vespa et al., n.d.). The data, measured in tens to one hundred terabytes, represent investigation on a scale that was not possible until just recently. It leverages state of the art analysis tools to track candidate biomarkers and their statistical associations.
Study Sites and Patient Population
The study sites include University of California, Los Angeles (UCLA – clinical coordinating center), University of California, Davis, Phoenix Childrens’ Hospital, Yale University, Harvard University/Massachusetts General Hospital, University of Pennsylvania, University of Cincinnati, University of Miami, University of Pittsburgh, Johns Hopkins University, Columbia University, Royal Melbourne Hospital, The Alfred, and Childrens National Hospital. 300 total patients will be enrolled over 4 years, and they will be followed longitudinally for 2 years after injury. Patients admitted into the ICU after an acute moderate-severe TBI involving a frontal and/or temporal lobe hemorrhagic contusion will be screened(Vespa et al., n.d.).
REDCap and LONI IDA Online Databases
ICU physiological data, demographic information, outcome measures, and prospective research data will be uploaded to the Research Electronic Data Capture (REDCap) data repository hosted by LONI. REDCap is a secure, web-based application designed to support data capture for research studies in a metadata-driven manner (Harris, et al., 2009). The online database contains 26 electronic case report forms (eCRFs) designed by the UCLA Brain Injury Research Center. Common data elements have been expanded to collect EEG, MRI, and biosample information. All continuous EEG and neuroimaging data across sites will be uploaded and managed by the USC LONI Online Image and Data Archive (IDA)(Vespa et al., n.d.).
Continuous Scalp and Depth EEG Monitoring to Detect Early Seizures
Enrolled patients will receive 24-hour continuous EEG (cEEG) for 72 hours minimum during the first 7 days after TBI. Scalp cEEG monitoring will be performed at the patient bedside using a 16-21 channel bipolar and referential composite montage implemented at each study center based on their established ICU EEG protocols. A subset of 100 patients will receive additional depth EEG monitoring using a 6-contact mini depth electrode during the first 7 days after TBI for higher resolution as well as pHFOs and repetitive high frequency oscillations and spikes (rHFOSs) detection. 24-hour cEEG files will be de-identified and uploaded to the LONI IDA(Vespa et al., n.d.).
The central analysis team reviews cEEG data using PERSYST Version 13 and MATLAB 8.1 to analyze spikes, pHFOs, and rHFOSs. A structured protocol will be performed to determine interictal epileptiform spike and seizure onset, location of epileptiform onset, spike morphology, spike repetition rate, clustering features, field size, and spread patterns. EEG data will be correlated with MRI data and metabolite plasma data over the first seven days of injury.
Multimodal MRI Analysis for Structural Biomarkers
The initial three standard of care clinical CT scans over the first 24 hours after trauma will be de-identified and uploaded to the LONI IDA to confirm initial injury characteristics. A high-resolution MRI, acquired on a 3T scanner, will be performed on Day 14 (± 4 days) post-injury. MRI sequences acquired include: 3D T1, 2D resting state bold oxygen level dependent imaging (rs-BOLD), 2D diffusion tensor imaging (DTI), 3D Gradient Echo/Susceptibility Weighted Imaging (GRE/SWI), 3D T2, and 3D T2-weighted fluid attenuated inversion recovery (FLAIR). MRI acquisition parameters will be optimized across sites and scanner types to reduce inter-scanner variability.
MRI injury location and total hemorrhagic lesion load will be correlated with post-traumatic epilepsy (PTE) occurrence. T1-weighted MRI scans will be used for 1) a regional volumetric analysis, using FMRIB Software Library (FSL) utilities, and a 2) subcortical morphometric shape analysis, using a validated open-source pipeline robust to brain pathology.
Biosample Collection and Analysis
Study sites will collect blood samples through central lines or venous punctures on post-injury day 1, 3, 5, 15, 30 ± 10, 90 ± 10, and 180 ± 10. Biosample collection, processing, and shipping information will be inputted into the REDCap database. cEEG and/or depth EEG data will be correlated to the biomarker results to characterize the relationship between EEG epileptiform activity and appearance and time course of selected biomarkers.
All longitudinal follow up evaluations will be uploaded to the REDCap database.
We have established the informatics and analytics framework of EpiBioS4Rx at LONI. Continually working closely with data collection sites, we assist in consolidating the data and providing analytic tools that show potential to lead to the identification of biomarkers of epileptogenesis. Well-established communication with the data collection sites ensures that we are constantly improving and optimizing our infrastructure as we receive feedback from our collaborators and outside investigators to make the process more efficient and user friendly. Furthermore, detailed documentation and annotations enable researchers to link different data types easily. For example, a researcher can look at EEG and find the corresponding patient’s imaging data to see spatially from where the EEG recordings were taken (and vice versa). Moreover, researchers will be able to compare clinical data over various time points with the EEG and MRI data. By combining these new data capabilities, which allow investigators to link various data modalities, we have been focusing on discovering quantitative methods, including dimensionality reduction and pattern recognition, of identifying epileptogenesis after TBI.
Moreover, biomarkers and models of epileptogenesis will help define preclinical trial populations, expedite interventions to prevent epilepsy after TBI, and document epilepsy before late seizures occur. Based on previous studies, it is likely that there are reproducible changes in biomarkers, such as occurrence of pathological high frequency oscillations (pHFOs) in the intracranial EEG, which identify the epileptogenic area before its overt clinical expression(Buja et al., 2009; Winden et al., 2015, 2011).
We have implemented new approaches for analyzing the collected data, including novel graphical methods to visualize multivariable interactions and to quantify patterns or variability in the data. Quantitative and data mining methods enable investigators to record and analyze gold-standard data and to create a shared bioinformatics resource for epilepsy research that will continue to exist after this study concludes. We are developing a wide variety of analytic tools for users and integrating multi-modal data in a way that transcends the capability of a single laboratory or center. We are providing a lasting and open platform for standardized biomarker research in both TBI and PTE. Furthermore, because of the existing data on the LONI IDA that have been collected from other projects, researchers may validate EpiBioS4Rx as specific for PTE or not.
Different data often have rich, high-dimensional levels of detail. As such, exploring and navigating through the full data set poses challenges, especially in providing investigators with tools that are easy to use and comprehend. Visualizing data helps to orient investigators and provides context to understand relationships and discover hidden insights within the underlying data. Although in recent years, database solutions have emerged to collect these types of data, they do not provide visualization or exploration functionalities. Applications such as tranSMART offer some analytic capability but require data to be laboriously imported and are not automatically updated when new data are archived. The LONI IDA data visualization interface unites the benefits of visual representations with a comprehensive, harmonized data set and allows creation of subject cohorts and to search, compare, and download data.
Data are being stored on the IDA, because LONI has the ability and experience to store and share petabytes of data (current data storage capacity is over 7PB and increasing each year). In terms of compute hours, we currently limit each user to 129,024 CPU hours on the LONI server per week; this is 768 slots at any given moment (for 24 hours and 7 days). This past year, the average monthly grid usage was approximately 800,000 hours. We intend to continue increasing storage availability as needed, allowing each user more CPU hours to ensure that users can process their data with little to no wait time.
Streamlined Data Consolidation
Users upload their raw data files (of various file types) directly to an extended version of the LONI infrastructure, where they are automatically classified, converted, and annotated (Figure 1). By automating much of this process, researchers uploading and downloading data are spared the time and effort previously involved in accessing and sharing epilepsy data. This streamlined data consolidation aims to increase the financial efficiency and scientific productivity of the broader epilepsy research community. In addition, physical samples are aggregated in a single biobank with our collaborators outside of LONI, further reducing coordination challenges. While LONI has many years of experience with large-scale neuroimaging studies and data integration(Crawford et al., 2016; Dinov et al., 2014; Torgerson et al., 2015; Van Horn and Toga, 2009), with data upload and download using the Image and Data Archive (IDA), we have expanded the types of data that can be uploaded to include EEG using the EDF+ format. Users have the option to select MRI or EEG when uploading or downloading data so that the process remains clear and easy to use. Furthermore, these features are being expanded for preclinical data so that users will be able to upload both preclinical and clinical data on the same platform. Besides the rich clinical data collected as part of EpiBioS4Rx, this will be the first prospective preclinical databank in the world.
Figure 1.

The EpiBioS4Rx portal supports heterogeneous data and provides essential features including upload/ingest, search/visualize, link/co-register, and analyze/annotate for users.
Data transformation software(Barker-Haliski et al., 2014) automatically detects new data, validates them, maps the data to a common data model (where applicable), and pre-indexes the clinical data by features and values to aid in search and co-registration. Through a federated architecture, key components of the data may be distributed across the LONI platform. Quality control and provenance information is maintained with all raw data.
Global Unique Identifiers (GUIDs)
An essential requirement when federating data from multiple research studies is to prevent data from subjects who participate in more than one research study from being multiply-represented in the federated system. Since personally-identifying information, such as first and last names, is regularly removed from the data collected and shared by research studies to protect each subject’s identity, determining what data belong to any given subject across datasets is a difficult, if not impossible, task for investigators who are analyzing the data. Global Unique Identifiers (GUIDs) are used to distinguish subjects uniquely across research studies while preventing the identities of the subjects from being discovered(Johnson et al., 2010).
We have designed and built a GUID system for the third phase of one of our other big data projects, the ADNI (Alzheimer’s Disease Neuroimaging Initiative)(Schneider et al., 2011; Toga and Crawford, 2010b), which is being used to create GUIDs for ADNI subjects. This system is also applied to EpiBioS4Rx. To ensure compatibility with the GUIDs created by the NIH, we have encapsulated the GUID algorithm used by the National Database for Autism Research (NDAR)(Payakachat et al., 2016) system into the Global Alzheimer’s Association Interactive Network (GAAIN) GUID generator, and it serves as our GUID generation engine. We have also developed an algorithm that allows for cross-comparisons of GUIDs between different GUID systems without revealing the internal hash codes used for GUID subject identification, including those stored in the NDAR and FITBIR systems.
Quality Control
The LONI Neuroimaging Quality Control (QC) System(Kim et al., n.d.) is used for all multi-modal data, checked for quality, and reviewed by participating investigators who are collecting the data (Figure 2). Since there is variability in data, annotation, and models among the various data collection sites, we have developed tools to normalize and harmonize signal, image, and other data. Images uploaded from participating centers are processed using LONI’s multicenter data review and assessment system (https://qc.loni.usc.edu). This system allows automated pre-processing that generates vector statistics and derived images to assess data quality. Data are run through automated artifact detection algorithms in preparation for initial biomarker processing(Brinkmann et al., 2009; LeVan et al., 2006). This system is web-accessible, user friendly, simple to navigate, and provides a long-term resource for this field.
Figure 2.
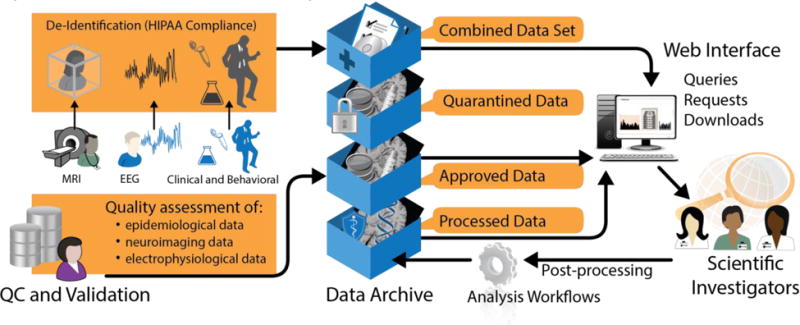
The major elements and functions of our data ingestion and archive.
User-friendly data search and navigation
Data are searchable and accessible through web clients as well as programmatic (MATLAB, C, Python, Java, and R) interfaces. By converting data to consistent file formats and tagging that data with metadata, we have enabled Google-style search of all available epilepsy data. However, since data are interlinked and co-registered across datasets and modalities, the search functionality does not simply match data against individual items as Google does— rather, it will find interlinked combinations of data (even across modalities and data sources) that match the desired criteria(Talukdar et al., 2010). This enables sophisticated custom searches that match the functionality of predefined query forms. Users can browse data in their most appropriate visual representation and pivot from one data view or modality to another. Based on our experience with previous big data studies, we have learned what access control and sharing mechanisms are required by the community and how to effectively enable inter-project as well as community-scale data sharing, which we have now implemented into EpiBioS4Rx. Key components include giving users explicit access control for their data and results as well as providing project groups for larger-scale permissions management.
LONI ANALYSIS METHODS
Automated analysis
The LONI Pipeline(Dinov et al., 2010, 2014; MacKenzie-Graham and Payan, 2008) contains a common framework for visual and programmatic construction of data-driven workflows for electrophysiology, imaging, and biosample data. With the aid of LONI’s workflow builder (Figure 3), complex analyses are represented visually, further supporting researchers’ investigations. Examples of LONI Pipeline applications include developing a unified coordinate space for seizure onset locations across various brains, including animal and human MRI, using string similarity and value overlap to predict that different contributor metadata fields are the same, and providing graphical interfaces for linking data. Co-registration algorithms are typically invoked at upload-time but may also be triggered later manually for further refinement. We provide MRI supervision and integration from different scanners and centers by supervising phantom studies, assessing quality, and fixing problems with heterogeneity. Robust workflow pipelines are provided for researchers to use on both humans and animal models. An example depicting steps of some MRI analysis using the LONI Pipeline is shown in Figure 3.
Figure 3.
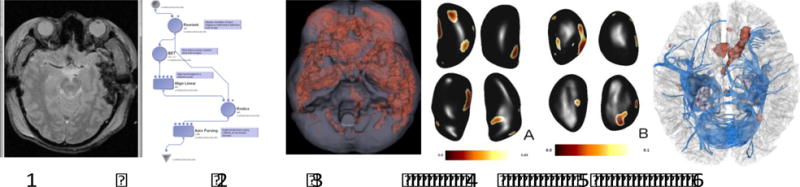
Example of analysis performed using the LONI Pipeline, including MRI and DTI data with group analysis over 46 patients depicting common locations of hemorrhages across these patients and physiological group differences in PTE, making use of both modular and automated LONI Pipeline techniques.
Standardized sample collection, shipping, and biobank storage protocols
We have developed protocols based on our previous studies to define methods for harvesting, freezing, and storing tissue and other biosamples (i.e., cerebrospinal fluid). Parallel storage protocols are followed to ensure that parallel human and animal samples are stored and treated in a similar fashion to facilitate comparison of findings. These protocols are also meant to be foundational to future sample collection and transport among the wider epilepsy research community.
We describe examples from the initial analysis on the MRI and EEG data collected in EpiBioS4Rx from the 16 currently enrolled patients and animal data from 10 rats.
MRI analysis
The collected human MRI data consist of structural, functional (resting state), and diffusion weighted measures(Vespa et al., n.d.). MRI analyses consist of structural analyses (performed in BrainSuite(Shattuck and Leahy, 2002)) to measure each subject’s intracranial volumes as well as gray matter volumes and other anatomical measures. Functional analyses are conducted using Statistical Parametric Mapping (SPM)(Ashburner, 2012), a software suite of MATLAB, to ascertain brain activation in different regions. Functional connectivity analyses are performed in the CONN toolbox of MATLAB to examine network connectivity in comparison to non-TBI data, to determine abnormally active/inactive networks. Lastly, the diffusion weighted analyses consist of constructing each subject’s fractional anisotropy (FA) maps, in addition to measuring each patient’s apparent diffusion coefficient (ADC) to assess white matter integrity and connectivity in FMRIB Software Library (FSL)(Jenkinson et al., 2012). These FA maps of TBI data are compared to five normal, non-TBI data in a group analysis via tract based spatial statistics (TBSS) in FSL.
The collected rat MRI consist of structural and diffusion weighted measures. MRI processing and analyses mainly consist of each rat’s diffusion weighted measures. Each rat’s FA map is constructed in FSL. Additionally, TBSS is performed on the TBI rats and non-TBI animal data to measure group differences. Example structural and DTI data for a control rat (Sprague-Dawley) and TBI rat (left lateral fluid percussion injury) are shown in Figure 4 using DSI Studio. In this example, the data were collected using a Bruker BioSpin MRI GmbH at the University of Eastern Finland, Kuopio using a dtiEpiT SpinEcho sequence.
Figure 4.

Rat (male, 2 month-old Sprague Dawley rat, 300g weight, courtesy of University of Eastern Finland, 7T/16cm Bruker Pharmascan) T1 MRI on the left and corresponding DTI on the right for a control rat in the first 2 images and a rat (left parietal LFPI model, 5mm, severe injury, on a male, 2 month-old Sprague Dawley rat, 300g weight, courtesy of University of Eastern Finland, 7T/16cm Bruker Pharmascan) in the third and fourth images (decreased FA map intensity circled in red); FA map used deterministic fiber tracking algorithm, anisotropy threshold was randomly selected, angular threshold was selected from 15-90, and fiber trajectories were smoothed by averaging propagation direction with percentage of previous direction. The images are in radiological orientation, so right and left are flipped. Colors correspond with the direction of the water/fluid flow in the WM tracts, in which blue is superior-inferior direction, red is right –left (lateral), and green is anterior-posterior.
We have introduced methods for TBI connectomics(Irimia et al., 2012a, 2012b; Torgerson et al., 2013; van Horn et al., 2012) to be used on the clinical data in this study. DTI is used to extract connectivity between all pairs of gyral and sulcal structures in the presence of brain trauma. Connectivity between all brain regions (165 in our scheme) is computed from DTI volumes acquired longitudinally from each patient. Diffusion tractography is used to determine connectivity properties (WM bundle length, connectivity density, and FA) and each subject’s weighted connectivity matrix. WM fiber tracking of inter-regional connectivity is conducted using TrackVis(Wedeen et al., 2008) or other tractography tools. Connectivity between regions (such as thalamo-cortical connections and hippocampal connections) is assessed systematically within each patient using purpose-built workflows for multi-modal co-registration of MRI. This will be followed by calculation of (i) inter-regional connectivity matrices and (ii) longitudinal changes in connectivity topology using network-theoretic descriptors of nodal and network-wide segregation (clustering coefficient, modularity, etc.) and integration (characteristic path length, global efficiency, etc.). Additional network-theoretic measures (scale freedom, small worldness, robustness, centrality, degree distribution, and communication efficiency)(Eguiluz et al., 2005; Salvador et al., 2005; Stam, 2004; Wilcox, 2012) will be computed. Results and changes over time in each patient are visualized and analyzed using connectograms(Irimia et al., 2012a). Workflows are fully integrated with the LONI Pipeline(Dinov et al., 2010; MacKenzie-Graham and Payan, 2008).
Translational aspect of analysis
The comparison of human and animal neuroimaging data presents anatomical challenges; we aim to compare the WM tracts’ characteristics and integrity of human and rat neuroimaging data. The TBI rats’ TBSS and the patients’ TBSS will be compared to examine WM tract similarities that could relate to network abnormalities in epileptic human patients. Specifically, we are analyzing the WM integrities in the animal model and how that may impact network performance or connectivity in humans. The translational aspect will be unique to EpiBioS4Rx with such a thorough dataset, including rats and patients, at LONI.
EEG Analysis
We use Persyst software,(Sierra-Marcos et al., 2015) linked to data from the IDA, for EEG data visualization over multiple channels, export of artifact reduced waveform data, seizure and spike detection, wavelets, matching pursuit, correlation, FFT phase, period evolution, and other EEG analysis tools. Due to the sheer volume of EEG data due to the continuous recordings and number of electrode contacts used, we have applied a variety of dimensionality reduction techniques to the EEG for both preclinical and clinical data. To increase the ease of understanding the high dimensional data and outline trends in these collected samples, we apply these methods, assuming the data can be reduced to lie on a nonlinear manifold of lower, intrinsic dimensionality. Furthermore, we use these methods to remove excessive noise in the data, which is particularly a problem with scalp EEG as well as to look for patterns or features of epileptogenesis.
We have applied and compared Principal Component Analysis (PCA), Diffusion Maps, Laplacian Eigenmaps, Kernel PCA, and Unsupervised Diffusion Component Analysis (UDCA), which are methods that can be used on both animal and human data. Each of these tools has its own benefits and weaknesses, so we are providing a variety of dimensionality reduction methods for researchers, because one method will not always be the best to use in every instance. PCA is a linear dimensionality reduction method(Wold et al., 1987); it is used by rotating data in a different orientation in the dimensional space by exposing the maximum variance. It detects and eliminates some noise and collects the redundancy of the data. Kernel PCA is an extension of PCA that uses techniques of kernel methods(Jade et al., 2003).
Laplacian Eigenmaps is a nonlinear dimensionality reduction method that assumes that data lie in a low dimensional manifold within the high dimensional space; it grabs information from nearest neighbors of each data point(Belkin and Niyogi, 2003). Thus, a low dimensional dataset is produced by preserving local properties of the manifold and minimizing the distance between a data point and its nearest neighbor. Diffusion Mapping is another nonlinear dimensionality reduction method(Coifman and Lafon, 2006; Duncan et al., 2013). A family of embeddings of a dataset is computed into a low-dimensional Euclidean space whose coordinates can be computed from the eigenvectors and corresponding eigenvalues of a diffusion operator on the data. We have developed UDCA(Duncan and Strohmer, 2016), which is an extension and adaptation of diffusion maps. In this algorithm, coordinates are constructed that generate efficient geometric representations of the complex data. Additionally, this algorithm performs well by removing noise from the data (using the Mahalanobis distance measure with inverse covariance matrices(Talmon et al., 2012)) and is completely automatic. EEGLAB(Delorme and Makeig, 2004), a MATLAB toolbox and graphic user interface, is used to open EDF+ EEG files in MATLAB.
Figure 5 shows UDCA applied to a sample of pre-ictal data, where the algorithm separates preseizure features that are not apparent from visually inspecting the raw data, and then a method of plotting the Euclidean distances of the points from the embedding to the origin is shown to demonstrate how we can set a threshold of a chosen amplitude that can be used to automatically extract features of epileptogenesis after TBI. This method is useful for noisy, complex data, such as EEG and allows researchers to extract the underlying brain activity that may be associated with biomarkers of epileptogenesis(Duncan et al., 2018).
Figure 5.
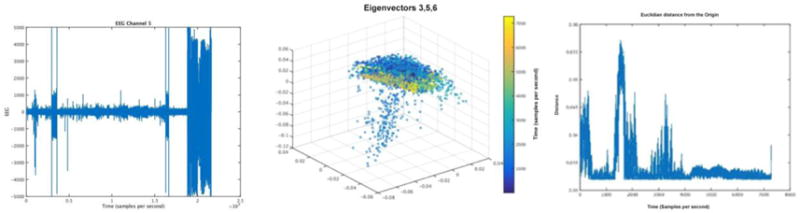
These images show an example of human pre-ictal scalp EEG raw data on the left, courtesy of UCLA with acquisition settings described above, an embedding into a 3-dimensional space using the 3rd, 5th, and 6th eigenvectors in the center (color represents time), and the Euclidean distance plotted of each point in the embedding to the origin on the right.
DISCUSSION
Our central data repository, the LONI IDA, has been configured to receive preclinical and clinical EpiBioS4Rx data, including MRI, CT, and EEG data submitted by the 16 participating clinical and 4 preclinical centers of the study. The data upload process (depicted in Figure 6) includes a data de-identification process that occurs before data are transferred to the central repository and is configured to work for multiple file formats, including DICOM, ECAT, HRRT, and EDF. Data arriving at the central repository are immediately and automatically checked in, and a subset of metadata attributes are extracted from the files and used to catalog and describe the data to support database searches. Check-in is typically completed within 3 minutes, at which time data become immediately available to investigators to import into the LONI QC and/or Pipeline workflow environments and/or to download for local analysis.
Figure 6.

Data flow process.
Some of the challenges that we have encountered are in managing the heterogeneity and scale of the potentially relevant data. This occurs (1) while integrating and interlinking the data such that it can be stored, accessed, searched, and analyzed; (2) while browsing and algorithmically analyzing the data in search of biomarkers, where relevant features are likely. We have worked to ensure rigorous experimental design for robust and unbiased results on our platform.
CONCLUSIONS
We have built upon decades of experience with big data projects at LONI to develop the informatics infrastructure needed for a large-scale study, such as EpiBioS4Rx.
We have created an infrastructure for EpiBioS4Rx investigators, collaborators, and the broader epilepsy clinical and research community. Additionally, we have established methods for mining the complex, multi-modal data collected in the study, and ultimately, to develop data-driven predictive mathematical models of the epileptogenic processes that represent sensitive and specific biomarkers to predict the development of epilepsy after TBI. We have described some of the analytic tools that we have developed and used in our search for biomarkers of epileptogenesis. Biomarkers that are discovered will be instrumental in our efforts to develop models aimed at predicting the probability of developing epilepsy in post-TBI subjects and identifying specific times, regions, and processes where intervention may be most beneficial. The online interface that we have established allows users to search across raw and processed data in a data mining way and enable visualization and analyses to test hypotheses and validate results. By sharing access to our data and analytic tools as well as our server for data processing, we hope to encourage collaborations among different centers around the world and to bring awareness to investigators about the data collected and analysis methods used by other teams. We have developed a data flow process that makes the data findable, accessible, interoperable and reusable for the epilepsy research community, thus following the Findable, Accessible, Interoperable, and Reusable (FAIR) guiding principles (Wilkinson et al., 2016). This infrastructure has the potential to spur the advancement and development of research directed toward translational or clinical development of additional disease-modifying or preventative therapies. The mechanisms that are revealed in our search for these biomarkers will be used as targets for pioneering antiepileptogenic treatments.
We will continue to extract features from neuroimaging, electrophysiologic, molecular, clinical, cognitive, and behavioral measures over time to identify candidate diagnostic biomarkers of epileptogenesis. Novel statistical tools, which we will continue to modify and improve, have been developed to visualize complex associations among multiple variables as they evolve over time during epileptogenesis. They will reveal processes, regions, and stages in epileptogenesis correlated with specific anatomical changes in imaging. Advanced statistical techniques will then be used with the goal of building models of epileptogenesis to predict the probability of epilepsy, based on biomarker inputs. The results of the predictive models will be validated by testing the robustness of the results in the presence of uncertainty.
Highlights.
We have created the infrastructure for a centralized data repository for multi-modal data
Innovative image and electrophysiology processing methods have been applied
Novel analytic tools are described to study epileptogenesis after traumatic brain injury
Acknowledgments
This research was supported by the National Institute of Neurological Disorders and Stroke (NINDS) of the National Institutes of Health (NIH) under Award Numbers U54NS100064 (EpiBioS4Rx), NIH P41-EB015922, and NIH U54-EB020406.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- A D, M LVQ, B S, G F, A S-B, S S. Epilepsiae - Evolving platform for improving living expectation of patients suffering from ictal events. Epilepsia 2009 [Google Scholar]
- Ashburner J. SPM: A history. Neuroimage. 2012 doi: 10.1016/j.neuroimage.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astakhov V, Gupta A, Santini S, Grethe JS. Data integration in the Biomedical Informatics Research Network (BIRN) Data Integr Life Sci Proc. 2005;3615:317–320. [Google Scholar]
- Barker-Haliski M, Friedman D, White HS, French JA. How clinical development can, and should, inform translational science. Neuron. 2014 doi: 10.1016/j.neuron.2014.10.029. [DOI] [PubMed] [Google Scholar]
- Belkin M, Niyogi P. Laplacian Eigenmaps for Dimensionality Reduction and Data Representation. Neural Comput. 2003;15:1373–1396. doi: 10.1162/089976603321780317. [DOI] [Google Scholar]
- Bourne PE, Bonazzi V, Dunn M, Green ED, Guyer M, Komatsoulis G, Larkin J, Russell B. The NIH big data to knowledge (BD2K) initiative. J Am Med Informatics Assoc. 2015;22:1114–1114. doi: 10.1093/jamia/ocv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann BH, Bower MR, Stengel KA, Worrell GA, Stead M. Multiscale electrophysiology format: An open-source electrophysiology format using data compression, encryption, and cyclic redundancy check, in: Proceedings of the 31st Annual International Conference of the IEEE Engineering in Medicine and Biology Society: Engineering the Future of Biomedicine. EMBC. 2009;2009:7083–7086. doi: 10.1109/IEMBS.2009.5332915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buja A, Cook D, Hofmann H, Lawrence M, Lee EK, Swayne DF, Wickham H. Statistical inference for exploratory data analysis and model diagnostics. Philos Trans R Soc A Math Phys Eng Sci. 2009;367:4361–4383. doi: 10.1098/rsta.2009.0120. [DOI] [PubMed] [Google Scholar]
- Coifman RR, Lafon S. Diffusion maps. Appl Comput Harmon Anal. 2006;21:5–30. doi: 10.1016/j.acha.2006.04.006. [DOI] [Google Scholar]
- Crawford KL, Neu SC, Toga AW. The Image and Data Archive at the Laboratory of Neuro Imaging. Neuroimage. 2016;124:1080–1083. doi: 10.1016/j.neuroimage.2015.04.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme A, Makeig S. EEGLAB: An open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J Neurosci Methods. 2004;134:9–21. doi: 10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Dinov I, Lozev K, Petrosyan P, Liu Z, Eggert P. Neuroimaging Study Designs, Computational Analyses and Data Provenance Using the LONI Pipeline. PLoS One. 2010;5:e13070. doi: 10.1371/journal.pone.0013070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinov ID, Petrosyan P, Liu Z, Eggert P, Zamanyan A, Torri F, Macciardi F, Hobel S, Moon SW, Sung YH, Jiang Z, Labus J, Kurth F, Ashe-McNalley C, Mayer E, Vespa PM, Van Horn JD, Toga AW. The perfect neuroimaging-genetics-computation storm: Collision of petabytes of data, millions of hardware devices and thousands of software tools. Brain Imaging Behav. 2014;8:311–322. doi: 10.1007/s11682-013-9248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan D, Strohmer T. Classification of Alzheimer’s disease using unsupervised diffusion component analysis. Math Biosci Eng. 2016;13:1119–1130. doi: 10.3934/mbe.2016033. [DOI] [PubMed] [Google Scholar]
- Duncan D, Talmon R, Zaveri HP, Coifman RR. Identifying preseizure state in intracranial EEG data using diffusion kernels. Math Biosci Eng. 2013;10:579–590. doi: 10.3934/mbe.2013.10.579. [DOI] [PubMed] [Google Scholar]
- Duncan D, Toga AW, Vespa PM. Detecting features of epileptogenesis in EEG after TBI using Unsupervised Diffusion Component Analysis. Math Biosci Eng. 2018;23 doi: 10.3934/dcdsb.2018010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguiluz VM, Chialvo D, Cecchi GA, Baliki M, Apkarian AV. Scale-free brain functional networks. Phys Rev Lett. 2005;94:018102. doi: 10.1103/PhysRevLett.94.018102. [DOI] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmer KG, Ambite JL, Ames J, Ananthakrishnan R, Burns G, Chervenak AL, Foster I, Liming L, Keator D, Macciardi F, Madduri R, Navarro JP, Potkin S, Rosen B, Ruffins S, Schuler R, Turner JA, Toga A, Williams C, Kesselman C. Enabling collaborative research using the Biomedical Informatics Research Network (BIRN) J Am Med Inform Assoc. 2011;18:416–22. doi: 10.1136/amiajnl-2010-000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle M, Feldwisch-Drentrup H, Teixeira CA, Witon A, Schelter B, Timmer J, Schulze-Bonhage A. EPILEPSIAE - A European epilepsy database. Comput Methods Programs Biomed. 2012;106:127–138. doi: 10.1016/j.cmpb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- Irimia A, Chambers MC, Torgerson CM, Van Horn JD. Circular representation of human cortical networks for subject and population-level connectomic visualization. Neuroimage. 2012a;60:1340–1351. doi: 10.1016/j.neuroimage.2012.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia A, Wang B, Aylward SR, Prastawa MW, Pace DF, Gerig G, Hovda DA, Kikinis R, Vespa PM, Van Horn JD. Neuroimaging of structural pathology and connectomics in traumatic brain injury: Toward personalized outcome prediction. NeuroImage Clin. 2012b doi: 10.1016/j.nicl.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jade AM, Srikanth B, Jayaraman VK, Kulkarni BD, Jog JP, Priya L. Feature extraction and denoising using kernel PCA. Chem Eng Sci. 2003;58:4441–4448. doi: 10.1016/S0009-2509(03)00340-3. [DOI] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62:782–790. doi: 10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Johnson SB, Whitney G, McAuliffe M, Wang H, McCreedy E, Rozenblit L, Evans CC. Using global unique identifiers to link autism collections. J Am Med Informatics Assoc. 2010;17:689–695. doi: 10.1136/jamia.2009.002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Irimia A, Hobel SM, Pogosyan M, Tang H, Petrosyan P, Esquivel Castelo-Blanco RI, Duffy BA, Zhao L, Crawford KL, Liew S-L, Clark K, Law M, Mukherjee P, Manley GT, Van Horn JD, Toga AW. LONI QC system: a semi-automated, web-based and freely-available environment for the comprehensive quality control of neuroimaging data. Front Neuroinform. doi: 10.3389/fninf.2019.00060. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kini LG, Davis KA, Wagenaar JB. Data integration: Combined imaging and electrophysiology data in the cloud. Neuroimage. 2016;124:1175–1181. doi: 10.1016/j.neuroimage.2015.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt J, Feldwisch-Drentrup H, Ihle M, Navarro V, Neufang M, Teixeira C, Adam C, Valderrama M, Alvarado-Rojas C, Witon A, Le Van Quyen M, Sales F, Dourado A, Timmer J, Schulze-Bonhage A, Schelter B. The EPILEPSIAE database: An extensive electroencephalography database of epilepsy patients. Epilepsia. 2012;53:1669–1676. doi: 10.1111/j.1528-1167.2012.03564.x. [DOI] [PubMed] [Google Scholar]
- LeVan P, Urrestarazu E, Gotman J. A system for automatic artifact removal in ictal scalp EEG based on independent component analysis and Bayesian classification. Clin Neurophysiol. 2006;117:912–927. doi: 10.1016/j.clinph.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Lim MD. Consortium sandbox: Building and sharing resources. Sci Transl Med. 2014 doi: 10.1126/scitranslmed.3009024. [DOI] [PubMed] [Google Scholar]
- MacKenzie-Graham A, Payan A. Neuroimaging data provenance using the LONI pipeline workflow environment. Proven. 2008:1–12. doi: 10.1007/978-3-540-89965-5_22. [DOI] [Google Scholar]
- Marcus DS, Harms MP, Snyder AZ, Jenkinson M, Wilson JA, Glasser MF, Barch DM, Archie KA, Burgess GC, Ramaratnam M, Hodge M, Horton W, Herrick R, Olsen T, McKay M, House M, Hileman M, Reid E, Harwell J, Coalson T, Schindler J, Elam JS, Curtiss SW, Van Essen DC. Human Connectome Project informatics: Quality control, database services, and data visualization. Neuroimage. 2013;80:202–219. doi: 10.1016/j.neuroimage.2013.05.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, Coffey C, Kieburtz K, Flagg E, Chowdhury S, Poewe W, Mollenhauer B, Sherer T, Frasier M, Meunier C, Rudolph A, Casaceli C, Seibyl J, Mendick S, Schuff N, Zhang Y, Toga A, Crawford K, Ansbach A, de Blasio P, Piovella M, Trojanowski J, Shaw L, Singleton A, Hawkins K, Eberling J, Russell D, Leary L, Factor S, Sommerfeld B, Hogarth P, Pighetti E, Williams K, Standaert D, Guthrie S, Hauser R, Delgado H, Jankovic J, Hunter C, Stern M, Tran B, Leverenz J, Baca M, Frank S, Thomas CA, Richard I, Deeley C, Rees L, Sprenger F, Lang E, Shill H, Obradov S, Fernandez H, Winters A, Berg D, Gauss K, Galasko D, Fontaine D, Mari Z, Gerstenhaber M, Brooks D, Malloy S, Barone P, Longo K, Comery T, Ravina B, Grachev I, Gallagher K, Collins M, Widnell KL, Ostrowizki S, Fontoura P, La-Roche FH, Ho T, Luthman J, van der Brug M, Reith AD, Taylor P. The Parkinson Progression Marker Initiative (PPMI) Prog Neurobiol. 2011 doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis R, Derr L, Dunn M, Huerta M, Larkin J, Sheehan J, Guyer M, Green ED. The National Institutes of Health’s big data to knowledge (BD2K) initiative: Capitalizing on biomedical big data. J Am Med Informatics Assoc. 2014;21:957–958. doi: 10.1136/amiajnl-2014-002974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payakachat NN, Tilford JM b M, Ungar WJ dWJWJd. National Database for Autism Research (NDAR): Big Data Opportunities for Health Services Research and Health Technology Assessment. Pharmacoeconomics. 2016;34:127–138. doi: 10.1007/s40273-015-0331-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakap S, Jones HA, Emery AK. Evaluation of a web-based professional development program (project ACE) for teachers of children with autism spectrum disorders. Teach Educ Spec Educ. 2015;38:221–239. doi: 10.1177/0888406414535821. [DOI] [Google Scholar]
- Salvador R, Suckling J, Schwarzbauer C, Bullmore E. Undirected graphs of frequency-dependent functional connectivity in whole brain networks. Philos Trans R Soc Lond B Biol Sci. 2005;360:937–946. doi: 10.1098/rstb.2005.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider LS, Insel PS, Weiner MW. Treatment with cholinesterase inhibitors and memantine of patients in the Alzheimer’s disease neuroimaging initiative. Arch Neurol. 2011;68:58–66. doi: 10.1001/archneurol.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Bonhage A, Ihle M, Sales F, Navarro V, Dourado A. A European EEG database of epilepsy patients EPILEPSIAE. Clin Neurophysiol. 2010;121:S200. [Google Scholar]
- Shattuck DW, Leahy RM. Brainsuite: An automated cortical surface identification tool. Med Image Anal. 2002;6:129–142. doi: 10.1016/S1361-8415(02)00054-3. [DOI] [PubMed] [Google Scholar]
- Sierra-Marcos A, Scheuer ML, Rossetti AO. Seizure detection with automated EEG analysis: A validation study focusing on periodic patterns. Clin Neurophysiol. 2015;126:456–462. doi: 10.1016/j.clinph.2014.06.025. [DOI] [PubMed] [Google Scholar]
- Stam CJ. Functional connectivity patterns of human magnetoencephalographic recordings: A “small-world” network? Neurosci Lett. 2004;355:25–28. doi: 10.1016/j.neulet.2003.10.063. [DOI] [PubMed] [Google Scholar]
- Talmon R, Kushnir D, Coifman RR, Cohen I, Gannot S. Parametrization of linear systems using diffusion kernels. IEEE Trans Signal Process. 2012;60:1159–1173. doi: 10.1109/TSP.2011.2177973. [DOI] [Google Scholar]
- Talukdar PP, Ives ZG, Pereira F. Automatically incorporating new sources in keyword search-based data integration. Proc 2010 Int Conf Manag data - SIGMOD ’10. 2010:387–398. doi: 10.1145/1807167.1807211. [DOI] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Toro R, Jahanshad N, Schumann G, Franke B, Wright MJ, Martin NG, Agartz I, Alda M, Alhusaini S, Almasy L, Almeida J, Alpert K, Andreasen NC, Andreassen OA, Apostolova LG, Appel K, Armstrong NJ, Aribisala B, Bastin ME, Bauer M, Bearden CE, Bergmann Ø, Binder EB, Blangero J, Bockholt HJ, Bøen E, Bois C, Boomsma DI, Booth T, Bowman IJ, Bralten J, Brouwer RM, Brunner HG, Brohawn DG, Buckner RL, Buitelaar J, Bulayeva K, Bustillo JR, Calhoun VD, Cannon DM, Cantor RM, Carless MA, Caseras X, Cavalleri GL, Chakravarty MM, Chang KD, Ching CRK, Christoforou A, Cichon S, Clark VP, Conrod P, Coppola G, Crespo-Facorro B, Curran JE, Czisch M, Deary IJ, de Geus EJC, den Braber A, Delvecchio G, Depondt C, de Haan L, de Zubicaray GI, Dima D, Dimitrova R, Djurovic S, Dong H, Donohoe G, Duggirala R, Dyer TD, Ehrlich S, Ekman CJ, Elvsåshagen T, Emsell L, Erk S, Espeseth T, Fagerness J, Fears S, Fedko I, Fernández G, Fisher SE, Foroud T, Fox PT, Francks C, Frangou S, Frey EM, Frodl T, Frouin V, Garavan H, Giddaluru S, Glahn DC, Godlewska B, Goldstein RZ, Gollub RL, Grabe HJ, Grimm O, Gruber O, Guadalupe T, Gur RE, Gur RC, Göring HHH, Hagenaars S, Hajek T, Hall GB, Hall J, Hardy J, Hartman CA, Hass J, Hatton SN, Haukvik UK, Hegenscheid K, Heinz A, Hickie IB, Ho BC, Hoehn D, Hoekstra PJ, Hollinshead M, Holmes AJ, Homuth G, Hoogman M, Hong LE, Hosten N, Hottenga JJ, Hulshoff Pol HE, Hwang KS, Jack CR, Jenkinson M, Johnston C, Jönsson EG, Kahn RS, Kasperaviciute D, Kelly S, Kim S, Kochunov P, Koenders L, Krämer B, Kwok JBJ, Lagopoulos J, Laje G, Landen M, Landman BA, Lauriello J, Lawrie SM, Lee PH, Le Hellard S, Lemaître H, Leonardo CD, Li C shan, Liberg B, Liewald DC, Liu X, Lopez LM, Loth E, Lourdusamy A, Luciano M, Macciardi F, Machielsen MWJ, MacQueen GM, Malt UF, Mandl R, Manoach DS, Martinot JL, Matarin M, Mather KA, Mattheisen M, Mattingsdal M, Meyer-Lindenberg A, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meisenzahl E, Melle I, Milaneschi Y, Mohnke S, Montgomery GW, Morris DW, Moses EK, Mueller BA, Muñoz Maniega S, Mühleisen TW, Müller-Myhsok B, Mwangi B, Nauck M, Nho K, Nichols TE, Nilsson LG, Nugent AC, Nyberg L, Olvera RL, Oosterlaan J, Ophoff RA, Pandolfo M, Papalampropoulou-Tsiridou M, Papmeyer M, Paus T, Pausova Z, Pearlson GD, Penninx BW, Peterson CP, Pfennig A, Phillips M, Pike GB, Poline JB, Potkin SG, Pütz B, Ramasamy A, Rasmussen J, Rietschel M, Rijpkema M, Risacher SL, Roffman JL, Roiz-Santiañez R, Romanczuk-Seiferth N, Rose EJ, Royle NA, Rujescu D, Ryten M, Sachdev PS, Salami A, Satterthwaite TD, Savitz J, Saykin AJ, Scanlon C, Schmaal L, Schnack HG, Schork AJ, Schulz SC, Schür R, Seidman L, Shen L, Shoemaker JM, Simmons A, Sisodiya SM, Smith C, Smoller JW, Soares JC, Sponheim SR, Sprooten E, Starr JM, Steen VM, Strakowski S, Strike L, Sussmann J, Sämann PG, Teumer A, Toga AW, Tordesillas-Gutierrez D, Trabzuni D, Trost S, Turner J, Van den Heuvel M, van der Wee NJ, van Eijk K, van Erp TGM, van Haren NEM, van ’t Ent D, van Tol MJ, Valdés Hernández MC, Veltman DJ, Versace A, Völzke H, Walker R, Walter H, Wang L, Wardlaw JM, Weale ME, Weiner MW, Wen W, Westlye LT, Whalley HC, Whelan CD, White T, Winkler AM, Wittfeld K, Woldehawariat G, Wolf C, Zilles D, Zwiers MP, Thalamuthu A, Schofield PR, Freimer NB, Lawrence NS, Drevets W. The ENIGMA Consortium: Large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Crawford KL. The informatics core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s Dement. 2010a doi: 10.1016/j.jalz.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Crawford KL. The informatics core of the Alzheimer’s Disease Neuroimaging Initiative. Alzheimer’s Dement. 2010b;6:247–256. doi: 10.1016/j.jalz.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toga AW, Neu SC, Bhatt P, Crawford KL, Ashish N. The Global Alzheimer’s Association Interactive Network. Alzheimer’s Dement. 2016;12:49–54. doi: 10.1016/j.jalz.2015.06.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson CM, Irimia A, Leow AD, Bartzokis G, Moody TD, Jennings RG, Alger JR, van Horn JD, Altshuler LL. DTI tractography and white matter fiber tract characteristics in euthymic bipolar I patients and healthy control subjects. Brain Imaging Behav. 2013;7:129–139. doi: 10.1007/s11682-012-9202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson CM, Quinn C, Dinov I, Liu Z, Petrosyan P, Pelphrey K, Haselgrove C, Kennedy DN, Toga AW, Van Horn JD. Interacting with the National Database for Autism Research (NDAR) via the LONI Pipeline workflow environment. Brain Imaging Behav. 2015;9:89–103. doi: 10.1007/s11682-015-9354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K. The WU-Minn Human Connectome Project: An overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Horn JD, Irimia A, Torgerson CM, Chambers MC, Kikinis R, Toga AW. Mapping connectivity damage in the case of phineas gage. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn JD, Toga AW. Is it time to re-prioritize neuroimaging databases and digital repositories? Neuroimage. 2009;47:1720–1734. doi: 10.1016/j.neuroimage.2009.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vespa PM, Shrestha V, Abend N, Agoston D, Au A, Bell MJ, Bleck TP, Buitrago Blanco M, Claassen J, Diaz-Arrastia R, Duncan D, Ellingson B, Foreman B, Gilmore EJ, Hirsch L, Hunn M, Kamnaksh A, McArthur D, Morokoff A, O’Brien T, O’Phelan K, Robertson CL, Rosenthal E, Staba R, Toga A, Willyerd FA, Zimmermann L, Real C, Martinez S, Yam E, Engel J, Jr, Group, F. the E.S. The Epilepsy Bioinformatics Epilepsy Study for Anti-Epileptogenic Therapy (Epibios4Rx) Clinical Biomarker: Study Design and Protocol. Neurobiol Dis. doi: 10.1016/j.nbd.2018.07.025. n.d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar JB, Worrell GA, Ives Z, Matthias D, Litt B, Schulze-Bonhage A. Collaborating and sharing data in epilepsy research. J Clin Neurophysiol. 2015 doi: 10.1097/WNP.0000000000000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WYI, Dai G, Pandya DN, Hagmann P, D’Arceuil H, de Crespigny AJ. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage. 2008;41:1267–1277. doi: 10.1016/j.neuroimage.2008.03.036. [DOI] [PubMed] [Google Scholar]
- Wilcox R. Introduction to Robust Estimation and Hypothesis Testing, Introduction to Robust Estimation and Hypothesis Testing. 2012 doi: 10.1016/B978-0-12-386983-8.00015-9. [DOI] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Brookes AJ, Clark T, Crosas M, Dillo I, Dumon O, Edmunds S, Evelo CT, Finkers R, Gonzalez-Beltran A, Gray AJ, Groth P, Goble C, Grethe JS, Heringa J, ‘t Hoen PA, Hooft R, Kuhn T, Kok R, Kok J, Lusher SJ, Martone ME, Mons A, Packer AL, Persson B, Rocca-Serra P, Roos M, van Schaik R, Sansone SA, Schultes E, Sengstag T, Slater T, Strawn G, Swertz MA, Thompson M, van der Lei J, van Mulligen E, Velterop J, Waagmeester A, Wittenburg P, Wolstencroft K, Zhao J, Mons B. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden KD, Bragin A, Engel J, Geschwind DH. Molecular alterations in areas generating fast ripples in an animal model of temporal lobe epilepsy. Neurobiol Dis. 2015;78:35–44. doi: 10.1016/j.nbd.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden KD, Karsten SL, Bragin A, Kudo LC, Gehman L, Ruidera J, Geschwind DH, Engel J. A systems level, functional genomics analysis of chronic epilepsy. PLoS One. 2011;6 doi: 10.1371/journal.pone.0020763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold S, Esbensen K, Geladi P. Principal component analysis. Chemom Intell Lab Syst. 1987;2:37–52. doi: 10.1016/0169-7439(87)80084-9. [DOI] [Google Scholar]


