Abstract
Recurrent mutations in the splicing factor SRSF2 are associated with poor clinical outcomes in myelodysplastic syndromes (MDS). Their high frequency suggests these mutations drive oncogenesis, yet the molecular explanation for this process is unclear. SRSF2 mutations could directly affect pre-mRNA splicing of a vital gene product; alternatively, a whole network of gene products could be affected. Here we determine how SRSF2 mutations globally affect RNA binding and splicing in vivo using HITS-CLIP. Remarkably, the majority of differential binding events do not translate into alternative splicing of exons with SRSF2P95H binding sites. Alternative splice alterations appear to be dominated by indirect effects. Importantly, SRSF2P95H targets are enriched in RNA processing and splicing genes, including several members of the hnRNP and SR families of proteins, suggesting a “splicing-cascade” phenotype wherein mutation of a single splicing factor leads to widespread modifications in multiple RNA processing and splicing proteins. We show that splice alteration of HNRNPA2B1, a splicing factor differentially bound and spliced by SRSF2P95H, impairs hematopoietic differentiation in vivo. Our data suggests a model whereby the recurrent mutations in splicing factors set off a cascade of gene regulatory events that together affect hematopoiesis and drive cancer.
Keywords: Splicing mutations, myelodysplasia, leukemia, myeloid neoplasia, SR proteins, SRSF2
INTRODUCTION
Recurrent mutations in key factors of the spliceosome, SRSF2, SF3B1, U2AF1 and ZRSR2, occur in over 50% of myelodysplastic syndromes (MDS) patients1,2. These mutations occur early in the disease, are present in the dominant clone, carry prognostic significance, and hold promise as new therapeutic targets1,3,4.
SRSF2 is a member of the serine/arginine rich (SR) class of splicing factors. SRSF2 contains a RNA binding domain (RBD, AA 1-101) that recognizes and binds exonic splicing enhancers (ESE) in a sequence specific manner and a SR domain that participates in protein-protein interactions to recruit the U2AF complex and the U1 snRNP to the 3′ and 5′ splice sites (SS), respectively5–9. Mutations in SRSF2 occur almost exclusively at proline 95 in the RNA binding domain and alone are sufficient to induce the hallmarks of MDS (leukopenia, macrocytic anemia, and dysplasia) in inducible Mx1-Cre Srsf2P95H/WT knock-in mice10.
We have previously shown that SRSF2 P95H/L/R mutations alter in vitro RNA binding affinity and specificity of the SRSF2 RNA binding domain resulting in higher affinity binding to the CCNG than to the GGNG consensus motif10, while the wild-type SRSF2 RBD recognizes both motifs equally well11. RNA sequencing of cell lines and patient samples expressing mutant SRSF2 identified differential splicing of CCNG-rich versus GGNG-rich exons; however, these cassette exon events only made up a small fraction of all alternative splicing (AS) events10,12,13.
SR proteins are essential regulators of constitutive and alternative pre-mRNA splicing and carry essential roles in other functions, such as transcriptional elongation, RNA export, RNA stability and translation5,14–19. The majority of AS events are orchestrated by a multitude of splicing factors, including members of the hnRNP family of splicing factors20–22. The versatile functions of SRSF2 and the complexity of regulation of AS emphasize the importance of studying splicing factor mutations in vivo within the context of the splicing machinery. Although wild-type SRSF2 RNA interactions have been characterized before20, no information is present for the mutant SRSF2 in vivo RNA interactome.
In this study, we performed high-throughput sequencing of RNA isolated by UV-crosslinking and immunoprecipitation (HITS-CLIP)23,24 to analyze and compare differential RNA binding and splicing between wild-type (WT) and mutant (P95H) SRSF2 in vivo in a human erythroid leukemia cell line, verified key splice events in primary patient samples and assessed their ability to affect hematopoiesis via colony formation assays.
Our data provide evidence for direct effects on RNA binding in vivo and in addition identify a “splicing-cascade” phenotype due to SRSF2-mediated functional regulation of cooperating and competing splicing factors. These detailed mechanistic studies have implications for the development of therapeutic approaches for SRSF2 mutant malignancies.
MATERIALS AND METHODS
SRSF2 mutant cell line construction and verification
Full length human SRSF2-Flag was cloned into the CS-TRE-Ubc-tTA-I2G plasmid1. Site-directed mutagenesis was performed to obtain SRSF2 mutants per standard protocol (Agilent Technologies). Lentivirus was produced by co-transfection of 293FT cells with psPAX2 (Addgene plasmid #12260) and pCMV-VSVG (Addgene plasmid #14888). HEL cells were transduced at MOI 1 and single cell clones established. Inducible expression of SRSF2 was verified by Sanger sequencing and western blotting with anti-Flag antibody (Sigma-Aldrich).
HITS-CLIP and RNA-Seq library generation
HITS-CLIP was performed in 4 replicates as previously published25 and according to Ule et al.23,24 with the following modifications: HEL/SRSF2WT and SRSF2P95H cells were treated with doxycycline [1 ug/ml] for 36 hours and UV-crosslinked (400mJ, Stratalinker 2400, Stratagene). RNA–protein complexes were immunoprecipitated with anti-Flag M2 Agarose beads (Sigma). RNA was partially digested with RNase A (Affymetrix) and P32-labeled (Perkin Elmer). SRSF2-RNA complexes were isolated by SDS-PAGE, treated with proteinase K, followed by RNA linker ligation using the NEB multiplex small RNA library prep set for Illumina (NEB). Libraries were deep sequenced (Illumina Genome Analyzer IIX System, single-end 50bp). Ribosomally depleted RNA from induced SRSF2WT and SRSF2P95H, as well as uninduced SRSF2P95H HEL cells, were used for paired-end 2x100bp RNA-Seq (Illumina HiSeq4000), performed in duplicate as per the ENCODE guidelines.
Computational procedures
HITS-CLIP reads were processed using the FASTX-Toolkit (removal of adapters and duplicated reads) and aligned to the human genome (GRCh38.p3) with Tophat (v2.0.14, --library-type fr-firststrand --no-coverage-search), using the Gencode 23 transcript annotation as transcriptome guide. Normalization was performed with the TMM method implemented in the edgeR Bioconductor package. Candidate binding sites were collected from genomic positions with at least 1 crosslinking induced deletion and coverage in at least two of the four replicates. For each binding site, the ratio between HITS-CLIP and RNA-seq normalized signals was calculated. Binding sites with HITS-CLIP to RNA-seq ratio < 5 were filtered out to remove potential aspecific interactions. Differential analysis of WT vs P95H binding sites was performed with edgeR exact negative binomial test (bcv = 0.4). Significant differentially bound sites were identified with the following thresholds: 1) mean normalized counts per million (CPM) > 1 in either WT or P95H samples, 2) absolute log2 Fold Change >1, 3) False Discovery Rate (FDR) < 0.05.
For RNA-seq reads, normalization and differential analysis between WT and P95H expression levels were performed with the edgeR package. Differential expression was evaluated with the same thresholds used for HITS-CLIP differential binding sites.
Differential alternative splicing (AS) analysis was performed with rMATS26 (v3.2.5, -t paired -len 100), capable of handling replicates. Differential AS events with FDR < 0.1 and absolute differential percentage of spliced in (delta PSI) > 5% were considered significant.
Functional annotation enrichment analysis with Gene Ontology terms, KEGG and REACTOME pathways and the DOSE ontology were performed using the clusterProfiler Bioconductor package.
RT-PCR validation
Splice isoforms were amplified by RT-PCR (see supplemental methods for primer sequences). cDNA from cell lines and primary patient samples cDNA was submitted to PCR with primers spanning target sequences alternatively bound by WT versus MUT SRSF2 (Supplementary Table S14). PCR products were resolved by agarose gel electrophoresis and visualized and quantified using Image Lab 3.0 software (BioRad). Splice isoforms were identified and validated by Sanger sequencing.
Primary human cell analysis and Colony Forming Unit (CFU) assays
All human primary cells were obtained with donor’s written consent or from commercial sources. All human studies were approved by the Yale University Human Investigation Committee. Colony formation assays of human fetal liver and adult mobilized peripheral blood CD34+ hematopoietic stem and progenitor cells were performed per standard protocol (Stem Cell Technologies) and as described in Sarma et al.27 Detailed information is provided in Supplementary Information.
Statistics
If not otherwise indicated, pairwise comparisons were analyzed using the unpaired two-sided t-test (Graphpad PRISM Software, version 7.0). P-values < 0.05 were considered significant with error bars representing the standard error of mean.
RESULTS
Defining the differential interactome of SRSF2 P95H in vivo
To characterize the in vivo effects of SRSF2P95H mutation in the hematopoietic context, we generated stable, isogenic Human Erythroid Leukemia (HEL) cell lines with lentivirally conferred inducible expression of Flag-tagged wild-type (WT) and mutant (P95H) SRSF2 (Figure 1a). Exogenous SRSF2 expression was approximately 2 fold higher than endogenous expression, with similar levels between WT and P95H (Figure 1b and Supplementary Figure S1a). We verified the absence of exogenous SRSF2 in uninduced cells (Supplementary Figure S1b) and we checked by proliferation assays that SRSF2 WT/P95H overexpressing cells did not show significant differences in growth (Supplementary Figure S1c), indicating that the Flag-tag does not alter the function of the WT/P95H SRSF2 protein as previously shown10,12,15,20, and confirming the viability of the widely used HEL cell model. We also verified localization of Flag-tagged SRSF2 to nuclear speckles by co-localization with SRSF1 in pcDNA transfected 293ft cells (Supplementary Figure S1d). Additionally, we tested our lentiviral construct in primary human CD34+ fetal liver cells, and performed Colony Forming Unit (CFU) assays to monitor the impact of SRSF2 mutations on hematopoiesis. The total number of colonies was significantly reduced upon SRSF2P95H expression compared to WT SRSF2, with a relative increase of monocytic lineage colonies (CFU-M) in colony composition (Figure 1c and Supplementary Figure S1e), consistent with the monocytic lineage skewing observed in previous studies10 and the preferential occurrence of SRSF2 mutations in CMML2,28.
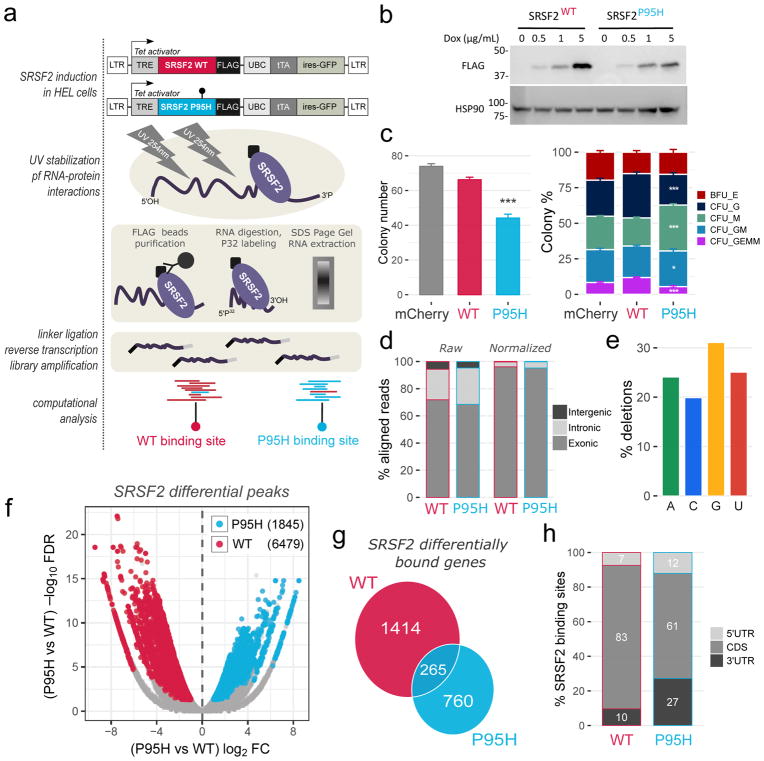
Figure 1. The SRSF2 P95H mutation alters SRSF2 in vivo RNA interactome.
(a) Overview of the HITS-CLIP procedure. Top: generation of lentiviral vector constructs expressing C-terminally Flag-tagged SRSF2 WT and P95H in a doxycycline inducible manner. Center: HITS-CLIP key experimental steps. Bottom: computational identification of differentially bound regions, preferentially bound by WT (in red) or by P95H (in cyan) SRSF2. (b) Dose dependent inducible expression of Flag-tagged SRSF2 (WT and P95H). (c) Colony Forming Unit Assay for control CD34+ cells (mCherry), and cells with transient induction of SRSF2 WT or P95H. Left panel: total number of colonies. Right panel: composition of colonies (mean values + SEM). Total colony numbers and colony type percentages were compared between WT and P95H by two-tailed t-test (*P<0.05, **P<0.01, ***P<0.001). (d) Percentage of HITS-CLIP reads aligned to exonic, intronic and intergenic regions. In the left panel, percentages were scaled using total region lengths as a normalizing factor. (e) Percentage of HITS-CLIP crosslinking induced deletions mapping to each RNA nucleotide. (f) Volcano plot displaying fold changes and false discovery rate values for each HITS-CLIP binding site. Significant differentially bound regions are highlighted in red and cyan (preferentially bound by WT or P95H SRSF2 respectively). (g) Venn diagram of genes with at least one SRSF2 differential binding site. (h) Percentage of SRSF2 differential binding sites located within 5′UTR, 3′UTR, and CDS regions of protein coding genes.
To analyze and compare in vivo differential RNA binding between SRSF2WT and SRSF2P95H HEL cells, we next performed high-throughput sequencing of RNA isolated by UV-crosslinking and immunoprecipitation (HITS-CLIP)23–25. Transgenes were induced for 36 hours and cells UV-crosslinked, followed by stringent RNA immunoprecipitation and high-throughput deep sequencing (see Figure 1a for an overview). The original protocol was modified to allow more efficient RNA-adapter ligation off-beads and increase yield (Figure 1a, see Materials and Methods for details). RNAse digestion was optimized (Supplementary Figure S1f) and bound RNAs isolated, reverse transcribed, amplified (Supplementary Figure S1g) and sequenced in four independent CLIP-replicates for both WT and P95H SRSF2 (Supplementary Tables S1–S2). The majority of reads for both WT and P95H SRSF2 aligned to exonic regions in protein coding genes (Figure 1d and Supplementary Table S3) consistent with SRSF2’s known binding to exonic splicing enhancers. Collectively, these results suggest preservation of the global RNA-binding function of SRSF2P95H.
UV crosslinking produces single nucleotide amino acid-RNA adducts, resulting in nucleotide deletions at the time of reverse transcription29,30 which mark bona fide protein binding sites with high specificity at single-nucleotide resolution31. We verified that uracil bias, a concern raised for crosslinking induced deletions in previous studies20,32, was absent in our experiments (Figure 1e). Therefore, we integrated read coverage and UV crosslinking induced deletion sites to identify differential in vivo RNA interactions between SRSF2WT and SRSF2P95H with high specificity (see Materials and Methods for details). We identified a total of 6479 and 1845 regions preferentially bound by SRSF2WT and SRSF2P95H respectively: differential binding is therefore unbalanced, with approximately 75% lost and 25% gained interactions for SRSF2P95H (Figure 1f, Supplementary Table S4). Differentially bound regions are located in 1679 and 1025 genes respectively, with 265 overlapping genes (Figure 1g), suggesting denser clustering of preferential sites in SRSF2WT than SRSF2P95H bound transcripts. The majority of genes with differential SRSF2 binding are protein coding (76–78%, Supplementary Figure S1h), with small percentages of antisense and long intergenic non-coding RNAs (2–4%). Interestingly, in protein coding transcripts, WT binding sites predominate within CDS regions (83%), while P95H preferential sites exhibit almost a three-fold percentage increase within 3′UTRs (Figure 1h). Further comparison highlighted that P95H preferential binding sites occur on longer (P=1.2x10−28) and less constitutive (P=8.3x10−13) exons than WT sites (Supplementary Figure S1i). An exemplary binding profile for SRSF2 is shown in Supplementary Figure S2a. Mutant SRSF2 has been reported to differentially splice its own CCNG-rich alternative 3′UTR exon, targeting the SRSF2 transcript for non-sense mediated decay (NMD)33. We confirmed this event in a minigene splicing assay34 (Supplementary Figure S2b).
Parallel RNA-Seq was performed in duplicate on ribosomally depleted total RNA harvested from the same cells used for HITS-CLIP, as well as from uninduced cells, allowing to quantify transcriptome-wide expression levels (Supplementary Tables S5-S6). Consistent with prior data10,12 overall expression changes between SRSF2WT and SRSF2P95H HEL cells were small with almost no significant changes (Supplementary Figure S2c). Importantly, direct comparison between HITS-CLIP and RNA-Seq signals showed a low positive correlation between binding site intensities and transcript expression levels (coefficient of determination = 0.05) (Supplementary Figure S2d). Therefore, binding affinity, rather than transcript expression levels, seems to be the main factor influencing binding peak intensity.
SRSF2 mutations skew RNA binding affinity and specificity in vivo
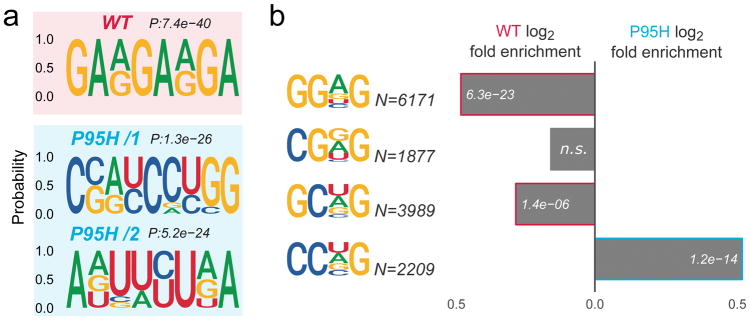
We have previously shown, that the 3–4-fold higher affinity of the RBD to the CCNG (Kd=0.06uM) compared to the GGNG (Kd=0.2uM) consensus motif of all three SRSF2 mutants (P95H/L/R) in vitro is attributable to mutation-induced structural changes in the RBD10. However, in vitro assays cannot study the full length protein that includes the SR domain nor its function within the complexity of the splicing machinery. Therefore, we analyzed whether in vitro binding preferences are maintained in vivo, by discriminative motif analysis between SRSF2WT and SRSF2P95H differential sites (see Supplementary Information). Preferentially WT-bound regions are strongly enriched in GA-rich motifs, while preferentially P95H-bound sites display an equal enrichment of two motifs, C-rich and U-rich respectively (Figure 2a). The U-rich motif is likely consistent with the increased frequency of P95H binding sites in 3′UTR (Figure 1h), although it can be found also in 5′UTR and CDS bound regions (Supplementary Figure S3). Since the first two emerging motifs in Figure 2a contain the canonical SSNG sequences associated with SRSF2 binding10,12,20, we compared their frequencies among in vivo differentially bound regions. Consistent with in vitro data, RNA regions preferentially bound by SRSF2P95H are enriched in CCNG motifs (P=1.2x10−14), while regions preferentially bound by SRSF2WT are enriched in GGNG (P=6.3x10−23) (Figure 2b). To a lesser extent, WT regions are also enriched in GCNG (P=1.4x10−6), while no significant differences were found for the CGNG motifs that were generally less frequent (Figure 2b).
Figure 2. The SRSF2 P95H mutation alters SRSF2 in vivo RNA motif specificity.
(a) Top enriched motifs for WT and P95H SRSF2 binding sites, identified by discriminative analysis of kmer composition. Corresponding p-values are displayed on top of each logo. (b) Relative enrichment of the SSNG (S=C/G, N=C/G/A/U) RNA consensus motifs in RNA regions preferentially bound by WT versus P95H SRSF2. The number of motif occurrences and differential enrichment p-values are displayed for each bar.
In summary, our data uncover skewed sequence specificity and affinity and increased relative binding in UTRs at the expense of binding in CDS exons. SRSF2 mutations thus confer a complex change of function consistent with the nature of hotspot mutations.
SRSF2 mutations globally affect splicing by targeting splicing factors
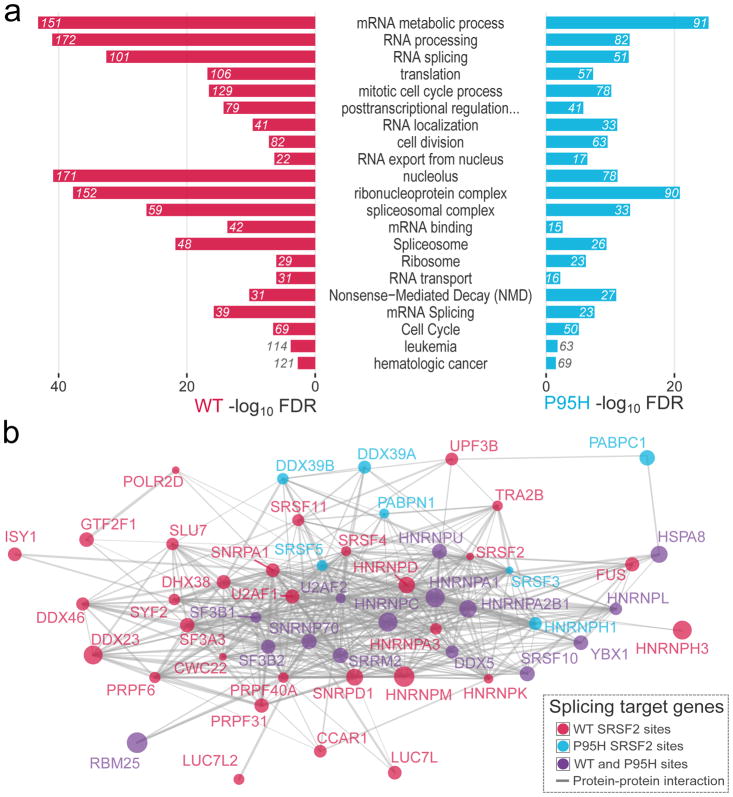
Since we identified differential SRSF2 binding in several hundred protein-coding transcripts, we applied functional annotation analysis to genes preferentially bound by SRSF2WT or SRSF2P95H. Interestingly, we identified significant enrichments for “RNA processing”, “RNA splicing”, “mRNA binding”, and “spliceosome” categories in both populations (Figure 3a, Supplementary Tables S7–S8). Prior studies have shown that SR proteins function in alternative splicing of related35 and other splicing factors36. Differential binding occurs in members of several mRNA splicing factor families, and in particular in the heterogeneous nuclear ribonucleoprotein (hnRNP) protein family, as well as in RNA binding motif (RBM) and other SR proteins known to participate in complex cooperative and competitive regulation of alternative splicing with SRSF221,37–41. This high degree of interplay is evident when overlaying known protein-protein interaction data, as all these factors emerge as a closely interconnected cluster (Figure 3b). These data suggest that SRSF2 mutations may affect a broad population of genes via alternative splicing of other RNA binding proteins and splicing factors, rather than solely via direct binding and splicing of its direct downstream targets.
Figure 3. Differentially bound SRSF2 targets are enriched in RNA binding and splicing genes.
(a) Functional annotation enrichment analysis of differentially bound transcripts by WT and P95H SRSF2. The number of genes belonging to each category is displayed. (b) Protein-protein interaction network of SRSF2 RNA interactors associated with splicing. The size of each node is proportional to the number of differential SRSF2 binding sites: WT (in red), P95H (in cyan) or both (in violet).
SRSF2P95H promotes inclusion of CCNG-rich exons and exclusion of GGNG-rich exons and results in differential splicing of splicing factors
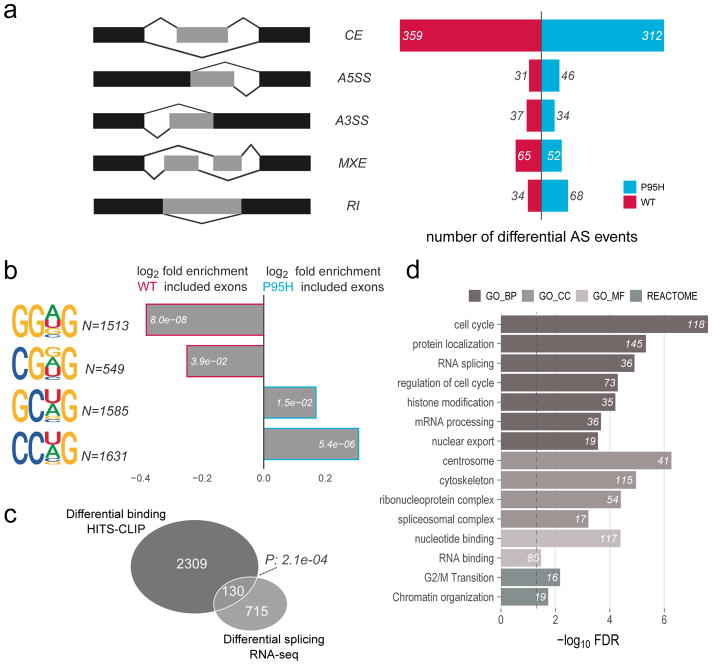
Differential splicing analysis between SRSF2P95H and SRSF2WT cells was performed with rMATS26, detecting 1038 significant alternative splice events (Figure 4a), mostly enriched in cassette exons (CE, n=671, 65%) and without a significant trend towards exon inclusion versus exclusion42 (Figure 4a, Supplementary Table S9). Consistent with the altered SRSF2P95H binding and previous reports10,12,13, cassette exons preferentially included in SRSF2WT cells were enriched in GGNG motifs (P=8.0x10−8) and CGNG motifs (P=3.9x10−2), while cassette exons preferentially included in SRSF2P95H cells were enriched in CCNG motifs (P=5.4x10−6) (Figure 4b). Curiously, GCNG motifs were enriched in cassette exons preferentially included in SRSF2P95H (P=1.5x10−2), while the same motif was enriched in SRSF2WT binding sites. A minor yet significant subset of 130 differentially bound targets, identified via HITS-CLIP, were also differentially spliced (P=2.1x10−4) (Figure 4c). Functional annotation analysis of genes with differential splicing confirmed the enrichment in “cell cycle”, “RNA splicing”, “RNA binding” and “spliceosomal complex” categories (Figure 4d, Supplementary Table S10).
Figure 4. SRSF2 P95H mutations promote alternative splicing with inclusion of CCNG rich exons and enrichment in RNA binding and splicing genes.
(a) Determination of differential alternative splice events via rMATS analysis in HEL cells engineered to express SRSF2 WT vs P95H. Left panel: five classes of alternative splicing events were considered: cassette exon (CE), alternative 5′ splice site (A5SS), alternative 3′ splice site (A3SS), mutually exclusive exons (MXE) and retained intron (RI). Right panel: the number of significant events with more inclusion in WT or P95H cells is displayed. (b) Relative enrichment of the SSNG (S=C/G, N=C/G/A/T) RNA consensus motifs in cassette exons preferentially spliced in WT vs preferentially spliced in P95H SRSF2 expressing cells. The number of motif occurrences and enrichment p-values are displayed for each bar. (c) Overlap between genes with differential binding and differential splicing in HEL cells expressing either WT or P95H SRSF2. The significance of the overlap is displayed. (d) Functional annotation enrichment analysis of differentially spliced genes in WT vs P95H HEL cells. The number of genes falling into each category is indicated beside each bar.
Since alternative splicing analysis of SRSF2 mutant versus wild-type has been performed in various systems, we integrated results from 9 published RNA-Seq datasets10,12,13,43,44 to determine whether our findings are applicable to SRSF2 mutant cell lines, mouse models and primary patient samples. Due to the high heterogeneity among biological samples, experimental procedures and computational pipelines, the comparison was performed by overlapping lists of genes reported to be differentially spliced in these datasets (Supplementary Table S11 and Supplementary Figure S4a). Strikingly, no gene was reported as differentially spliced in all the datasets (Supplementary Figure S4b). Among the 446 genes recurrently reported as differentially spliced in at least 3 datasets, 28% contain differential SRSF2P95H and SRSF2WT binding sites (Supplementary Figure S4b and Supplementary Table S12). Functional annotation analysis of these recurrent genes again once more highlighted strong enrichments for RNA splicing and processing pathways, as well as cell cycle and leukemia signatures (Supplementary Figure S4c and Supplementary Table S13). Interaction annotation analysis revealed a cluster of interacting RNA binding proteins among these genes, including multiple differentially bound hnRNPs such as HNRNPA2B1, HNRNPM, HNRNPH1 and HNRNPH3 (Supplementary Figure S4d).
Together, these data suggest that differential binding does not invariably result in differential splicing and that RNA binding and splicing are likely context dependent, as also shown by the small percentage of genes found to be recurrently mis-spliced in primary patient samples and cell line models. SRSF2 mutations result in context dependent alternative splicing of direct targets as well as indirect targets via differential binding and splicing of RNA processing and splicing partners.
SRSF2 mutations modulate the splicing network by altering splicing of members of the hnRNP and SR protein families
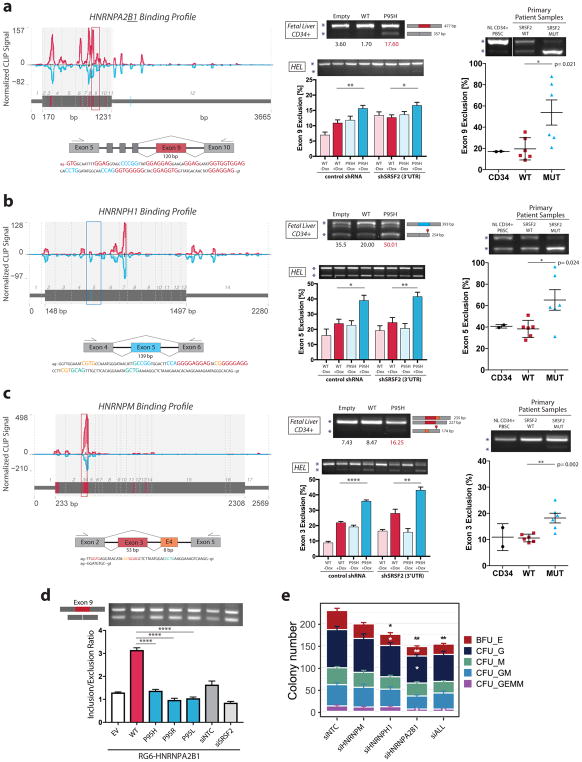
hnRNP proteins are known to antagonize SR protein function45 and were highly represented in our HITS-CLIP and in alternative splicing datasets (Figure 3b and Supplementary Figure S4d). To validate our results, we determined differential splicing via exon-specific RT-PCR (see Supplementary Table S14 for primer design) in dox-induced HEL cells (72h Dox), in virally-transduced CD34+ cells and in primary patient samples with SRSF2 mutations (Supplementary Table S15). To remove potential splicing effects due to SRSF2 overexpression in our HEL system, we also combined knockdown of endogenous SRSF2 (targeted against the 3′UTR) with the expression of WT or P95H SRSF2. Importantly, we confirmed differential splicing of HNRNPA2B1, HNRNPH1, HNRNPH3, and HNRNPM in both our cell systems and in primary patient samples (Figure 5a–c, Supplementary Figure S5a–c, Supplementary Table S16).
Figure 5. SRSF2 mutations results in differential binding and splicing of HNRNP proteins.
(a–c) Differential binding and splicing in HNRNP proteins is shown for HNRNPA2B1 (a), HNRNPH1 (b), and HNRNPM (c). Left panels: transcript maps showing WT (red) and P95H (cyan) SRSF2 binding profiles. The maps display mean normalized HITS-CLIP signal with nucleotide resolution. Standard errors for each position are shown as ribbons under mean lines. Crosslinking-induced deletions are marked in black. Exon boundaries are represented as vertical dotted lines. Differential interaction sites are highlighted on the transcript. Center panels: RT-PCR capturing differentially bound exons was performed in 3–4 replicates in HEL cells with or without doxycycline induction of SRSF2 WT or P95H expression, and with or without knockdown of endogenous SRSF2, and in human fetal liver CD34+ cells transduced with empty, SRSF2 WT or SRSF2 P95H expressing lentivirus. Right panels: primary patient derived samples - alternative splice events were quantified in normal CD34+ (n=2), WT MDS/AML (n=6), and MUT MDS/AML (n=6) samples. The magnitude of the alternative splice event in % was calculated as ratio of alternative splice event to total expression. Considered bands are marked by an asterisk (*). Predicted band sizes and transcript sizes are indicated to the right of the gel. (d) Direct differential splicing of the HNRNPA2B1 exon 9 by WT vs P95H SRSF2 verified by a minigene splicing assay. RG6-HNRNPA2B1 plasmid was co-transfected with empty vector, SRSF2 WT, SRSF2 P95H/L/R, and control or SRSF2 siRNA. Alternative splicing of exon 9 was determined via semi-quantitative PCR by measuring the ratio of alternative exon exclusion over total (exclusion+inclusion) band intensities (n=3). (e) Colony Forming Unit Assay for control cells (siNTC), cells silenced for HNRNPM or HNRNPH1, cells induced to splice out HNRNPA2B1 exon 9, and cells treated for all three modifications together (siALL). The same number of cells was plated in all experiments. The total number and the composition of colonies is displayed. Total colony numbers and colony type percentages were compared to the control (siNTC). Standard errors (SE) for colony numbers are displayed. In panels (a–e), significance values were determined by one way ANOVA with Sidák Post Hoc Test (*P<0.05, **P<0.01, ***P<0.001).
In HNRNPA2B1, reduced binding of MUT SRSF2 to the GGNG-rich exon 9 results in exon skipping (Figure 5a; P=2.1x10−2 and Supplementary Figure S5a–b). Exon 9 encodes parts of the glycine-rich and low complexity region of the protein and affects nuclear-to-cytoplasmic mRNA trafficking in neuronal cells46. We confirmed in a minigene splicing assay that HNRNPA2B1 exon 9 is a direct target of SRSF2 and differentially spliced by mutant SRSF2 (Figure 5d). In HNRNPH1, increased mutant SRSF2 binding to exon 5 surprisingly correlates with exon 5 exclusion (Figure 5b). Exon 5 represents a frame preserving exon and its loss results in a premature stop codon predicted to induce NMD-mediated degradation of the alternatively spliced transcript. While HNRNPM (Figure 5c) and HNRNPH3 (Supplementary Figure S5c) both display decreased SRSF2P95H binding, this difference leads to divergent dysregulation of splicing. Decreased binding of mutant SRSF2 to exon 5 in HNRNPH3 facilitates the use of a proximal 5′SS in exon 4, while decreased binding of mutant SRSF2 to exons 3 and 4 of HNRNPM results either in inclusion of both exons, or the skipping of exon 3 that is included in the predominant isoform in wild-type cells. Interestingly, exogenous expression of SRSF2WT can induce similar alternative splice changes in these target genes, albeit at lower levels compared to the P95H mutant, corroborating previous reports that changes in SRSF2 expression levels can induce alternative splicing10,46 (Figure 5a–c). Reduced mutant SRSF2 binding also resulted in variable splice outcomes in the splicing factors SRSF10 (intron retention; Supplementary Figure S6a) and in RBM25 (exon skipping, Supplementary Figure S6b), both predicted to result in NMD of the alternatively spliced transcripts.
To prove the indirect impact of SRSF2 mutations on hematopoiesis via alternative splicing of hnRNP proteins, we mimicked the effect of dysregulated HNRNP splicing observed in P95H mutant cells by performing single and combined siRNA treatment of HNRNPM, HNRNPH1 and HNRNPA2B1 in human CD34+ hematopoietic stem and progenitor cells. Since alternative splicing in HNRNPH1 and HNRNPM is predicted to result in NMD, we designed siRNAs predicted to result in knockdown of their respective targets (Supplementary Figure S5d). To mimic the low delta PSI splice changes, we titrated siRNAs to achieve knockdown by < 50%. On the other hand, the siRNA against HNRNPA2B1 was specifically targeted against the alternatively spliced exon 9 with the goal to affect overall expression as little as possible while still achieving exclusion of exon 9 (Supplementary Figure S5e). Importantly, we detected significant alterations in hematopoietic differentiation for two of the tested hnRNPs: in fact, our CFU assay shows a reduced number of total colonies with respect to controls for HNRNPA2B1 alternatively spliced and HNRNPH1 silenced cells (Figure 5e), reciprocating the finding in SRSF2P95H cells (Figure 1c). The decrease particularly affects the class of erythroid burst-forming units (Figure 5e). These results demonstrate that isoform regulation of downstream targets of SRSF2P95H can alter hematopoiesis, underlining the biological impact of splicing changes in splicing factors arising from the alteration of the SRSF2 binding fingerprint.
In summary, our data confirm the complexity of binding – to – splicing outcomes and suggest that SRSF2 mutations affect splicing of other RNA binding proteins such as hnRNPs, known to modulate splicing in conjunction with SRSF2 and other SR proteins.
DISCUSSION
SRSF2 mutations, associated with myelodysplastic syndromes, portend a poor prognosis. In depth understanding of how mutations alter SRSF2 function is essential for the development of novel therapeutics. We here provide the first transcriptome-wide, unbiased characterization of SRSF2P95H differential RNA-binding in vivo in a syngeneic hematopoietic cell context.
We have previously shown via in vitro structure function studies that compared to SRSF2WT, SRSF2P95H has an approximately 4-fold higher affinity for the CCNG consensus motif, while binding affinity to the GGNG motif remains unchanged10. While these in vitro RNA binding data explain the preferential inclusion of CCNG-rich exons by SRSF2P95H, they do not explain the preferential exclusion of GGNG-rich exons identified in RNA sequencing data presented here or previously published10,12,13. Our in vivo HITS-CLIP RNA binding data on the other hand reveal preferential binding of SRSF2P95H to CCNG-rich regions and reduced binding to GGNG-rich regions, thus providing the mechanistic basis for the observed differential splicing of CCNG- and GGNG-rich exons. The P95 residue lies in the linker region between the canonical RRM (aa16-90) and the SR domain (aa117-211) and may have dual function in determining RNA binding specificity, as part of the RNA binding domain (aa1-101)10,11, and the conformation between the RRM and the SR domains2, thereby altering protein-protein interactions in a sequence specific manner. We identify overall more events with reduced binding of SRSF2P95H (Figure 1f). This loss of binding may explain the absolute requirement for the WT allele to avoid rapid bone marrow failure when only the mutant allele is expressed10.
By checking whether the SRSF2P95H mutation affects distinct pathways that may contribute to its role in MDS and leukemogenesis, we identified strong enrichment in RNA processing and splicing functions among genes differentially bound and spliced by wild-type and mutant SRSF2, confirmed in several published datasets analyzing alternative splicing10,12,43,44. Autoregulation of splicing factors is widely recognized as a means to control expression via a negative feedback loop33,34,47–50. Reciprocal regulation of splicing and RNA binding factors through alternative splicing adds additional regulatory complexity to the splicing network36,48. SRSF2 mutations thus may not only affect targets by direct binding, but via alternative splicing of other splicing factors. This finding is particularly interesting, given the differential sensitivity of splicing factor mutant cells to global splicing inhibitors4,51,52. Downstream accumulation of aberrant yet functional splice transcripts might also lead to the creation of neo-antigens susceptible to immune modulator therapy in AML with agents such as Pembrolizumab, as currently investigated in clinical trials (NCI-2016-01287).
SR proteins are critical in exon definition and regulate a variety of AS events, with an overall preference towards repression of intron retention events and activation of cassette exon events22. We validated binding-to-splicing effects in cell lines with extended transgene induction (72h), in primary CD34+ cells expressing exogenous WT or P95H SRSF2, and most importantly in primary patient-derived cells. We focused our analysis on hnRNP proteins, overrepresented in our HITS-CLIP data and known to antagonize SR proteins, including SRSF248. We confirmed differential splicing of several HNRNP proteins, including HNRNPA2B1, HNRNPH1, HNRNPM, and HNRNPH3. HNRNPA2B1 exon 9 encodes parts of the glycine-rich and low complexity region (LCR), which has been implicated in liquid droplet organelle formation53. Most recently, HNRNPA2B1 was identified as a potential reader of the N6-methyladenosine mark on pre-mRNA and primary miRNA transcripts, facilitating splicing and pri-miRNA processing54. Splice site mutations in HNRNPA2B1 have been described in T-cell leukemia/lymphoma55. Importantly, we detected impairment in hematopoietic differentiation in human primary human CD34+ fetal liver cells when HNRNPA2B1 is alternatively spliced, mimicking the effect of SRSF2 P95H mutation. This result is a proof of concept, demonstrating that isoform regulation of downstream targets of SRSF2 P95H alter hematopoiesis, highlighting the biological impact of splicing changes in splicing factors stemming from the alteration of the SRSF2 binding fingerprint.
Differential splicing of other splicing factors may also explain the marked heterogeneity observed between different datasets and also between individual SRSF2 mutant patient samples. We have previously identified EZH2 as differentially spliced in SRSF2 mutant MDS10. However, EZH2 differential splicing is present only in 3 out of the 9 published data sets and we do not detect significant differences in EZH2 binding in our inducible cell lines (Supplementary Figure S7), similar to findings by Zhang et al.12. We can thus not determine whether EZH2 differential splicing detected in patient cells is a direct splice event due to increased SRSF2P95H binding. Several improvements in CLIP techniques will be necessary to allow determination of direct splicing factor targets in primary patient cells to shed further light on mechanisms of splicing factor mutation-dependent oncogenesis.
The work presented here represents a significant step towards identifying the mechanism of SRSF2 mutation-induced alternative splicing in hematologic malignancies. Our studies highlight the complexity of AS induced via differential splicing of other RNA processing and splicing genes, resulting in a “splicing-cascade” phenotype. This phenomenon may underlie the vulnerability of SRSF2 mutant MDS to global splicing inhibitors. Subtle, but broad disruption of splicing may not only lie at the root of how splicing factor mutations cause MDS/AML, but also represent their Achilles heel. The challenge ahead is to fully resolve the combinatorial texture of alternative splicing events leading to hematologic malignancies.
Supplementary Material
Acknowledgments
We thank all our patients. We thank all clinicians and clinical staff for their help with patient recruitment. Work was funded in part by the Edward P. Evans Foundation, by the NIH/NIDDK R01DK102792, departmental funds from the Yale Comprehensive Cancer Center (YCCC), and a YCCC pilot grant (to SH). This material is based in part upon work supported by the State of Connecticut under the Regenerative Medicine Research Fund (GS, SH). Its contents are solelythe responsibility of the authors and do not necessarily represent the official views of the State ofConnecticut or Connecticut Innovations, Incorporated. Research reported in this publication was in part supported by the NIDDK under Grant U54DK106857. YL was partially supported by the National Natural Science Foundation of China (Grant No. 81660682). We thank Diane Krause, Manoj Pillai, and Karla Neugebauer (Yale University) for helpful suggestions. We thank the Yale Stem Cell Center Genomics Core and the Yale Center for Genome Analysis (YCGA) for high-throughput sequencing and the Yale University High Performance Computing Center for use of clusters to run bioinformatics analysis. We thank Dr. Tomoyuki Yamaguchi at the Japan Science and Technology Agency for the kind gift of the CS-TRE-Ubc-tTA-I2G plasmid [56]. We also thank Didier Trono for the psPAX2 plasmid (Addgene plasmid # 12260) and Tannishtha Reya for the pCMV-VSVG plasmid (Addgene plasmid # 14888).
Footnotes
AUTHOR CONTRIBUTIONS
TT analyzed next-generation sequencing data and wrote the manuscript. YL, KR, PJ, and GS performed experiments, analyzed the data, and wrote the manuscript. AT, YS, JM, KB, RV, AA, and AD performed experiments. AQ provided essential input for the manuscript. SH initiated the study, performed experiments, analyzed the data, provided supervision and wrote the manuscript with input from the other authors.
DISCLOSURE DECLARATION
The authors declare no conflict of interest
DATA ACCESS
Sequencing files generated from this work have been deposited in the GEO database and are available under the accession number GSE111900.
References
- 1.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–9. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 2.Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML) Blood. 2012;120:3080–8. doi: 10.1182/blood-2012-01-404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122:3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SC-W, Dvinge H, Kim E, Cho H, Micol J-B, Chung YR, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22:672–678. doi: 10.1038/nm.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard JM, Sanford JR. The RNAissance family: SR proteins as multifaceted regulators of gene expression. Wiley Interdiscip Rev RNA. 2015;6:93–110. doi: 10.1002/wrna.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–71. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 7.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–70. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 8.Mayeda A, Screaton GR, Chandler SD, Fu XD, Krainer AR. Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol Cell Biol. 1999;19:1853–63. doi: 10.1128/mcb.19.3.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 10.Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC-W, Ramakrishnan A, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell. 2015;27:617–30. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daubner GM, Cléry A, Jayne S, Stevenin J, Allain FH-T. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012;31:162–74. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Lieu YK, Ali AM, Penson A, Reggio KS, Rabadan R, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A. 2015;112:E4726–34. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kon A, Yamazaki S, Nannya Y, Kataoka K, Ota Y, Nakagawa MM, et al. Physiological Srsf2 P95H expression causes impaired hematopoietic stem cell functions and aberrant RNA splicing in mice. Blood. 2018;131:621–635. doi: 10.1182/blood-2017-01-762393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu X-D. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Müller-McNicoll M, Botti V, de Jesus Domingues AM, Brandl H, Schwich OD, Steiner MC, et al. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016;30:553–66. doi: 10.1101/gad.276477.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Änkö M-L, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, et al. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Änkö M-L. Regulation of gene expression programmes by serine–arginine rich splicing factors. Semin Cell Dev Biol. 2014;32:11–21. doi: 10.1016/j.semcdb.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 18.Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol Cell Biol. 2000;20:1063–71. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chandler SD, Mayeda A, Yeakley JM, Krainer AR, Fu XD. RNA splicing specificity determined by the coordinated action of RNA recognition motifs in SR proteins. Proc Natl Acad Sci U S A. 1997;94:3596–601. doi: 10.1073/pnas.94.8.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, et al. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–35. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J, Mayeda A, Krainer AR. Exon identity established through differential antagonism between exonic splicing silencer-bound hnRNP A1 and enhancer-bound SR proteins. Mol Cell. 2001;8:1351–61. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 22.Bradley T, Cook ME, Blanchette M. SR proteins control a complex network of RNA-processing events. RNA. 2015;21:75–92. doi: 10.1261/rna.043893.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37:376–86. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–5. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 25.Stefani G, Chen X, Zhao H, Slack FJ. A novel mechanism of LIN-28 regulation of let-7 microRNA expression revealed by in vivo HITS-CLIP in C. elegans. RNA. 2015;21:985–96. doi: 10.1261/rna.045542.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen S, Park JW, Lu Z, Lin L, Henry MD, Wu YN, et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarma NJ, Takeda A, Yaseen NR. Colony forming cell (CFC) assay for human hematopoietic cells. J Vis Exp. 2010 doi: 10.3791/2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121:2186–2198. doi: 10.1182/blood-2012-06-440347. [DOI] [PubMed] [Google Scholar]
- 29.Kishore S, Jaskiewicz L, Burger L, Hausser J, Khorshid M, Zavolan M. A quantitative analysis of CLIP methods for identifying binding sites of RNA-binding proteins. Nat Methods. 2011;8:559–64. doi: 10.1038/nmeth.1608. [DOI] [PubMed] [Google Scholar]
- 30.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci U S A. 2009;106:9613–8. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol. 2011;29:607–14. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugimoto Y, König J, Hussain S, Zupan B, Curk T, Frye M, et al. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sureau A, Gattoni R, Dooghe Y, Stévenin J, Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–96. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dreumont N, Hardy S, Behm-Ansmant I, Kister L, Branlant C, Stévenin J, et al. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2010;38:1353–66. doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 36.Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, et al. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 2009;19:381–94. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ankö M-L, Neugebauer KM. RNA-protein interactions in vivo: global gets specific. Trends Biochem Sci. 2012;37:255–62. doi: 10.1016/j.tibs.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Guil S, Gattoni R, Carrascal M, Abián J, Stévenin J, Bach-Elias M. Roles of hnRNP A1, SR proteins, and p68 helicase in c-H-ras alternative splicing regulation. Mol Cell Biol. 2003;23:2927–41. doi: 10.1128/MCB.23.8.2927-2941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooke N, Markovtsov V, Cagavi E, Black DL. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol Cell Biol. 2003;23:1874–84. doi: 10.1128/MCB.23.6.1874-1884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Expert-Bezançon A, Sureau A, Durosay P, Salesse R, Groeneveld H, Lecaer JP, et al. hnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of beta-tropomyosin exon 6B. J Biol Chem. 2004;279:38249–59. doi: 10.1074/jbc.M405377200. [DOI] [PubMed] [Google Scholar]
- 41.Zahler AM, Damgaard CK, Kjems J, Caputi M. SC35 and heterogeneous nuclear ribonucleoprotein A/B proteins bind to a juxtaposed exonic splicing enhancer/exonic splicing silencer element to regulate HIV-1 tat exon 2 splicing. J Biol Chem. 2004;279:10077–84. doi: 10.1074/jbc.M312743200. [DOI] [PubMed] [Google Scholar]
- 42.Han J, Ding J-H, Byeon CW, Kim JH, Hertel KJ, Jeong S, et al. SR Proteins Induce Alternative Exon Skipping through Their Activities on the Flanking Constitutive Exons. Mol Cell Biol. 2011;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komeno Y, Huang Y-J, Qiu J, Lin L, Xu Y, Zhou Y, et al. SRSF2 Is Essential for Hematopoiesis, and Its Myelodysplastic Syndrome-Related Mutations Dysregulate Alternative Pre-mRNA Splicing. Mol Cell Biol. 2015;35:3071–82. doi: 10.1128/MCB.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu J, Zhou B, Thol F, Zhou Y, Chen L, Shao C, et al. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA. 2016;22:1535–49. doi: 10.1261/rna.056101.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busch A, Hertel KJ. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han SP, Friend LR, Carson JH, Korza G, Barbarese E, Maggipinto M, et al. Differential subcellular distributions and trafficking functions of hnRNP A2/B1 spliceoforms. Traffic. 2010;11:886–898. doi: 10.1111/j.1600-0854.2010.01072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGlincy NJ, Tan L-Y, Paul N, Zavolan M, Lilley KS, Smith CWJ. Expression proteomics of UPF1 knockdown in HeLa cells reveals autoregulation of hnRNP A2/B1 mediated by alternative splicing resulting in nonsense-mediated mRNA decay. BMC Genomics. 2010;11:565. doi: 10.1186/1471-2164-11-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–78. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bergeron D, Pal G, Beaulieu YB, Chabot B, Bachand F. Regulated Intron Retention and Nuclear Pre-mRNA Decay Contribute to PABPN1 Autoregulation. Mol Cell Biol. 2015;35:2503–17. doi: 10.1128/MCB.00070-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinez FJ, Pratt GA, Van Nostrand EL, Batra R, Huelga SC, Kapeli K, et al. Protein-RNA Networks Regulated by Normal and ALS-Associated Mutant HNRNPA2B1 in the Nervous System. Neuron. 2016;92:780–795. doi: 10.1016/j.neuron.2016.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell. 2016;30:404–417. doi: 10.1016/j.ccell.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J, et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer Cell. 2015;27:631–43. doi: 10.1016/j.ccell.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–33. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 Is a Mediator of m(6)A-Dependent Nuclear RNA Processing Events. Cell. 2015;162:1299–308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47:1304–1315. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.