Abstract
Recurrence of Clostridium difficile infection (CDI) places a major burden on the healthcare system. Previous studies have suggested that specific C. difficile strains, or ribotypes, are associated with severe disease and/or recurrence. However, in some patients a new strain is detected in subsequent infections, complicating longitudinal studies focused on strain differences that may contribute to disease outcome. We examined ribotype composition over time in patients who did or did not develop recurrence to examine infection with multiple C. difficile ribotypes (mixed infection), during the course of infection. Using a retrospective patient cohort, we isolated and ribotyped a median of 36 C. difficile colonies from 61 patients (105 total samples) at initial infection, recurrence (a second case of CDI within 15–56 days of initial infection), and reinfection (a second case of CDI after 56 days of initial infection). We observed mixed infection in 78.6% of samples at initial infection in patients who went on to develop recurrence compared to 18.1% of patients who did not, and mixed infection remained associated with subsequent recurrence after adjusting for gender and prior antibiotic exposure (OR 3.5, 95% CI 1.3–9.4, P =.015). In patients who were sampled longitudinally (44 consecutive events in 32 patients), the dominant ribotype changed in 31.8% of consecutive samples and the newly dominant ribotype was not detected in prior samples from that patient. Our results suggest that mixed C. difficile infection is more prevalent than previously demonstrated and potentially a marker of susceptibility to recurrence.
Keywords: Clostridium difficile infection, C. difficile ribotypes, recurrent infection
Introduction
Clostridium difficile infection (CDI) is the most common hospital associated infection in the United States, contributing to an estimated $1.5 billion in healthcare costs (1, 2). Of the half million individuals diagnosed with CDI each year, nearly 83,000 patients will develop a second infection after the completion of treatment (1). Recurrent cases of CDI, which occur after treatment and initial symptomatic recovery, have been associated with more severe health outcomes and can significantly increase disease burden and healthcare costs (3).
Susceptibility to CDI and the development of recurrence is impacted by a combination of environmental, host, and pathogen factors. Antibiotic use and increased age, which both have the potential to cause disruption of the microbiota, the indigenous microbial community of the host, are known risk factors for developing CDI (4). Both the host immune response (5) and a lack of microbiota recovery have been implicated in the development of recurrent CDI (6). In the early 2000s, increased rates of CDI were observed in in North American and European hospitals and attributed to epidemics caused by a specific strain of C. difficile, the PCR ribotype F027 (7). This strain has subsequently been associated with more severe infection and recurrence (8). In recent epidemiological studies, hospital transmission and outbreaks have been attributed to other emerging strains of concern (9, 10).
One aspect of C. difficile epidemiology that complicates the identification of the role of pathogenic factors or strain type in CDI is the presence of multiple strains during infection (referred to as “mixed infection”), throughout recurrence/reinfection. Previous studies have suggested that anywhere from 16% to 50% of individuals who experience a second incidence of CDI are due to infection with a new strain (11–13). Furthermore, the presence of multiple C. difficile strains has been estimated to be as high as 16% in a single sample from an individual (12, 14). The relationship between mixed infection and disease outcome such as disease severity or recurrence has not been examined.
In a previous retrospective, longitudinal study conducted in patients with recurrent CDI, we observed that 20/61 patients exhibited more than one ribotype throughout reinfection or recurrence when a single ribotype was sampled at a given timepoint (38%) (6). In this current study, we isolated multiple colonies of C. difficile from fecal samples collected from patients who did or did not experience disease recurrence over time for ribotyping to evaluate mixed C. difficile infection at diagnosis of primary CDI and throughout recurrence/reinfection. Our main objectives were 1) to evaluate the relationship between mixed infection at initial disease diagnosis and recurrence and 2) determine the presence of detected strains throughout infection in individuals reinfected with a new strain.
Materials and Methods
Sample collection and patient definitions
Fecal specimens used in this study were selected from a biorepository of samples collected from patients receiving care at the University of Michigan Health System (UMHS) from October 2010 to June 2014 as a part of the NIH Enterics Research Investigational Network (ERIN) study (6, 15). Fecal specimens were collected from discarded fecal material used by the clinical microbiology lab at UMHS to diagnose CDI. These specimens were collected in Cary-Blair Transport Media and subsequently stored at −80C. We identified a cohort of patients who were sampled longitudinally who did or did not develop recurrent CDI, as previously described by Seekatz et al. (6).
CDI diagnosis was defined following the Infectious Disease Society of America (IDSA) national guidelines at the discretion of the patients’ inpatient care team, who are advised to test only symptomatic patients (16). The UMHS clinical lab follows a two-step algorithm to test for presence of toxigenic C. difficile: 1) detection of glutamate dehydrogenase antigen and toxins A or B using the C. diff Quik Chek Complete (TechLab, Blacksburg, Virginia, USA), followed by reflex testing for tcdB using polymerase chain reaction (PCR) after a discordant test. Chart reviews of the patients’ medical history were conducted to confirm the patients’ first diagnosis of CDI (index) and categorize patients into the following categories: nonrecurrent (no subsequent CDI diagnoses following index infection), recurrent (another positive diagnosis within 15–56 days of infection), and reinfected (another CDI diagnosis > 56 days from index infection), as per CDC surveillance guidelines (17). Additional patient characteristics are described in Table S1.
C. difficile enrichment and isolation
We aimed to isolate 40 colonies of C. difficile from every sample in the study. Initial isolation (without enrichment) was conducted as follows: an aliquot (40 μL) of the thawed fecal sample on ice was placed into an anaerobic chamber (Coy), and 10μL of this was streaked directly onto 0.01 % taurocholate 0.025% cycloserine 0.0016% cefoxitine fructose agar (TCCFA) media and grown overnight at 37°C. The remaining 30μL of the sample was enriched prior to plating in two ways: 1) using ethanol shock (1:1 dilution of fecal material and 70% EtOH) and/or 2) inoculated into 1ml of TCCF-broth, grown overnight at 37°C, diluted 10−4 and 10−5 with PBS, and plated onto TCCFA plates for overnight growth of colonies at 37°C. Samples that initially failed were processed using both forms of enrichment. Isolated C. difficile colonies for PCR ribotyping were picked from the plates and inoculated into 800μL of brain heart infusion (BHI) media for an additional 18 hours of growth at 37°C. A 1:10 dilution of this culture in UltraPure water (Gibco) was boiled at 95°C for 20 min for a DNA template for PCR stored at −20°C. The remaining culture isolates were placed in 40% glycerol and stored at −80°C for future use if necessary.
PCR and ribotype identification
We identified C. difficile ribotypes using PCR-ribotyping of the 16S and 23S rRNA intergenic spacer region as described previously (14, 18). The PCR reaction was carried out as follows: 12 μL of Promega PCR Master Mix (#M7502), 0.5 μL of the forward primer (GTGCGGCTGGATCACCTCCT) and reverse primer (56-FAM/CCCTGCACCCTTAATAACTTGACC) at 10 pmol/μL, 10.5 μL of nuclease free water, and 1μL of DNA template, adjusting template DNA volume to 3 μL to optimize conditions if necessary. PCR cycling conditions were as follows: 95°C for 10 minutes followed by 35 cycles of (30 sec at 95°C, 30 sec at 55°C, and 90 sec at 72°C), and 10 minutes at 72°C.
The PCR product was diluted 1:1000 with nuclease-free water and then loaded into a capillary electrophoresis (CE) plate provided by the University of Michigan DNA Sequencing Core containing a 1:240 ratio of ROX 1000 size standard and Hi-Di Formamide (18). Resulting chromatograms were analyzed using Applied Biosystems Peak Scanner Software (v. 1.0), generating the peak sizes between 200–1000 bp. Peak files were uploaded for analysis where the presence or absence of peaks and their relative abundance was used for comparison to a ribotype database containing known ribotypes (characterized and validated by Martinson et al., http://walklab.rcg.montana.edu/ribo.html) (18). Peak comparisons with a Bray-Curtis similarity < 0.2 were considered a match to the database. Sequences unmatched to database were included in analyses, marked as ‘unmatched’. The F designation differentiates the method used to determine the ribotype (fluorescent PCR) compared to ribotypes determined using traditional gel electrophoresis (generally designated with an R).
Data analysis and statistics
Samples with > 1 successfully isolated colonies were included in our final analyses. Statistical analyses, including counts, proportions, means and inter-quartile ranges, were conducted using R version 3.4.3 (R Foundation for Statistical Computing, Vienna, Austria) and the tidyverse collection of R packages (19). Graphs of relative ribotype abundance were created using R package ggplot2 (20). Alluvial plots were created using R package ggalluvial (21). For initial bivariable analyses, categorical variables were compared among the three patient groups (nonrecurrent, recurrent, and reinfected) using Chi-squared tests, and continuous variables were compared using ANOVA. For subsequent comparisons of mixed infection between only two of the groups (only performed if initial group-wise Chi-squared or ANOVA were significant), simple logistic regression was used to derive the odds ratio (OR), confidence interval (CI), and P value. We further modeled whether mixed infection during a particular episode was associated with subsequent recurrence. Since many patients had more than one qualifying episode for this analysis, we used generalized estimating equation (GEE) models to account for intra-patient correlation, using the R package geepack (22). Multiple covariance structures for the GEE model were considered and the one with the best fit was carried forward for the rest of the analyses. Covariates considered for adjustment were age, gender, history of prior CDI, prior or concurrent antibiotic use, and proton pump inhibitor use, based on review of the literature on clinical risk factors for recurrent CDI (23). Covariates with a P value of <.2 on bivariable analysis were included in the final adjusted model.
Results
At primary infection, patients who developed recurrence are more likely to harbor multiple C. difficile strains
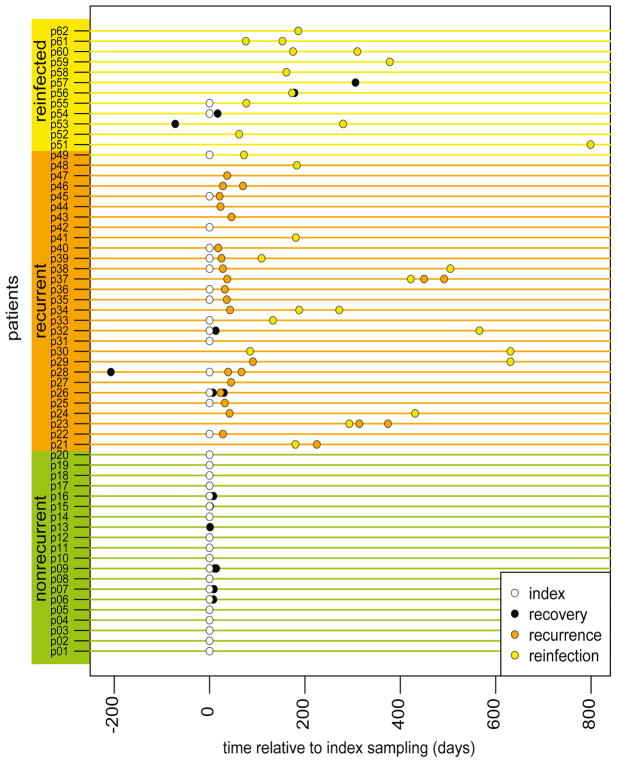
We successfully processed 2,880 C. difficile colonies from 105 total fecal samples. Samples were collected from 61 individuals at CDI diagnosis who experienced a single episode of CDI (nonrecurrent group, patient n = 20, sample n = 26), experienced recurrence (recurrent group, defined as another episode of CDI 14–56 days of initial, or index, infection, patient n = 28, sample n = 59), or reinfection (reinfected, defined as another episode of CDI > 8 weeks after index, patient n = 13, sample n = 20) (Figure 1, Table 1). In examining Figure 1, one notes that we were not able to collect a sample during the index CDI episode for every patient, as some of the first samples we had available on a patient were later during treatment/recovery from an index episode, during a recurrence, or during a reinfection. Thus, we report our data below on all samples but also separately on those samples collected during an index episode.
Figure 1. Timeline of fecal samples collected and processed for ribotyping from patients with C. difficile infection (CDI).
Relative time (days) to CDI index (primary diagnosed episode of CDI) in nonrecurrent patients (single diagnosis of CDI; green), recurrent patients (patients with at least one subsequent diagnosis of CDI within 15–56 days of another CDI episode; orange), and reinfected patients (patients with subsequent episode(s) of CDI > 8 weeks apart; yellow). Sample status based on CDI diagnosis at time of sampling is color-coded by legend (‘recovery’ status = no CDI diagnosis at that timepoint). Only available and successfully processed samples are displayed.
Table 1.
Comparison of mixed infection and dominant ribotype across 1) all samples collected and 2) index samples (primary C. difficile infection) in patients who did not recur (nonrecurrent), patients who recurred (subsequent CDI episode in 15–56 days), and patients who were reinfected (subsequent CDI episode > 8 weeks).
| Across all samples | Nonrecurrent Patients | Recurrent Patients | Reinfected Patients | All Patients | P |
|---|---|---|---|---|---|
| # Patients (% of Total Patients) | 20 (32.8) | 28 (45.9) | 13 (21.3) | 61 | - |
| Age, mean (±SD) | 58.3 (±20.3) | 56.4 (±17.4) | 58.6 (±15.7) | 55.2 (±18.7) | .89 |
| Female gender, n (%) | 10 (50) | 18 (64.2) | 6 (46.1) | 34 (55.7) | .5 |
| PPI use, n (%) | 7 (35) | 5 (17.9) | 3 (25) | 15 (24.6) | .4 |
| Prior antibiotic use | 7 (35) | 6 (21.4) | 2 (15.4) | 15 (24.6) | .4 |
| Concurrent antibiotics, n (%) | 16 (80) | 15 (53.6) | 7 (53.8) | 38 (62) | .1 |
| Mean # samples per patient [IQR] | 1.3 [1,1.25] | 2.11 [1,3] | 1.54 [1,2] | 1.72 [1,2] | .00023 |
| Mean # colonies isolated | 33.6 [26.3, 40] | 35.1 [25, 44] | 29.1 [13.5, 40] | 33.6 [21, 43] | .48 |
| # samples with mixed infection (% samples in group) | 12/26 (46.2) | 42/59 (71) | 8/20 (40) | 62/105 (59) | .015 |
| Mean # of unique ribotypes in samples with mixed infection [IQR] | 2.67 [2,3] | 2.71 [2,3] | 2.25 [2,2] | 2.65 [2,3] | .49 |
| Most common dominant ribotype | F014-020 | F027 | F078-126 | F014-020 | .38 |
|
| |||||
| At Index Sampling (1st documented CDI episode) | Nonrecurrent Patients | Recurrent Patients | Reinfected Patients | All Patients | |
|
| |||||
| # Index samples available (% samples in group) | 19 (18.1) | 14 (13.3) | 3 (2.9%) | 36 (34.3) | - |
| # Index samples with mixed infection (% index samples in group) | 7/19 (36.8) | 11/14 (78.6) | 1/3 (33.3) | 19/36 (52.8) | .047 |
| Mean # of ribotypes in index samples with mixed infection [IQR] | 3 [2,3] | 2.38 [2,3] | 2 [2,2] | 2.57 [2,3] | .59 |
| Most common dominant ribotype (index) | F002 | F027 | F019 | F027 | - |
P values: Chi-squared test for categorical variables; ANOVA for continuous variables
Our dataset represents samples collected during initial CDI diagnosis and throughout recurrence and reinfection (Figure 1). A mean of 33.6 colonies per fecal sample (median = 36) were isolated, processed, and ribotyped, revealing the presence of 32 unique ribotypes. Our ribotyping results demonstrated that 52.8% of all samples contained more than one unique ribotype, reflective of mixed infection. A mean of 2.65 unique ribotypes (median = 2) were observed in these mixed samples. For 19 nonrecurrent and 14 recurrent patients, a sample was collected during the patient’s first CDI diagnosis (index sampling). Mixed infection incidence differed among patients in all three groups (P =.047) (Table 1), and in particular for patients who did not recur, only 36.8% of samples collected at index sampling were mixed compared to 78.6% of index samples collected from patients who developed recurrence (OR 6.3, 95% CI 1.4–35.6, P =.023). Of the three index samples collected from reinfected patients (patients who experienced another CDI episode later), only one sample harbored more than one strain.
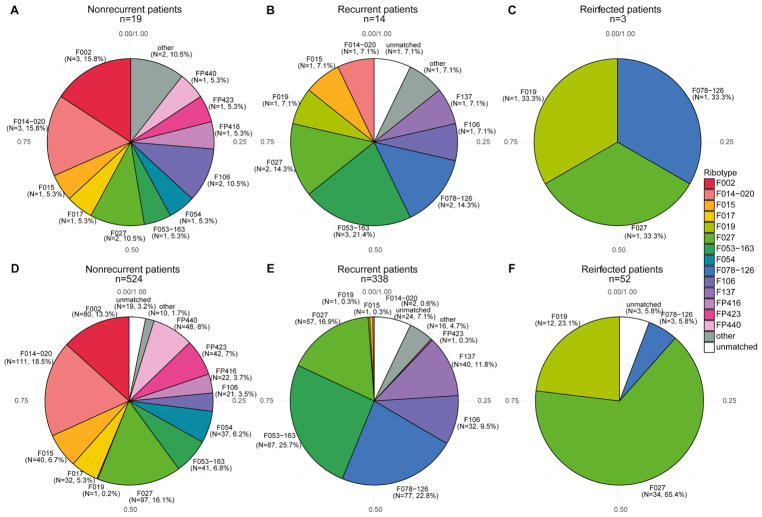
The most dominant ribotypes in index samples collected from nonrecurrent patients were F002 (15.8%), F014-020 (15.8%), and F027 (10.5%). The most dominant ribotypes observed in index samples from recurrent and reinfected patients were F027 (14.3%/33.3%), F053-163 (21.4%/NA), and F078-126 (14.3%/33.3%) (Figure 2A–C). Similar ratios were observed for the total number of C. difficile isolates processed across index samples in each patient group (Figure 2D–F).
Figure 2. Ribotypes observed at index sampling show variation among patient groups.
Proportion of dominant ribotypes observed at index sampling in A) nonrecurrent patients, B) recurrent patients, and C) reinfected patients. The proportion of total number of isolated colonies in each respective group is shown in D–F.
Variable and mixed C. difficile colonization was observed throughout recurrence and reinfection
Given the high percentage of patients presenting with a new ribotype during recurrence or reinfection observed in previous studies (12), we sought to delineate how mixed colonization might change over time in our patient population. A total of 32 patients had > 1 sample available (nonrecurrent sample n = 5, recurrent n = 20, reinfected n = 7), with a mean of 2.5 samples per patient collected, and a mean of 106 days between samplings (Table 2). Of the samples collected subsequent to primary infection and throughout reinfection or recurrence, 66.1% were mixed, and the proportion was not significantly different across patient groups (71.8%% in recurrent patients vs. 66.6%% in nonrecurrent or 66.1% in reinfected patients; P = 0.26).
Table 2.
Comparison of mixed infection and dominant ribotype in post-index samples in nonrecurrent, recurrent, and reinfected patient groups.
| For Patients with > 1 Samples | Nonrecurrent Patients (n = 5) | Recurrent Patients (n = 20) | Reinfected Patients (n = 7) | All Patients (n = 32) | P |
|---|---|---|---|---|---|
| Mean # of days between samples per patient [IQR] | 5.7 [2,8.5] |
119.2* [27.5,96.25]* |
94.7 [44.5,106] |
98.2 [13.75,82.25] |
.21 |
| Most common dominant ribotypes in post-index samplings | F014-020 | F027 | F078-126 | F014-020 | - |
| # samples with mixed infection, post-index (%post-index samples in each group) | 4/6 (66.6) | 28/39 (71.8) a16/25 (64) b11/14 (78.6) |
5/11 (45.5) | 37/56 (66.1) | .26 a/b1 |
| # changes in dominant ribotype across consecutive samplings (% of total consecutive samplings) | 2/6 (40) | 10/31 (32.3) | 3/7 (42.9) | 14/44 (31.8) | .65 |
time reflective of samples collected in recurrent patients who experienced recurrence and reinfection
recurrent episodes in patients recurrent patients
reinfection episodes in recurrent patients
Chi-squared test in recurrent patients alone, comparing recurrent vs. reinfection episodes
In patients who experienced recurrence, a subset of patients was also reinfected at later time points (11/20 patients). To account for this phenomenon, we analyzed the proportion of mixed C. difficile presence at a recurrence timepoint (within 15–56 days of the patient’s last CDI episode) and at a reinfected timepoint (> 8 weeks after the patient’s last CDI episode). The percent of mixed colonization observed in both of these episodes (64% during recurrent episodes and 78.6% during reinfection episodes in recurrent patients) was slightly higher but not statistically significant compared to patients who had only experienced reinfection episodes (45.1%; P = 1) (Table 2).
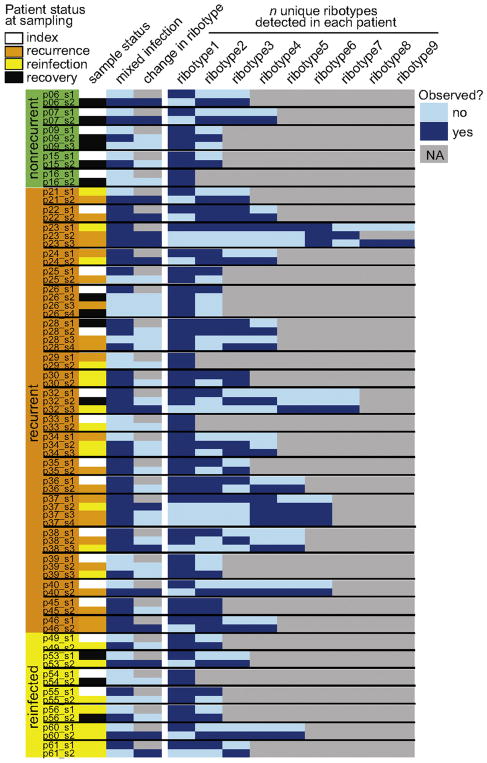
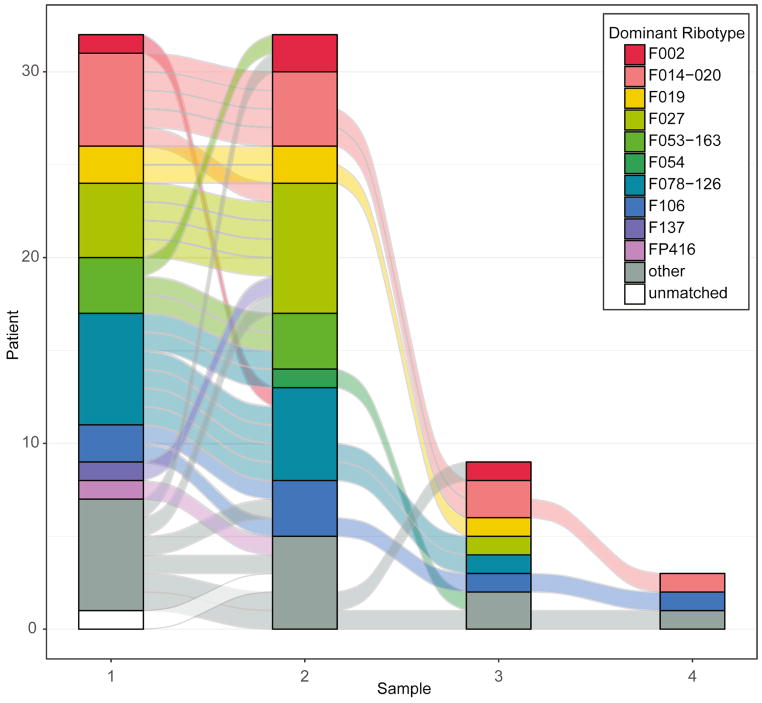
We observed dynamic changes in ribotype dominance and detection in longitudinally sampled patients (Figure 3, Figure 4). The most dominant ribotype observed in non-index samples collected from nonrecurrent, recurrent, and reinfected patients were F014-020, F027, and F078-126 (Table 2, Figure 4). In all longitudinally sampled patients, we observed a change in the dominant ribotype in 34% (14/44) of consecutive samplings (in 14/32 patients, or 43.8% of patients) (Figure 3). In all patients where a change in the dominant ribotype was observed between consecutive samplings, we did not detect the newly dominant ribotype in a previous sample, independent of whether the sample was mixed or not (Figure 3, Table S1).
Figure 3. Patients exhibited variable status in mixed infection over time.
Sample status of diagnosed CDI (index, during recovery, recurrence, or reinfection episode) is shown in column 1 for all samples collected from nonrecurrent, recurrent, and reinfected patients (samples > 1 per patient). Mixed infection (> 1 ribotype observed in sample), change in dominant ribotype (based on sample immediately prior), and co-occurrence of the number of unique ribotypes observed in each patient is shown for each sample (yes/no).
Figure 4. Ribotype stability was variable across time in all patients.
Changes observed in the dominant ribotype observed in samples collected from patients across consecutive samplings (color-coded by ribotype). The connecting flow lines are color coded to match the prior sample’s dominant ribotype.
Detection of multiple strains during an episode of CDI predisposed patients to subsequent recurrence
When modeling subsequent recurrence for each episode of CDI, excluding episodes where samples were collected late during treatment/recovery, we noted an increased odds of subsequent recurrence when mixed infection was present during the initial episode (OR 2.7, 95% CI 1.1–6.7, P =.032), using an exchangeable correlation structure for the GEE model. After adjustment for gender and prior antibiotic exposure (within 1 month of the episode), this increased odds of subsequent recurrence was still statistically significant (OR 3.5, 95% CI 1.3–9.4, P =.015).
Discussion
We observed a high rate of mixed C. difficile infection in fecal samples collected from patients over time at the time of CDI diagnosis and throughout recurrence. In particular, patients who later developed a recurrent episode of CDI were more likely to be infected with more than one C. difficile ribotype during primary CDI diagnosis, even after adjusting for other clinical predictors. Across time, multiple patients demonstrated the ability to carry multiple strains at a given timepoint, independent from dynamics of which dominant ribotype was observed at initial diagnosis.
If mixed infection is confirmed in future studies to be independently predictive of subsequent recurrence, it could be used as a marker to aid clinical decision-making. Treatments that minimize risk of recurrence currently exist, such as monoclonal antibodies (24), probiotics (25), fidaxomicin in lieu of vancomycin (26), or a vancomycin taper (27) but are not practical to use in all patients due to expense, safety concerns, and/or tolerability. Although our methods were too time consuming for clinical deployment, advances and cost reductions in other methods to detect mixed C. difficile presence, such as a bioinformatic approach like metagenomics, may make detection of mixed infection more practical in a clinical setting.
The proportion of mixed infection observed in our study is higher than what has been observed in previous studies (12, 14, 28, 29). One possible explanation is that we isolated more colonies per sample than previous studies (up to 40), making the identification of mixed infection more likely. Arguing against this is that Behroozian et al isolated 100 C. difficile colonies from each of their 95 samples, and yet only observed mixed colonization in 16% (14). A more likely possibility is that our high proportion of mixed infection may be a reflection of selection bias of the patients sampled in our study. Since one of our initial aims was to identify ribotype dynamics overtime, we used a convenience sample of inpatients that were tested for and/or developed CDI multiple times (6). It may be that such patients are at higher baseline risk of multiple infection compared to the general CDI population due to a higher burden of comorbid disease, greater antibiotic exposure, or other risk factors that may alter the host response and lead to alterations in the microbiota that permit infection and/or colonization by multiple C. difficile strains. Thus, our measured proportion of mixed infection may not reflect general epidemiological patterns for all patients diagnosed with CDI. It is possible that our patient selection unmasks the rates at which we are environmentally exposed to C. difficile. Rates of community-acquired CDI (CDI acquired in the absence of a healthcare-facility overnight stay) are increasingly reported in surveillance data (30). It was recently reported that 45% of C. difficile strains isolated from four geographically defined hospitals over the course of three years could not be genetically linked, suggesting diverse sources of C. difficile exposure (31). Although the rate of mixed infection in our study may represent a niche hospital population, it could also unmask already changing C. difficile epidemiology that is reflective of the general population.
Some of these factors are also supported by the proportion of samples with changes in dominant ribotype within a patient over time in our study (40%), which is consistent with what has been observed in other longitudinal studies (12, 13, 32, 33). Other studies have distinguished relapse (infection with the same strain) from reinfection (infection with a new strain) in patients over time (34). We defined recurrence vs. reinfection using CDC surveillance definition guidance, with the hypothesis that recurrent episodes would be more likely to be consistently dominated by the same ribotype. However, the proportion of samples with changes in dominant ribotype was the same in both recurrent and reinfected patient populations. What was perhaps more surprising is that in a patient where a change in the dominant ribotype was observed, we did not detect its presence in a previous sample for that patient. Although this seems unlikely, it is an observation consistent with the variable dynamics of mixed infection generally observed in our study.
Our study has several limitations. Of the mixtures of C. difficile ribotypes observed in our samples, we did not observe any relationship between combinations of C. difficile, i.e. that one ribotype was more likely to exist in a mixed population compared to another. However, our study was not powered to identify whether specific ribotypes were more likely to persist with other specific ribotypes or co-occur with any other ribotype in a given sample. We did however observe that patients who went on to recur were more likely to be dominated by certain strains (F027, F078-126, and F053-163) that have been previously associated with recurrence or severe disease. We also did not account for which enrichment methods (heat, ethanol shock, broth growth) were more likely to lead to the identification of specific strains, which could possibly bias towards strains that are more robust in a frozen fecal sample; i.e., more tolerant to heat, freeze/thaw cycles, or more likely to grow in liquid culture. Future studies could be designed to use samples where a specific strain has already been identified (likely representing a dominant strain), accounting for enrichment methods at each step, to investigate some of these relationships and control for specific enrichment methods. Finally, though we did perform adjusted modeling of subsequent recurrence with mixed infection as the primary risk factor, this was limited by sample size. It is possible that unmeasured confounders could explain this association and future studies should validate the observation in a larger sample size and with adjustment for a broad array of putative clinical confounders.
Conclusions
In summary, our study suggests that mixed C. difficile colonization may be more prevalent than previously thought, particularly in patient populations more susceptible to recurrence. These observations have implications for C. difficile epidemiology and our understanding of asymptomatic and symptomatic colonization across different populations.
Supplementary Material
Highlights.
More than one Clostridium difficile strain (mixed infection) at primary C. difficile infection diagnosis was associated with the development of recurrent disease
A high number (40%) of individuals developed recurrence or reinfection with a new strain of C. difficile
Newly infecting strains were not detected in prior samples, independent of mixed infection status
Acknowledgments
Funding Information
AMS and KR were supported by a Clinical and Translational Science Award grant number from the Michigan Institute for Clinical and Health Research UL1TR000433. This work was also supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases, grant numbers U19AI090871 and U01AI124255.
The authors would like to thank the University of Michigan DNA Sequencing Core for rapid processing of ribotype PCR products.
Abbreviations
- CDI
Clostridium difficile infection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, Farley MM, Holzbauer SM, Meek JI, Phipps EC, Wilson LE, Winston LG, Cohen JA, Limbago BM, Fridkin SK, Gerding DN, McDonald LC. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–34. doi: 10.1056/NEJMoa1408913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zimlichman E, Henderson D, Tamir O, Franz C, Song P, Yamin CK, Keohane C, Denham CR, Bates DW. Health care-associated infections: a meta-analysis of costs and financial impact on the US health care system. JAMA Intern Med. 2013;173:2039–46. doi: 10.1001/jamainternmed.2013.9763. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Prabhu VS, Marcella SW. Attributable Healthcare Resource Utilization and Costs for Patients With Primary and Recurrent Clostridium difficile Infection in the United States. Clin Infect Dis. 2018 doi: 10.1093/cid/cix1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson S. Recurrent Clostridium difficile infection: a review of risk factors, treatments, and outcomes. J Infect. 2009;58:403–10. doi: 10.1016/j.jinf.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Kyne L, Warny M, Qamar A, Kelly CP. Association between antibody response to toxin A and protection against recurrent Clostridium difficile diarrhoea. Lancet. 2001;357:189–93. doi: 10.1016/S0140-6736(00)03592-3. [DOI] [PubMed] [Google Scholar]
- 6.Seekatz AM, Rao K, Santhosh K, Young VB. Dynamics of the fecal microbiome in patients with recurrent and nonrecurrent Clostridium difficile infection. Genome Medicine. 2016;8:1–11. doi: 10.1186/s13073-016-0298-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDonald LC, Killgore GE, Thompson A, Owens RC, Jr, Kazakova SV, Sambol SP, Johnson S, Gerding DN. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–41. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 8.Petrella LA, Sambol SP, Cheknis A, Nagaro K, Kean Y, Sears PS, Babakhani F, Johnson S, Gerding DN. Decreased cure and increased recurrence rates for Clostridium difficile infection caused by the epidemic C. difficile BI strain. Clin Infect Dis. 2012;55:351–7. doi: 10.1093/cid/cis430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crobach MJT, Voor In ‘t Holt AF, Knetsch CW, van Dorp SM, Bras W, Harmanus C, Kuijper EJ, Vos MC. An outbreak of Clostridium difficile infections due to new PCR ribotype 826: epidemiologic and microbiologic analyses. Clin Microbiol Infect. 2017 doi: 10.1016/j.cmi.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 10.Fawley WN, Davies KA, Morris T, Parnell P, Howe R, Wilcox MH. Enhanced surveillance of Clostridium difficile infection occurring outside hospital, England, 2011 to 2013. Euro Surveill. 2016:21. doi: 10.2807/1560-7917.ES.2016.21.29.30295. [DOI] [PubMed] [Google Scholar]
- 11.Johnson S, Adelmann A, Clabots CR, Peterson LR, Gerding DN. Recurrences of Clostridium difficile diarrhea not caused by the original infecting organism. J Infect Dis. 1989;159:340–3. doi: 10.1093/infdis/159.2.340. [DOI] [PubMed] [Google Scholar]
- 12.Eyre DW, Walker AS, Griffiths D, Wilcox MH, Wyllie DH, Dingle KE, Crook DW, Peto TE. Clostridium difficile mixed infection and reinfection. J Clin Microbiol. 2012;50:142–4. doi: 10.1128/JCM.05177-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oka K, Osaki T, Hanawa T, Kurata S, Okazaki M, Manzoku T, Takahashi M, Tanaka M, Taguchi H, Watanabe T, Inamatsu T, Kamiya S. Molecular and microbiological characterization of Clostridium difficile isolates from single, relapse, and reinfection cases. J Clin Microbiol. 2012;50:915–21. doi: 10.1128/JCM.05588-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Behroozian AA, Chludzinski JP, Lo ES, Ewing SA, Waslawski S, Newton DW, Young VB, Aronoff DM, Walk ST. Detection of mixed populations of Clostridium difficile from symptomatic patients using capillary-based polymerase chain reaction ribotyping. Infect Control Hosp Epidemiol. 2013;34:961–966. doi: 10.1086/671728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao K, Micic D, Natarajan M, Winters S, Kiel MJ, Walk ST, Santhosh K, Mogle JA, Galecki AT, LeBar W, Higgins PD, Young VB, Aronoff DM. Clostridium difficile ribotype 027: relationship to age, detectability of toxins A or B in stool with rapid testing, severe infection, and mortality. Clin Infect Dis. 2015;61:233–41. doi: 10.1093/cid/civ254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA) Infect Control Hosp Epidemiol. 2010;31:431–55. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 17.McDonald LC, Coignard B, Dubberke E, Song X, Horan T, Kutty PK. Recommendations for surveillance of Clostridium difficile-associated disease. Infect Control Hosp Epidemiol. 2007;28:140–5. doi: 10.1086/511798. [DOI] [PubMed] [Google Scholar]
- 18.Martinson JN, Broadaway S, Lohman E, Johnson C, Alam MJ, Khaleduzzaman M, Garey KW, Schlackman J, Young VB, Santhosh K, Rao K, Lyons RH, Jr, Walk ST. Evaluation of portability and cost of a fluorescent PCR ribotyping protocol for Clostridium difficile epidemiology. J Clin Microbiol. 2015;53:1192–7. doi: 10.1128/JCM.03591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wickham H. tidyverse: Easily Install and Load the ‘Tidyverse’. vR package version 1.2.1. 2017 https://CRAN.R-project.org/package=tidyverse.
- 20.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Google Scholar]
- 21.Brunson JC. Alluvial Diagrams in ‘ggplot2’. vR package version 0.6.0. 2018 https://CRAN.R-project.org/package=ggalluvial.
- 22.Hojsgaard S, Halekoh U, Yan J. [Accessed Jan 2016];Package ‘geepack’: Generalized Estimating Equation Package. 2014 https://cran.r-project.org/web/packages/geepack/geepack.pdf.
- 23.Abou Chakra CN, Pepin J, Sirard S, Valiquette L. Risk factors for recurrence, complications and mortality in Clostridium difficile infection: a systematic review. PLoS One. 2014;9:e98400. doi: 10.1371/journal.pone.0098400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilcox MH, Gerding DN, Poxton IR, Kelly C, Nathan R, Birch T, Cornely OA, Rahav G, Bouza E, Lee C, Jenkin G, Jensen W, Kim YS, Yoshida J, Gabryelski L, Pedley A, Eves K, Tipping R, Guris D, Kartsonis N, Dorr MB. Bezlotoxumab for Prevention of Recurrent Clostridium difficile Infection. N Engl J Med. 2017;376:305–317. doi: 10.1056/NEJMoa1602615. [DOI] [PubMed] [Google Scholar]
- 25.Shen NT, Maw A, Tmanova LL, Pino A, Ancy K, Crawford CV, Simon MS, Evans AT. Timely Use of Probiotics in Hospitalized Adults Prevents Clostridium difficile Infection: A Systematic Review With Meta-Regression Analysis. Gastroenterology. 2017;152:1889–1900. e9. doi: 10.1053/j.gastro.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Crook DW, Walker AS, Kean Y, Weiss K, Cornely OA, Miller MA, Esposito R, Louie TJ, Stoesser NE, Young BC, Angus BJ, Gorbach SL, Peto TE. Fidaxomicin versus vancomycin for Clostridium difficile infection: meta-analysis of pivotal randomized controlled trials. Clin Infect Dis. 2012;55(Suppl 2):S93–103. doi: 10.1093/cid/cis499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin Infect Dis. 2018 doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Berg RJ, Ameen HA, Furusawa T, Claas EC, van der Vorm ER, Kuijper EJ. Coexistence of multiple PCR-ribotype strains of Clostridium difficile in faecal samples limits epidemiological studies. J Med Microbiol. 2005;54:173–9. doi: 10.1099/jmm.0.45825-0. [DOI] [PubMed] [Google Scholar]
- 29.Wroblewski D, Hannett GE, Bopp DJ, Dumyati GK, Halse TA, Dumas NB, Musser KA. Rapid molecular characterization of Clostridium difficile and assessment of populations of C. difficile in stool specimens. J Clin Microbiol. 2009;47:2142–8. doi: 10.1128/JCM.02498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chitnis AS, Holzbauer SM, Belflower RM, Winston LG, Bamberg WM, Lyons C, Farley MM, Dumyati GK, Wilson LE, Beldavs ZG, Dunn JR, Gould LH, MacCannell DR, Gerding DN, McDonald LC, Lessa FC. Epidemiology of community-associated Clostridium difficile infection, 2009 through 2011. JAMA Intern Med. 2013;173:1359–67. doi: 10.1001/jamainternmed.2013.7056. [DOI] [PubMed] [Google Scholar]
- 31.Eyre DW, Wilcox MH, Walker AS. Diverse sources of C. difficile infection. N Engl J Med. 2014;370:183–4. doi: 10.1056/NEJMc1313601. [DOI] [PubMed] [Google Scholar]
- 32.Kamboj M, Khosa P, Kaltsas A, Babady NE, Son C, Sepkowitz KA. Relapse versus reinfection: surveillance of Clostridium difficile infection. Clin Infect Dis. 2011;53:1003–6. doi: 10.1093/cid/cir643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilcox MH, Fawley WN, Settle CD, Davidson A. Recurrence of symptoms in Clostridium difficile infection--relapse or reinfection? J Hosp Infect. 1998;38:93–100. doi: 10.1016/s0195-6701(98)90062-7. [DOI] [PubMed] [Google Scholar]
- 34.Figueroa I, Johnson S, Sambol SP, Goldstein EJ, Citron DM, Gerding DN. Relapse versus reinfection: recurrent Clostridium difficile infection following treatment with fidaxomicin or vancomycin. Clin Infect Dis. 2012;55(Suppl 2):S104–9. doi: 10.1093/cid/cis357. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.