Abstract
Long non-coding RNAs (lncRNAs) are emerging as important regulators of cellular processes and are extensively involved in the development of different cancers; including leukemias. As one of the accepted methods of lncRNA function is affecting chromatin structure; lncRNA binding has been shown for different chromatin modifiers. Histone lysine methyltransferases (HKMTs) are also subject of lncRNA regulation as demonstrated for example in the case of Polycomb Repressive Complex 2 (PRC2). Mixed Lineage Leukemia (MLL) proteins that catalyze the methylation of H3K4 have been implicated in several different cancers; yet many details of their regulation and targeting remain elusive. In this work we explored the RNA binding capability of two; so far uncharacterized regions of MLL4; with the aim of shedding light to the existence of possible regulatory lncRNA interactions of the protein. We demonstrated that both regions; one that contains a predicted RNA binding sequence and one that does not; are capable of binding to different RNA constructs in vitro. To our knowledge, these findings are the first to indicate that an MLL protein itself is capable of lncRNA binding.
Keywords: MLL proteins, MLL4, lncRNA, HOTAIR, MEG3, leukemia, histone lysine methyltransferase, RNA binding, intrinsically disordered protein
1. Introduction
Long non-coding RNAs (lncRNAs) are transcribed RNA molecules longer than 200 nucleotides that do not code for translated proteins. The human genome is estimated to code for about 58,000 lncRNAs [1], that are being more and more recognized as central players in a plethora of biological processes. They can act as flexible scaffolds providing binding platforms for different proteins, they can interfere with other endogenous RNAs acting as microRNA “sponges” and they can modify chromatin state [2], thus regulating the expression of various proteins. LncRNAs have also been shown to play a role in several layers of epigenetic regulation: they are involved in DNA methylation and demethylation, they can modify chromatin conformation through binding to remodelers [3] and many of them interact with histone modifier enzyme complexes such as PRC2, coREST or SMCX [4].
The physiological processes where lncRNA regulation have been suggested involve cell cycle regulation, epithelial mesenchymal transition (EMT) [5], cancer progression [6] and maintenance of cancer stem cells [5], hypoxia [7] and leukemia [8].
Various lncRNAs are shown to have altered expression levels in different leukemias, resulting in a crucial influence on cellular transformation [9], chromosomal translocation [10], apoptosis [11] and on drug resistance [12]. Accumulating evidence regarding the involvement of lncRNAs in leukemic processes prompted the suggestion to use them as prognostic and classification factors. It was found that lncRNA expression has prognostic value in AML patients [13] and multiple pathways were involved in lncRNA expression, including chromosome organization and trans-membrane receptor protein tyrosine kinase signalling pathway.
As lncRNAs are also considered valuable drug targets, it is essential that the molecular details of their functions are uncovered.
Polycomb repressive complex (PRC2) is the most studied histone modifier that relies on lncRNA binding in its function, being able to bind several lncRNAs including HOTAIR, Xist, RepA, Braveheart, MALAT1 and MEG3 [14]. In vitro experiments revealed that not only EZH2, but other PRC2 subunits are also capable of lncRNA binding [15], thus providing a pattern of binding regions distributed along the surface of the complex. Even though there remain open questions regarding the specificity of the RNA binding by PRC2 [16], it is widely accepted that lncRNA binding plays a defining role in PRC2 targeting and the ensuing gene silencing [14]. It is interesting to note that despite the numerous experimental results that show EZH2 to be an RNA binding protein, it cannot be found in databases that list RNA binding proteins, furthermore no RNA binding site is predicted to be located in the region that is shown to be responsible for the RNA-protein interaction [17].
Apart from PRC2, other histone lysine methyltransferases (HKMTs) or HKMT complex components also appear to bind lncRNAs with a relevant physiological outcome.
LncRNA EZR-AS1 enhances EZR expression through recruiting SMYD2 to the upstream region of its promoter region and elevating the activating H3K4 methylation [18].
G9a interacts with lncRNA PARTICLE to regulate MATA2 expression upon mild irradiation [19]. The interaction was shown using ChIP assay and apart from G9a, the PRC2 subunit Suz12 was also pulled down. In a later experiment, it was found that PARTICLE can also interact with DNA methylase DNMT1 and that it increases H3K27 methylation as well as EZH2 expression. It was suggested that PARTICLE may serve as a functional platform that enables the specific targeting of chromatin modifiers, such as PRC2 [20].
WDR5, a component of the MLL1-4 and SET1a/1b complexes was proven to interact with lncRNAs NeST and HOTTIP with an effect on microbial susceptibility through the enhancement of interferon-γ expression [21]. Further investigation of the WDR5-HOTTIP interaction led to the recognition that lncRNA binding by WDR5 is essential in maintaining embryonic stem cell pluripotency [22]. However, not this work nor any previous studies investigated the possibility that the enzymatic component of the methyltransferase complex may also be capable of lncRNA binding.
The family of mammalian MLL (Mixed Lineage Leukemia) proteins consist of Set1a, Set1b and four MLL proteins, MLL1, MLL2, MLL3 and MLL4. They work in COMPASS-like complexes and catalyze H3K4 mono-, di- or tri-methylation, each complex having different specificity and methylase activity [23]. MLL3 and MLL4 are responsible for the monomethylation of H3K4 at enhancer regions [24] and has been linked to a high number of different cancers. Properly functioning MLL3 and MLL4 act as tumor suppressors [23], therefore mutations affecting their activity or stability can result in cancer development. Despite their central role in several types of cancers, many open questions regarding the regulation of the activity and the targeting of the MLL complexes remain unanswered. The exact molecular details of how MLL3 and MLL4 are targeting enhancer regions [23] as well as the specific molecular effects of the interactions of their different regulatory domains [25] are largely unknown. It is also worth noting that the known structured domains represent only 15–21% of the sequences of MLL proteins, leaving the vast majority of these proteins uncharacterized both structurally and functionally.
In a previous work [26] we suggested that the disordered regions of HKMTs may harbor so far unrecognized interaction sites, adding more layers of the regulation of their activity. Based on the observation that many lncRNAs are involved in processes governed by HKMTs, we hypothesized that lncRNA binding might be one of the functions of these regions.
Since multiple evidence point in the direction that leukemic processes are fundamentally affected by lncRNAs and MLL complexes are involved in this regulation, we concentrated on MLL proteins. Taken the analogy of the PRC2 complex, where more than one complex subunits are capable of lncRNA binding, we aimed at testing the ability of MLL4 to bind different RNA molecules.
2. Results
2.1. In Silico Analysis of the RNA Binding Capacity of MLL Proteins
As a first step, we mapped the predicted RNA binding motifs on the sequence of four MLL proteins. We used DisoRDPbind, an RNA interaction prediction tool specifically designed to find RNA interaction sites in the disordered regions of proteins. Results shown in Table 1 indicate that all MLL proteins contain several putative RNA interaction motifs in their disordered regions. These regions are found at various positions in the proteins and vary in length from a couple of amino acids to almost a hundred residues, suggesting that RNA binding might be a common feature in MLL proteins.
Table 1.
Predicted RNA binding regions in the disordered regions of Mixed Lineage Leukemia (MLL) proteins (aa positions).
| MLL1 | MLL2 | MLL3 | MLL4 |
|---|---|---|---|
| 296–327 | 84–107 | 1068–1079 | 1559–1567 |
| 348–408 | 184–234 | 1678–1695 | 3526–3581 |
| 415–418 | 241–244 | 1701–1709 | 3899–3983 |
| 1155–1194 | 536–560 | 1715–1737 | 4960–5014 |
| 1977–1992 | 783–806 | 2406–2409 | 5147–5165 |
| 3854–3861 | 820–828 | 3052–3073 | 5227–5251 |
| 1753–1778 | 3246–3250 | ||
| 2600–2616 | 3394–3427 | ||
| 2685–2709 | 4330–4356 | ||
| 4514–4524 | |||
| 4586–4625 |
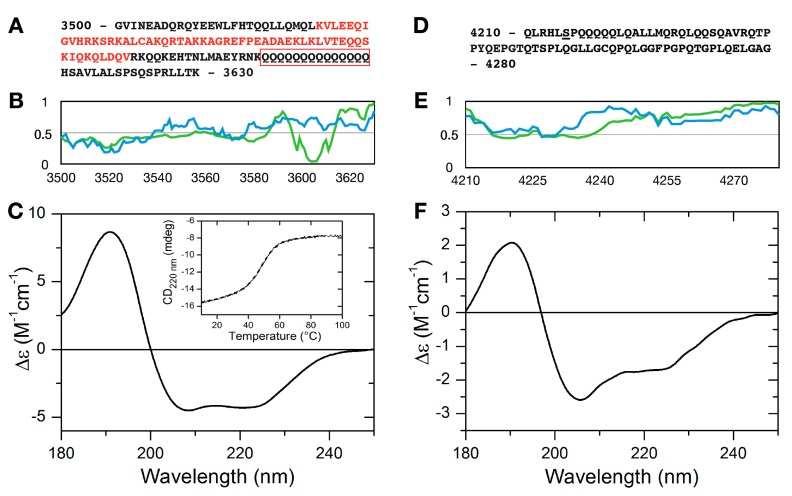
A comparison with our earlier studies [26] revealed that two conserved disordered binding sites (residues 3537–3545 and 3560–3567) reside within one of the predicted RNA binding regions (residues 3526–3581, Figure 1A) of MLL4, underlining the reliability of the predictions. This region also harbors several cancer-related point mutations, two of them corresponding to a predicted binding site at positions 3560 (D-N) and 3561 (A-D). All these evidences point to the physiological importance of this protein region, making its structural and functional study worthwhile. ANCHOR prediction [27] shows that within the C-terminal border of the predicted RNA binding region there is a region with a strong tendency of the protein chain to form protein-protein interactions (residues 3597–3613, Figure 1A) that corresponds to a run of 14 glutamine residues. Since polyQ repeats in RNA binding proteins have been linked to protein-RNA droplet formation [28], this raises the intriguing possibility of granule formation potency of this segment. Therefore, we chose to test the RNA binding capacity of the MLL4 region between residues 3500–3630 (Figure 1A). As an internal control, another disordered region with no predicted RNA or protein binding sites was selected between residues 4210–4280 of MLL4 (Figure 1D).
Figure 1.
Structural characterization of the MLL4 regions. Sequences of MLL43500–3630 (A) and MLL44210–4280 (D). Predicted RNA binding region is indicated by red letters and the polyQ stretch is framed with red. IUPRed (blue) and Anchor (green) prediction of MLL43500–3630 (B) and MLL44210–4280 (E). Residues having an IUPred score above 0.5 are considered to be disordered, while residues with an Anchor score below 0.5 constitute predicted binding sites. Far-UV CD spectra of MLL43500–3630 (C) and MLL44210–4280 (F). Inset: temperature-dependent changes in the structure of MLL43500–3630 as observed by monitoring the changes in the absorbance at 220 nm.
As for binding RNAs, we opted to test two different lncRNA constructs, both having been reported to play a role in leukemias. The first is HOTAIR, that has the ability to bind EZH2 (PRC2). The 5’ 300 nucleotides of HOTAIR are thought to mediate its binding to PRC2 complex subunits, but the latest annotation in the NCBI database contains an additional 140 bases at the beginning of HOTAIR sequence, compared to the one reported earlier. Therefore, we prepared the longer version (HOTAIR440) that encompasses the 300 nucleotides already known to be involved in protein-RNA interactions and also the nucleotides that has not been studied yet. Since there is no information available about the region of MEG3 that is able to bind proteins, we used the full length MEG3 for our experiments.
2.2. Secondary Structure of MLL43500–3630 and MLL44210–4280
Disorder prediction profiles (Figure 1B,E) indicated that both protein regions have a significant disorder tendency. Disorder profile of MLL43500–3630 indicates a rather ambiguous disorder state, with prediction scores fluctuating around the 0.5 limit between ordered and disordered states. This disorder prediction might indicate a disordered region that has an elevated tendency to fold or a relatively unstable folded segment as well. Far-UV CD measurements revealed that MLL43500–3630 has a helical structure in isolation (Figure 1C). The CD spectrum of this region of MLL4 showed a typical alpha helical conformation with a pronounced double minimum at 208 and 220 nm. Secondary structure content calculation using the BeStSel algorithm [29,30] gave an α-helix content of ~36.2%, while another ~36% of the secondary structure content was characterized as “Others”, which mainly corresponds to the disordered structure. Thermal unfolding of the observed helical structure was followed by gradually heating the sample to 100 °C while recording the absorbance at 220 nm (Figure 1C inset). The melting curve indicated a cooperative unfolding of the structure with a melting point of 48 °C. The CD spectrum of the thermal denatured state is shown in Supplementary Figure S1, demonstrating a complete loss of structure at high temperatures.
MLL44210–4280 has a more pronounced disorder tendency, as demonstrated by the IUPred profile and is devoid of any predicted ANCHOR binding sites (Figure 1E). Its sequence contains a significant portion of glutamines (Figure 1D), but it does not contain Q stretches longer than 4 residues. Far-UV CD measurements confirmed the disorder predictions, indicating that the protein is mostly disordered in solution, with a considerable α-helical tendency. Secondary structure calculations gave a result of 16% α-helix and ~45% “Others” content, underlining that this segment of MLL4 is not fully disordered and contrary to interaction site predictions, might be involved in molecular recognition.
2.3. RNA Binding of MLL43500–3630 and MLL44210–4280
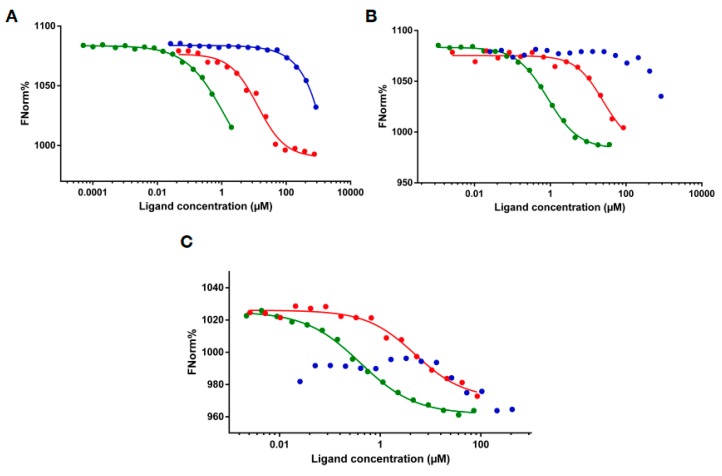
Microscale thermophoresis measurements were performed to characterize the RNA binding of the expressed protein regions. We used two lncRNA constructs, HOTAIR440, a segment of HOTAIR that contains the region involved in binding to EZH2 [31], MEG3, a lncRNA involved in leukemias [32] and a 50 nt long RNA with random nucleotide sequence. Contradicting to the lack of predicted binding sites, MLL44210–4280 showed a relatively strong binding to HOTAIR440 with an apparent Kd of 13.05 μM (Figure 2A), while the negative control Thymosin beta 4 (Tβ4) did not bind to the RNA, showing any sign of interaction at only the highest concentrations applied.
Figure 2.
RNA binding detected by microscale thermophoresis. MST binding curves of MLL43500–3630 (green), MLL44210–4280 (red) and thymosin beta 4 (blue) to different RNAs: HOTAIR440 (A), MEG3 (B) and 50 nt RNA (C).
In the case of MLL43500–3630, saturation of the reaction could not be reached because of marked aggregation above 1:20 RNA:protein ratio (Supplementary Figure S2) but using the T-jump values of the MST measurement (Supplementary Figure S3) an approximate binding constant of 0.1 μM could be determined. The appearance of large particles in the solution, generally considered to be aggregates, is indicated by a “wavy” MST curve and a randomly fluctuating normalized fluorescent percentage as shown on Supplementary Figures S2 and S5. The observed aggregation was dependent on the RNA species, since it was not seen with either of the other tested RNAs (Figure 3B,C), or with a shorter, 300 nt long HOTAIR construct (Supplementary Figure S4). The HOTAIR300 construct overlaps with HOTAIR440 in the 3′ 300 nucleotides but lacks the first 140 nucleotides of the latter. This shorter HOTAIR construct bound to MLL43500–3630 with a Kd of 0.97 μM, with no sign of irregular behavior. Centrifugation (15 min at 13,000× g) of the samples resulted in the loss of fluorescent signal in a protein concentration-dependent manner (Supplementary Figure S5), indicating a formation of structures containing both RNA and protein. Such phenomenon was not observed with MLL44210–4280, or Tβ4 upon mixing them with HOTAIR440, even at significantly higher protein concentrations than MLL43500–3630. Also, MLL43500–3630 did not show aggregation-prone behavior in the absence of RNA.
Figure 3.
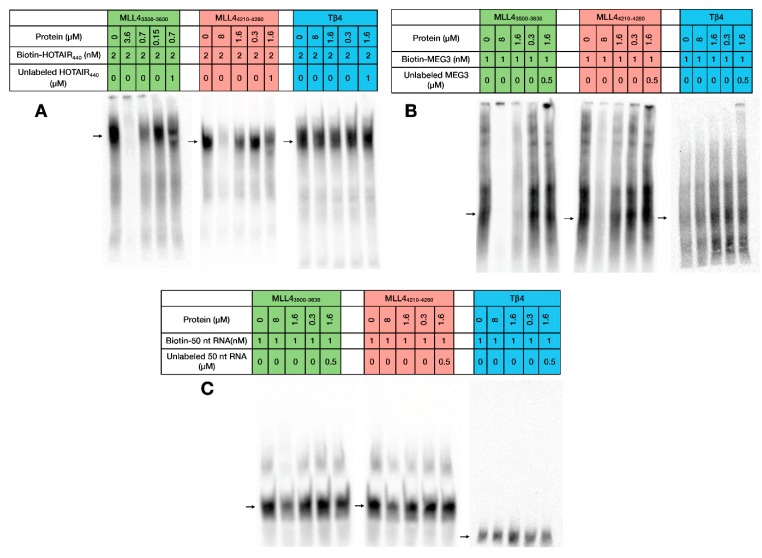
Electrophoretic Mobility Shift Assay. Interaction of MLL43500–3630, MLL44210–4280 and Tβ4 with HOTAIR440 (A), MEG3 (B) and 50 nt RNA (C). For easier understanding, the coloring scheme of Figure 2 is followed (MLL43500–3630: green MLL44210–4280: red, Tβ4: blue). Free RNA is indicated by arrows.
As we experienced no anomaly in the behavior of MLL43500–3630 when titrated to MEG3, determination of a binding constant was straightforward for this interaction. As shown in Figure 2B, affinity to MEG3 of this region of MLL4 was higher than that of MLL44210–4280. The Kd of MLL43500–3630 binding to MEG3 was calculated to be 0.722 μM, while Kd calculation for MLL44210–4280 was not reliable since saturation of the reaction could not be reached throughout the protein concentration range tested. Tβ4 did not show significant affinity to MEG3, resulting in a failure of binding curve fitting.
To check for any specificity of binding that the expressed MLL4 regions may possess, we also tested a physiologically non-relevant 50 nt RNA construct. Binding curves presented in Figure 2C indicate that both MLL43500–3630 and MLL44210–4280 are capable of binding to this RNA species, but with a remarkably lower affinity than to the lncRNA constructs, while Tβ4 could not bind to it at all. The extended shape of the binding curve and the absence of saturation in the case of both MLL4 constructs indicate weak binding that resulted in an inability to reliably determine the binding constants. Nevertheless, MLL43500–3630 still displayed a stronger affinity towards the RNA than MLL44210–4280.
Electrophoretic Mobility Shift Assay (EMSA) experiments confirmed the findings of the MST measurements (Figure 3) as both MLL4 regions caused a significant change in RNA mobility in the case of HOTAIR440 and MEG3 (Figure 3A,B) RNAs. This shift was drastically less pronounced with the 50 nt RNA sample (Figure 3C), resulting only in a minor weakening of the RNA signal in the lane with the highest protein concentration. This observation corresponds to the outcome of the MST experiments, underlining the existence of a certain level of specificity in the RNA recognition by these two MLL4 regions. The negative control Tβ4 failed to cause any visible change in the RNA mobility, indicating a lack of interaction with any of the tested RNAs. Competitive RNA binding (Figure 3, compare the 3rd and 5th lanes) demonstrated that the observed shift in mobility was indeed a result of RNA-protein interaction, since the shift could be prevented at least to some extent by adding excess unlabeled RNA to the reaction mixtures.
The anomalous behavior of the MLL43500–3630:HOTAIR440 interaction observed in MST was seen in the EMSA experiments as well, since at high protein:RNA ratios the samples obtained a highly viscous quality and completely remained in the wells during the electrophoretic run. Successful experiments could only be carried out by lowering the applied protein concentration, but the interaction was clearly observable even under these circumstances.
In all of the tested interactions, MLL43500–3630, which contains a predicted RNA binding region presented higher affinities to RNAs than the other MLL4 segment, indicating the validity of the prediction. On the other hand, binding of MLL44210–4280 could also be detected in all cases, raising the possibility of the existence of RNA binding sequences differing from the already described interaction motifs. EZH2, a known RNA binding HKTM also interacts with RNAs through a region [17] that has no recognizable RNA binding sequence, emphasizing our lack of complete knowledge of the sequential determinants of protein-RNA interactions.
3. Discussion
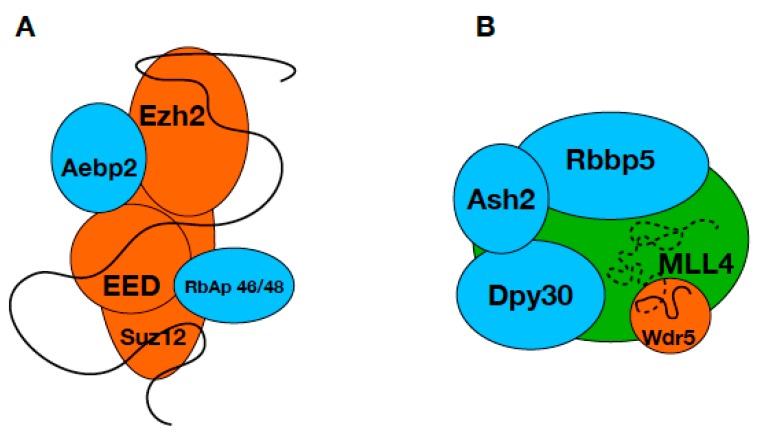
Histone methylation is one of the most studied and best-characterized histone modifications that drive the regulation of complete genetic programs in the cells. However, many details of the regulation and targeting of the enzyme complexes mediating histone methylation remain elusive and a subject of debate [23]. One possible regulatory pathway is represented by the ability of certain HKMT complexes to bind different lncRNAs that serve as a targeting platform, bridging transcription factors and HKMT complexes [20,33] at the promoter regions of target genes. PRC2 is one example where it was shown by multiple experiments that it’s binding to different lncRNAs results in different physiological outcomes [34]. lncRNAs are involved in many other processes connected to histone modification and there are examples in the literature of direct interaction between lncRNAs and histone modifier complexes [4,22]. Experimental evidence supports the direct binding of WDR5, a canonical MLL complex subunit, to different lncRNAs in cells [22] indicating the involvement of lncRNAs in the regulation of MLL complexes. Taken the analogy of the PRC2, where multiple subunits are shown to be involved in lncRNA binding (Figure 4A) [15], we hypothesized that MLL proteins might also interact with lncRNAs. This hypothesis was supported by our earlier bioinformatics studies that suggested the existence of several interaction sites in the so far uncharacterized, mostly disordered regions of HKMTs [26] and our prediction presented here that the disordered segments of MLL proteins contain several putative RNA binding sequences. We chose to test the RNA binding capability of one such region of MLL4 that also contains a polyQ stretch and is affected by mutations in different cancers. As an internal control, we also tested a different region of MLL4 that contains no such predicted RNA interaction site.
Figure 4.
lncRNA binding of PRC2 and MLL4/COMPASS complex. Schematic representation of the PRC2 (A) and MLL4/COMPASS (B) complexes, where the known RNA binding subunits are shown in orange and the suggested lncRNA binding subunit MLL4 is green. Subunits currently not known to be involved in lncRNA binding are blue and the lncRNA is represented by a black line. Suggested lncRNA-MLL4 interaction is indicated by dashed line.
Our expectation was that the isolated small regions of the MLL4 protein would bind RNAs in a nonspecific manner, such as was observed for the isolated PRC2 complex components [34]. Surprisingly, we found that MLL44210–4280 bound MEG3 stronger than HOTAIR440 or the 50 nt random RNA, even though the determination of the exact Kd-s was not successful in all cases.
More interesting was the behavior of the MLL43500–3630 region that showed dramatically different behavior with the different RNAs. Binding to MEG3 gave a Kd of 0.722 μM, while the binding to the 50 nt random RNAs proved to be so weak that a Kd calculation was not successful. Binding to HOTAIR440 seemed to be the strongest with an apparent Kd of 0.1 μM, but it led to the aggregation of the protein-RNA complex. The aggregation was dependent on protein-RNA ratio and could be detected through a wide protein concentration range. The same aggregation could not be observed with a shorter HOTAIR construct that consisted of 300 bases (Supplementary Figure S3). The fact that we could not induce such aggregation by the addition of MEG3, which is much longer than HOTAIR440, points to specific recognition rather than a side-effect of RNA length. We also observed the aggregation at low protein concentrations, but only in the presence of an appropriate amount of HOTAIR440, indicating that the process is not driven by the protein in itself and is not a derivative of sample preparation errors.
It has been recently revealed that many proteins can go through liquid-liquid phase separation when interacting with RNAs, leading to the formation of membraneless organelles that have a significant importance in cellular processes [35]. Experimental evidence supports the involvement of polyQ regions of proteins in the RNA mediated phase separation [28], sometimes in an RNA secondary structure-dependent manner [36]. Since MLL43500–3630 sequence contains 22.9% glutamine residues and a continuous run of 15 glutamines (Figure 1A), it is not unfounded to speculate that this specific region plays a role in the observed anomaly but the fact that it only occurs with one of the tested RNA constructs, indicates that the process is coordinated by the RNA itself. One possibility is that the longer HOTAIR construct contains more than one binding sites for MLL43500–3630, thus facilitating the formation of higher order protein-RNA structures. Alternatively, HOTAIR440 may have the ability to form secondary structures not found in HOTAIR300 or MEG3, which would also provide an explanation for the different behavior of the three systems. As MLL4 is the only HKMT that contains long polyglutamine repeat stretches [26], phase separation might be a regulatory step specific for this protein. Therefore, it is certainly promising to investigate this peculiar phenomenon in more detail.
Since both tested lncRNAs are implicated in different cancers [5,37,38] involving leukemias, our finding that MLL4 has a capacity to bind them raises the possibility that lncRNAs play a role in MLL/COMPASS complex targeting and regulation to a larger extent than currently recognized.
Although cellular experiments are necessary to prove the validity of the observed interactions, our findings provide the first insights into the structure and function of two regions of MLL4 that have been uncharacterized so far. We were able to show that these regions are capable of RNA binding and may be involved in the lncRNA mediated regulation of the MLL4 complexes. Based on our results, we suggest that and MLL4 complexes utilize different regions on their surface to bind lncRNAs (Figure 4B), similarly to the way PRC2 subunits take part in lncRNA binding. As it was shown that lncRNA binding to WDR5 increases the dwelling time of the protein on the chromatin surface [22], binding of the same RNA to MLL4 might facilitate and accelerate the assembly of a functional methyltransferase complex. Since lncRNAs are large molecules that can adopt various secondary structures and interact with many different partners simultaneously, it is plausible to speculate that a specific and high-affinity interaction can be achieved by the combination of different binding sites distributed along the large surfaces of multi-subunit complexes. Given the central role of histone modifications in gene regulation, it is essential to understand the mechanisms that regulate this process. Mounting evidence supports the involvement of lncRNAs in the coordination of histone modifying enzymes but the exact molecular details of their interactions with proteins are yet to be discovered. Recognizing the importance of the disordered/structurally uncharacterized regions of HKMTs in these interactions might be the first step towards a more complete picture regarding the regulation of histone methylation.
4. Materials and Methods
4.1. Bioinformatics Analysis
Disorder and disordered binding site predictions were performed with the IUPred2A online prediction tool (https://iupred2a.elte.hu/) [39] which incorporates the IUPred and Anchor predictors. RNA binding regions located in disordered regions were predicted using the DisoRDPbind tool (http://biomine.cs.vcu.edu/servers/DisoRDPbind/) [40]. Cancer-related single nucleotide polymorphisms in the long conserved IDR regions were collected from the BioMuta v2.0 [41] and COSMIC databases [42].
4.2. Accession Numbers
HOTAIR: Gene ID: 100124700
MEG3: Gene ID: 55384
MLL1: Uniprot: Q03164
MLL2: Uniprot: Q9UMN6
MLL3: Uniprot: Q8NEZ4
MLL4: Uniprot: O14686
4.3. Overexpression and Purification of MLL4 Protein Regions
The same methods of protein overexpression and purification were used for both protein constructs, MLL43500–3630 and MLL44210–4280. DNA sequences coding for each protein were cloned into pET22b cloning vector. Induction was done for 4 h at 28 °C by 0.1 M IPTG, cells were pelleted by centrifugation (4000 rpm, 20 min, 4 °C) then lysed by sonication in lysis buffer (50 mM Tris, 200 mM NaCl, 0.5% Triton X-100 pH 8.0 and EDTA-free SIGMAFAST Protease Inhibitor Cocktail Tablets), cell debris was removed by centrifugation (12,100 rpm, 40 min, 4 °C). The supernatant was filtered through 0.2 μm nitrocellulose filter then purified over HisTrap HP column on an AKTA Explorer system using a gradient elution of two buffers (Buffer A: 20 mM imidazole, 200 mM NaCl, 20 mM Tris. pH 7.5. Buffer B: 1 M imidazole, 200 mM NaCl, 20 mM Tris, pH 7.5). Representative purification results are shown on Supplementary Figure S7. The mostly disordered nature of the MLL44210–4280 region was highlighted by its appearance at a larger size than its actual molecular weight (17 kDa vs. 7 kDa). Elution fractions containing sufficiently pure proteins were dialyzed against distilled water then lyophilized and stored at −20 °C. Lyophilized proteins were dissolved before use in ultrapure water or the appropriate assay buffer. The identity of the purified proteins was confirmed by mass spectrometry.
4.4. RNA Preparation
HOX transcript antisense RNA (HOTAIR):
HOTAIR300 (140–440 nt) and HOTAIR440 (1–440 nt) DNA sequences cloned into pEX-A128 vector were purchased from Eurofins Genomics (Ebersberg, Germany). After 2 h digestion with EcoRV restriction enzyme at 37 °C, the gel-purified, linearized DNA templates were used to synthesize RNA by in-vitro transcription.
Maternally Expressed 3 (MEG3) lncRNA:
pCI-MEG3 was a gift from Anne Klibanski (Addgene plasmid #44727, Watertown, MA, USA) [43]. Primers to obtain the DNA template for in vitro transcription were as follows:
T7 RNA promoter region followed by:
MEG3 forward primer:
TAATACGACTCACTATAGGGGCAGAGAGGGAGCGCGCCTTGG
MEG3 reverse primer:
GATATCTTTTTGTTAAGACAGGAAACACATTTATTGAGAGC
50 nt RNA:
50 nucleotide RNA was an artificial randomized RNA sequence.
DNA templates were T7 promoter region followed by:
50 nt forward oligo:
TAATACGACTCACTATAGAAGAATGGCCTCGCGGAGGCATGCGTCATGCTAGCGTGCGGGGTACTCTT and
50 nt reverse oligo:
AAGAGTACCCCGCACGCTAGCATGACGCATGCCTCCGCGAGGCCATTCTTCTATAGTGAGTCGTATTA
Transcribed RNA:
GAAGAAUGGCCUCGCGGAGGCAUGCGUCAUGCUAGCGUGCGGGGUACUCUU
All primers and oligonucleotides were purchased from Sigma-Aldrich Ltd. (St. Louis, MO, USA).
Tested RNAs were synthesized by in vitro transcription carried out with New England BioLabs HiScribe™ T7 Quick High Yield RNA Synthesis Kit (Ipswich, MA, USA). Fluorescein-labelled, single-stranded RNA probes were generated by using Roche (Basel, Switzerland) Fluorescein RNA Labeling Mix (11685619910) and NEB 10× T7 reaction buffer (#B2041A). After transcription, remaining DNA templates were eliminated with DNaseI treatment. RNA sample purification was carried out using Macherey-Nagel NucleoSpin® RNA Clean-up XS Kit (Düren, Germany). The quality and intactness of the purified transcription products were analysed by native and formaldehyde agarose gel electrophoresis.
Biotinylation of the RNAs was performed using Pierce™ RNA 3′ End Biotinylation Kit (Cat. Number 20160, Thermo Fisher Scientific, Waltham, UK) according to the instructions of the manufacturer. Overnight incubation at 16 °C was applied for the ligation of the biotin label. Final RNA concentrations were determined using NanoDrop™ 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, UK).
Purified RNAs were stored −80 °C until usage in the presence of RNAINH-RO Roche Protector RNase Inhibitor (20U).
4.5. Far-UV CD Measurements
CD measurements were performed in quartz cells of 0.1 mm pathlengths using a Jasco J-810 (Jasco, Tokyo, Japan) spectropolarimeter. Far-UV CD spectra were recorded in the range of 180–260 nm with a scanning speed of 20 nm/min, bandwidth of 1 nm and integration time of 4 s. 6 scans were accumulated. Thermal denaturation was recorded in a 1 mm cell at 220 nm from 10 to 100 °C with scanning rate of 120 °C/h. The temperature was controlled using a PTE Peltier unit. The thermal denaturation profile was fitted according to the Gibbs-Helmholtz equation assuming a two-state model, which is represented by a sigmoidal curve [44]. CD spectra were quantitatively analyzed by the BeStSel method [29,30] (http://bestsel.elte.hu).
4.6. Microscale Thermophoresis
RNA-protein binding assays were carried out on a Microscale Thermophoresis system (Monolith NT. 115 from NanoTemper Technologies, München, Germany). Standard treated capillaries (Cat. Number: MO-K002) were used for measurements. Instrument settings are presented in Table 2.
Table 2.
Instrument settings for MST.
| Title | LED Power (%) | MST Power (%) | Before MST (s) | MST on (s) | After MST (s) | Delay (s) |
|---|---|---|---|---|---|---|
| Round 1 | 10 or 40 | 20 | 5 | 30 | 5 | 25 |
| Round 2 | 10 or 40 | 40 | 5 | 30 | 5 | 25 |
Normalized fluorescence values after 1.25 s after turning on the IR laser were used as T-jump values.
RNA concentrations were set to give an initial raw fluorescence between 300 and 1000 counts and varied between 30 and 100 nM. All experiments were done at room temperature. DEPC-treated PBS buffer containing 0.05% NP-40 was used as assay buffer.
4.7. Electrophoretic Mobility Shift Assay (EMSA)
LightShift® Chemiluminescent RNA EMSA Kit (Thermo Scientific, Cat. No. 20158, Thermo Fisher Scientific, Waltham, UK) was used for the EMSA experiments. Assay control was performed according to the instructions of the manufacturer with the control reaction provided with the kit. In short, 6.25 nM biotin-labeled IRE RNA was incubated with 2 μg of cytosolic liver extract with or without 1 μM of unlabeled IRE RNA. The result of the assay control is presented on Supplementary Figure S6. Binding, electrophoresis and detection of the tested RNAs with the proteins were carried out following the protocol of the kit. Briefly, proteins of varying concentrations were incubated with 1 or 2 nM of RNAs for 30 min at room temperature, then loaded on 4 or 6% native polyacrylamide gels. RNA was transferred to nitrocellulose membranes using Trans-Blot® Turbo™ Transfer System (Bio-Rad, Hercules, CA, USA) and crosslinked to the membrane by UV-light crosslinking. After proper washing and blocking, biotin labeled RNA was detected by chemiluminescence using Streptavidin-Horseradish Peroxidase Conjugate.
Abbreviations
| EMSA | Electrophoretic mobility shift assay |
| EZH2 | Enhancer of zeste homolog 2 |
| HKMT | Histone lysin methyltransferase |
| HOTAIR | HOX transcript antisense RNA |
| lncRNA | Long non-coding RNA |
| MEG3 | Maternally Expressed 3 |
| MLL | Mixed lineage leukemia |
| MST | Microscale thermophoresis |
| PRC2 | Polycomb repressive complex |
| WDR5 | WD repeat-containing protein 5 |
Supplementary Materials
The following are available online at http://www.mdpi.com/1422-0067/19/11/3478/s1.
Author Contributions
Conceptualization, A.T.; Data curation, B.S. (Beáta Szabó), E.S. and B.S. (Bálint Szeder); Formal analysis, J.K.; Funding acquisition, J.K., L.B. and A.T.; Investigation, B.S. (Beáta Szabó), N.M., R.A. and J.K.; Methodology, B.S. (Beáta Szabó) and A.T.; Project administration, B.S. (Beáta Szabó) and A.T.; Resources, B.S. (Beáta Szabó) and N.M.; Software, E.S.; Supervision, L.B. and A.T.; Validation, J.K. and L.B.; Visualization, A.T.; Writing—original draft, A.T.; Writing—review & editing, B.S. (Beáta Szabó), N.M., R.A., E.S., J.K., B.S. (Bálint Szeder) and L.B.
Funding
This research was funded by the National Research, Development and Innovation Office, Hungary (grants: K-125340 (A.T.), K-120391 (J.K.), KH-125597 (J.K. and A.T.), TÉT_16-1-2016-0134 (J.K.), 2017-1.2.1-NKP-2017-00002 (J.K.), Medinprot synergy grant (A.T. and J.K.)) and a Korea-Pan-European Research Agreement (A.T.).
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S., et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartonicek N., Maag J.L.V., Dinger M.E. Long noncoding RNAs in cancer: Mechanisms of action and technological advancements. Mol. Cancer. 2016;15:43. doi: 10.1186/s12943-016-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang F., Deng X., Ma W., Berletch J.B., Rabaia N., Wei G., Moore J.M., Filippova G.N., Xu J., Liu Y., et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015;16:52. doi: 10.1186/s13059-015-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Morales D.R., Thomas K., Presser A., Bernstein B.E., Van Oudenaarden A., et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heery R., Finn S.P., Cuffe S., Gray S.G. Long Non-Coding RNAs: Key Regulators of Epithelial-Mesenchymal Transition, Tumour Drug Resistance and Cancer Stem Cells. Cancers. 2017;9:38. doi: 10.3390/cancers9040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z., Chen Z., Fan R., Jiang B., Chen X., Chen Q., Nie F., Lu K., Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol. Cancer. 2017;16:82. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhan A., Deb P., Shihabeddin N., Ansari K.I., Brotto M., Mandal S.S. Histone methylase MLL1 coordinates with HIF and regulate lncRNA HOTAIR expression under hypoxia. Gene. 2017;629:16–28. doi: 10.1016/j.gene.2017.07.069. [DOI] [PubMed] [Google Scholar]
- 8.Chen S., Liang H., Yang H., Zhou K., Xu L., Liu J., Lai B., Song L., Luo H., Peng J., et al. Long non-coding RNAs: The novel diagnostic biomarkers for leukemia. Environ. Toxicol. Pharmacol. 2017;55:81–86. doi: 10.1016/j.etap.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 9.Guo G., Kang Q., Zhu X., Chen Q., Wang X., Chen Y., Ouyang J., Zhang L., Tan H., Chen R., et al. A long noncoding RNA critically regulates Bcr-Abl-mediated cellular transformation by acting as a competitive endogenous RNA. Oncogene. 2015;34:1768–1779. doi: 10.1038/onc.2014.131. [DOI] [PubMed] [Google Scholar]
- 10.Wang P., Ren Z., Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J. Cell. Biochem. 2012;113:1868–1874. doi: 10.1002/jcb.24055. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L., Xu H.-G., Lu C. A novel long non-coding RNA T-ALL-R-LncR1 knockdown and Par-4 cooperate to induce cellular apoptosis in T-cell acute lymphoblastic leukemia cells. Leuk. Lymphoma. 2014;55:1373–1382. doi: 10.3109/10428194.2013.829574. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X., Yuan P., Liu Q., Liu Z. LncRNA MEG3 Regulates Imatinib Resistance in Chronic Myeloid Leukemia via Suppressing MicroRNA-21. Biomol. Ther. 2017;25:490–496. doi: 10.4062/biomolther.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mer A.S., Lindberg J., Nilsson C., Klevebring D., Wang M., Grönberg H., Lehmann S., Rantalainen M. Expression levels of long non-coding RNAs are prognostic for AML outcome. J. Hematol. Oncol. 2018;11:52. doi: 10.1186/s13045-018-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidovich C., Cech T.R. The recruitment of chromatin modifiers by long noncoding RNAs: Lessons from PRC2. RNA. 2015;21:2007–2022. doi: 10.1261/rna.053918.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cifuentes-Rojas C., Hernandez A.J., Sarma K., Lee J.T. Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell. 2014;55:171–185. doi: 10.1016/j.molcel.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davidovich C., Wang X., Cifuentes-Rojas C., Goodrich K.J., Gooding A.R., Lee J.T., Cech T.R. Toward a Consensus on the Binding Specificity and Promiscuity of PRC2 for RNA. Mol. Cell. 2015;57:552–558. doi: 10.1016/j.molcel.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko S., Li G., Son J., Xu C.F., Margueron R., Neubert T.A., Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24:2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X.D., Huang G.W., Xie Y.H., He J.Z., Guo J.C., Xu X.E., Liao L.D., Xie Y.M., Song Y.M., Li E.M., et al. The interaction of lncRNA EZR-AS1 with SMYD3 maintains overexpression of EZR in ESCC cells. Nucleic Acids Res. 2018;46:1793–1809. doi: 10.1093/nar/gkx1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Leary V.B., Ovsepian S.V., Carrascosa L.G., Buske F.A., Radulovic V., Niyazi M., Moertl S., Trau M., Atkinson M.J., Anastasov N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015;11:474–485. doi: 10.1016/j.celrep.2015.03.043. [DOI] [PubMed] [Google Scholar]
- 20.O’Leary V.B., Hain S., Maugg D., Smida J., Azimzadeh O., Tapio S., Ovsepian S.V., Atkinson M.J. Long non-coding RNA PARTICLE bridges histone and DNA methylation. Sci. Rep. 2017;7:1790. doi: 10.1038/s41598-017-01875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez J.A., Wapinski O.L., Yang Y.W., Bureau J.F., Gopinath S., Monack D.M., Chang H.Y., Brahic M., Kirkegaard K. The NeST Long ncRNA Controls Microbial Susceptibility and Epigenetic Activation of the Interferon-γ Locus. Cell. 2013;152:743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y.W., Flynn R.A., Chen Y., Qu K., Wan B., Wang K.C., Lei M., Chang H.Y. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. Elife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sze C.C., Shilatifard A. MLL3/MLL4/COMPASS Family on Epigenetic Regulation of Enhancer Function and Cancer. Cold Spring Harb. Perspect. Med. 2016;6:a026427. doi: 10.1101/cshperspect.a026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herz H.M., Mohan M., Garruss A.S., Liang K., Takahashi Y.H., Mickey K., Voets O., Verrijzer C.P., Shilatifard A. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26:2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henikoff S., Shilatifard A. Histone modification: Cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Lazar T., Schad E., Szabo B., Horvath T., Meszaros A., Tompa P., Tantos A. Intrinsic protein disorder in histone lysine methylation. Biol. Dir. 2016;11:30. doi: 10.1186/s13062-016-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mészáros B., Simon I., Dosztányi Z. Prediction of Protein Binding Regions in Disordered Proteins. PLoS Comput. Biol. 2009;5:e1000376. doi: 10.1371/journal.pcbi.1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., Gladfelter A.S. RNA Controls PolyQ Protein Phase Transitions. Mol. Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Micsonai A., Wien F., Bulyáki É., Kun J., Moussong É., Lee Y.H., Goto Y., Réfrégiers M., Kardos J. BeStSel: A web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46:W315–W322. doi: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Micsonai A., Wien F., Kernya L., Lee Y.H., Goto Y., Réfrégiers M., Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. USA. 2015;112:E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwarzer A., Emmrich S., Schmidt F., Beck D., Ng M., Reimer C., Adams F.F., Grasedieck S., Witte D., Käbler S., et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nat. Commun. 2017;8:218. doi: 10.1038/s41467-017-00212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Battistelli C., Cicchini C., Santangelo L., Tramontano A., Grassi L., Gonzalez F.J., de Nonno V., Grassi G., Amicone L., Tripodi M. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2016;36:942–955. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davidovich C., Zheng L., Goodrich K.J., Cech T.R. Promiscuous RNA binding by Polycomb repressive complex 2. Nat. Struct. Mol. Biol. 2013;20:1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banani S.F., Lee H.O., Hyman A.A., Rosen M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langdon E.M., Qiu Y., Niaki A.G., McLaughlin G.A., Weidmann C., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M., et al. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018;360:922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z.-Y., Yang L., Liu X.-J., Wang X.-Z., Pan Y.-X., Luo J.-M. The Long Noncoding RNA MEG3 and its Target miR-147 Regulate JAK/STAT Pathway in Advanced Chronic Myeloid Leukemia. EBioMedicine. 2018;34:61–75. doi: 10.1016/j.ebiom.2018.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y.-Y., Huang S.-H., Zhou H.-R., Chen C.-J., Tian L.-H., Shen J.-Z. Role of HOTAIR in the diagnosis and prognosis of acute leukemia. Oncol. Rep. 2016;36:3113–3122. doi: 10.3892/or.2016.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mészáros B., Erdos G., Dosztányi Z. IUPred2A: Context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018;46:W329–W337. doi: 10.1093/nar/gky384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng Z., Kurgan L. High-throughput prediction of RNA, DNA and protein binding regions mediated by intrinsic disorder. Nucleic Acids Res. 2015;43:e121. doi: 10.1093/nar/gkv585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu T.J., Shamsaddini A., Pan Y., Smith K., Crichton D.J., Simonyan V., Mazumder R. A framework for organizing cancer-related variations from existing databases, publications and NGS data using a High-performance Integrated Virtual Environment (HIVE) Database. 2014;2014:bau022. doi: 10.1093/database/bau022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2014;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y., Zhong Y., Wang Y., Zhang X., Batista D.L., Gejman R., Ansell P.J., Zhao J., Weng C., Klibanski A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 44.Shih P., Holland D.R., Kirsch J.F. Thermal stability determinants of chicken egg-white lysozyme core mutants: Hydrophobicity, packing volume, and conserved buried water molecules. Protein Sci. 1995;4:2050–2062. doi: 10.1002/pro.5560041010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.