Abstract
Stacked traits have become an important trend in the current development of genomically modified crops. The bidirectional promoter can not only prevent the co-suppression of multigene expression, but also increase the efficiency of the cultivation of transgenic plants with multigenes. In Gossypium hirsutum, Ghrack1 and Ghuhrf1 are head-to-head gene pairs located on chromosome D09. We cloned the 1429-bp intergenic region between the Ghrack1 and Ghuhrf1 genes from Gossypium hirsutum. The cloned DNA fragment GhZU had the characteristics of a bidirectional promoter, with 38.7% G+C content, three CpG islands and no TATA-box. Using gfp and gus as reporter genes, a series of expression vectors were constructed into young leaves of tobacco. The histochemical GUS (Beta-glucuronidase) assay and GFP (green fluorescence protein) detection results indicated that GhZU could drive the expression of the reporter genes gus and gfp simultaneously in both orientations. Furthermore, we transformed the expression vectors into Arabidopsis and found that GUS was concentrated at vigorous growth sites, such as the leaf tip, the base of the leaves and pod, and the stigma. GFP was also mainly expressed in the epidermis of young leaves. In summary, we determined that the intergenic region GhZU was an orientation-dependent bidirectional promoter, and this is the first report on the bidirectional promoter from Gossypium hirsutum. Our findings in this study are likely to enhance understanding on the regulatory mechanisms of plant bidirectional promoters.
Keywords: bidirectional promoter, Gossypium hirsutum, cloning, transient expression, stable expression
1. Introduction
Transgenic plants with stacked traits harboring two or more foreign genes can meet the diverse needs of growers and provide multiple benefits; thus, stacked traits have become an important trend in the current development of genomically modified crops. In 2017, the planting area devoted to stacked traits was 77.8 million hectares and covered 41% of the biotech crop planting area globally, representing a 2.5% increase in planting area compared with that in 2016 [1].
Co-transformation is an effective strategy for the cultivation of transgenic plants with stacked traits. The selection of a promoter is an important factor for the successful expression of multigenes during co-transformation [2]. Multi-gene engineering strategies are often hampered by sub-optimal expression levels or improper tissue-specificity of particular promoters, or rely on the use of multiple copies of the same promoter, which can result in DNA instability or transgene silencing [3]. A homology of 90 bp in promoter sequences has been reported to be sufficient for the co-suppression of gene expression and transgene silencing [4]. The use of a bidirectional promoter can avoid this disadvantage and increase the efficiency of co-transformation.
A bidirectional promoter is the intergenic region between divergent or head-to-head gene pairs, and bidirectional promoters can drive the simultaneous transcription of divergent gene pairs. With the development of sequencing technology and genome-wide annotations, it has been shown that divergent or head-to-head gene pairs are very common in eukaryotes [5,6]. In the human genome, more than 10% of the genes are arranged in a head-to-head manner with an intervening sequence of less than 1000 bp, and these gene pairs are driven by 1352 bidirectional promoters [5,7,8,9]. However, in the plant genome, researchers have found many head-to-head genes with an intervening sequence of more than 1000 bp [10,11,12]. In the Arabidopsis genome, a large proportion (13.3%) of bidirectional gene pairs have been observed, and some of these pairs share an intergenic region of 1 to 1.5 kb [13]. Using the same selection standards as those used for human bidirectional promoters (length less than 1 kb), 8742, 5763, and 8823 divergent gene pairs in total have been identified, accounting for 30.9%, 24%, and 39% of the genomes of rice, Arabidopsis thaliana, and Populus, and they have confirmed 2106, 1242, and 613 bidirectional promoters in rice, Arabidopsis, and Populus, respectively [14,15].
In plants, since Shwarz et al. [16] reported the first bidirectional promoter of maize chloroplast genes, several bidirectional promoters have been reported in maize, hot pepper, Arabidopsis thaliana, rice, soybean, and Populus [11,15,17,18,19]. In 2003, Shin et al. identified that the CaTin 1 and CaTin1-2 genes in hot pepper had an opposite transcriptional direction and that a 955-bp DNA sequence between the two genes could drive gus gene expression in tobacco from two directions, and also regulate the expression of the two genes in response to biotic stress due to pathogen infection, representing the first endogenous bidirectional promoter identified in plants [17]. In analyses of Arabidopsis thaliana genomic data, Banerjee found that the At4g35987 and At4g35985 genes were bidirectionally transcribed gene pairs, and cloned a sequence of 1258 bp between the two adjacent genes. The authors connected the gus and gfp genes at both orientations to transform Arabidopsis thaliana and tobacco, respectively. The functional verification of the GUS and GFP proteins was performed in both Arabidopsis thaliana and tobacco, confirming that the sequence was indeed a bidirectional promoter [11]. In Arabidopsis thaliana, the caseinolytic protease B-cytoplasmic (ClpB-C)/heat shock protein 100 protein (AtClpB-C) gene (At1g74310) and the choline kinase (AtCK2) gene (At1g74320) are located in divergent orientations, and their 1329 bp serves as a heat-inducible bidirectional promoter [19]. In 2014, Liu et al. conducted a genome-wide search in maize using genome sequencing results from the inbred line, B73. In total, 1696 bidirectional transcript pairs were identified using a modified search model. The authors functionally characterized the promoter activity of the intergenic regions of most of the bidirectional transcript pairs that were expressed in embryos using a maize embryo transient expression system [18]. In 2015, Liu et al. used the promoter::GUS transgenic approach and revealed that the intergenic region of the Arabidopsis thaliana divergent genes At1g71850 and At1g71860 was an asymmetric bidirectional promoter, exhibiting an orientation-dependent expression profile. The authors demonstrated that the activity of the At1g71850-At1g71860 bidirectional promoter was modulated by complex interactions between both positive and negative cis-acting elements [20]. In 2016, Wang et al. initiatively combined RNA-seq data and cDNA microarray data to discover the potential bidirectional promoters in rice genomes. Based on the expression level and correlation of each adjacent and oppositely transcribed gene pair, they selected four candidate gene pairs, and subsequently found that GUS and GFP assays of the transgenic plants indicated that all the intergenic regions showed bidirectional expression activity in various tissues of rice [12]. However, to date, there are no reports of a bidirectional promoter from Gossypium hirsutum.
In this paper, we report the isolation and characterization of an intergenic region (1429 bp) shared by Ghrack1 (receptor for activated C kinase 1, Gohir.D09G171000.1) and Ghuhrf1 (E3 ubiquitin-protein ligase gene, Gohir.D09G171100.1) divergent genes on chromosome D09 from Gossypium hirsutum. The transient expression analysis showed that the intergenic sequence could simultaneously drive the expression of the reporter genes gus and gfp from the forward and reverse orientations. Subsequently, we transformed Arabidopsis thaliana to achieve a stable expression, which fully demonstrated that the sequence had bidirectional promoter activity, and was an orientation-dependent bidirectional promoter in Arabidopsis thaliana. This study provides the first report of a bidirectional promoter from Gossypium hirsutum in Arabidopsis thaliana.
2. Results
2.1. Genomic Organization of the Head-to-Head Gene Pairs, Ghrack1 and Ghuhrf1
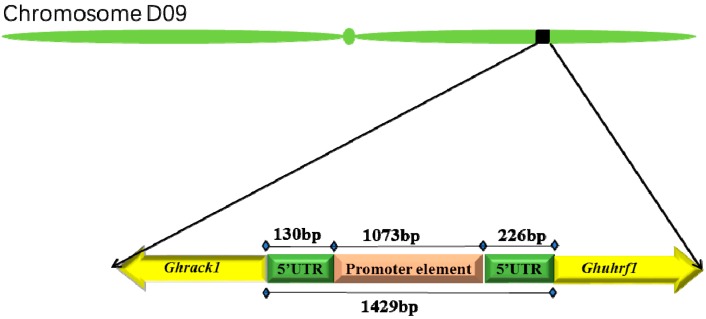
Analysis of the genomic organization of the Ghrack1 gene in the phytozome database indicated that a Ghuhrf1 gene was located immediately upstream. The two genes were head-to-head on chromosome D09 of the Gossypium hirsutum genome. In silico analysis revealed that the translation initiation sites of these two adjacent genes were 1429 bp apart (Figure S1). Precise identification of transcription start sites (TSS) in Ghrack1 and Ghuhrf1 was through 5′-rapid amplification of cDNA ends (5′-RACE) (Figure S4). Ghrack1 had a 130-bp 5′-UTR, and the Ghuhrf1 gene contained a 5‘-untranslated region (5′-UTR) of 226 bp. The distance between the transcription start sites of the two genes was 1073 bp (Figure 1).
Figure 1.
Schematic representation of the organization of the Ghrack1 and Ghuhrf1 genes on chromosome D09 of Gossypium hirsutum. The 1429 bp of the intergenic region and the core 1073 bp promoter are marked. In total, 226 bp and 130 bp 5′-UTRs were upstream of Ghuhrf1 and Ghrack1, respectively.
2.2. Relative Expression (Transcript) of the Ghrack1 and Ghuhrf1 Genes in Various Upland Cotton Tissues
The relative transcript abundance of these two (Ghrack1 and Ghuhrf1) adjacent genes in the root, leaf, anther, stigma, and fiber tissues from different periods of the upland cotton K312 was assayed by semiquantitative PCR (Polymerase Chain Reaction) and quantitative real-time PCR (qRT-PCR) to evaluate their possible functional roles during development and growth.
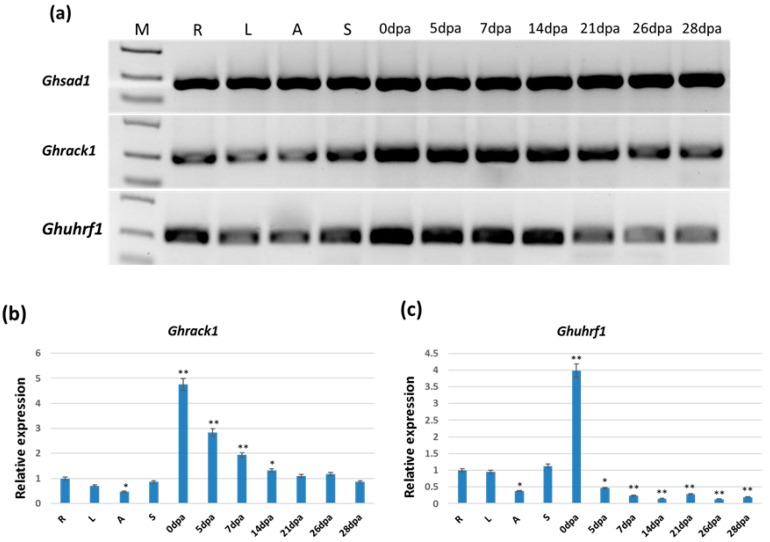
The semiquantitative PCR results showed that the Ghrack1 gene had a higher expression level in the fiber tissues of various periods, but a lower expression level in other tissues. The transcript abundance of Ghuhrf1 was highest in the initiation differentiation and elongation stage of fiber. For the semiquantitative PCR analysis, the Ghsad1 gene was used as an internal control, and its expression level was found to be stable in all tissues (Figure 2a).
Figure 2.
Semiquantitative PCR and qRT-PCR (quantitative real-time PCR) data of the transcript levels of the Ghrack1 and Ghuhrf1 genes in different tissues of Gossypium hirsutum. (a) Semiquantitative PCR survey of various cotton tissues for the detection of Ghrack1 and Ghuhrf1 transcripts. Ghsad1 was used as a control. (b,c) Relative transcript abundance of two Gossypium hirsutum genes, that is, Ghrack1 (a) and Ghuhrf1 (c), detected in various cotton tissues by qRT-PCR. The data represent the relative expression of the Ghrack1 and Ghuhrf1 transcripts ± SD of three biological replicates in each tissue (n = 3). Asterisks and double asterisks indicate significant deviations from the root at p < 0.05 and p < 0.01, respectively, using the Student’s t-test for comparisons between the root and other tissues separately for both genes. R (root), L (leaf), A (anther), S (stigma), 0 dpa (0 dpa fiber), 5 dpa (5 dpa fiber), 7 dpa (7 dpa fiber), 14 dpa (14 dpa fiber), 21 dpa (21 dpa fiber), 26 dpa (26 dpa fiber), and 28 dpa (28 dpa fiber).
The qRT-PCR results showed that the high expression level of the Ghrack1 gene was exhibited in the initiation-differentiation stage of fiber (Figure 2b). Compared with the relative expression of Ghrack1 in the root, the relative transcript abundance was significantly higher (p < 0.01) in the 0 dpa (days post-anthesis), 5 dpa, and 7 dpa fibers and moderately higher (p < 0.05) in the 14 dpa fiber. This result was consistent with the semiquantitative PCR results. The Ghuhrf1 transcript abundance was highest in the 0 dpa fiber, but there was a lower expression level in other periods of fiber and tissues (Figure 2c). Compared with the relative expression of Ghuhrf1 in the root, the relative transcript abundance was significantly lower (p < 0.01) in the 7–28 dpa fibers and moderately lower (p < 0.05) in the anther and 5 dpa fiber, but significantly higher in the 0 dpa fiber (p < 0.01). These results somewhat differed from the semiquantitative PCR results, potentially due to the lower sensitivity of semiquantitative PCR. Thus, because of they shared an identical DNA region, the genes in both orientations which were driven by the same promoter exhibited similar expression profiles.
2.3. Sequence Analysis of the Cloned Promoter GhZU
The intergenic region between Ghrack1 and Ghuhrf1 was cloned from Gossypium hirsutum K312 and named GhZU. The DNA sequencing analysis showed the high G+C content (38.7%) and three CpG islands in GhZU. The GhZU promoter fragment was submitted to PLACE (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?lang=en&pj=640&action=page&page=newplace) and PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict putative cis-elements involved in the regulation of gene expression (Figure 3). Potential regulatory elements were identified within GhZU (Table S2), including core elements, such as the CAAT-Box and GC-Box, but no TATA-Box. Some cis-elements were known to be involved in the growth and development of cotton fiber, such as MYB2AT, L1BOXATPDF1, and MYB2CONSENSUSAT. In addition, many elements in GhZU had been shown to participate in tissue-specific expression, such as ROOTMOTIFTAPOX1 (organ-specific gene expression), POLLEN1LELAT52 (anther-specific gene expression), and AACACOREOSGLUB1 and CANBNNAPA (the expression of endosperm-specific genes). A few elements were related to the ethylene response induced by cis-regulatory elements (ERELEE4). The predicted results of the 5’UTRs of Ghrack1 and Ghuhrf1 are presented in the supplementary materials (Figure S2). We defined the transcriptional orientation of the Ghuhrf1 gene as forward and named it GhZUf, and the transcriptional orientation of Ghrack1 as reverse and named it GhZUr.
Figure 3.
Sequence analysis of the cloned promoter GhZU. The putative cis-acting elements in both orientations of the promoter GhZU determined by the softwares PLACE and PlantCARE.
2.4. GhZU Drives the Transient Expression of the Reporter Genes in Both Orientations
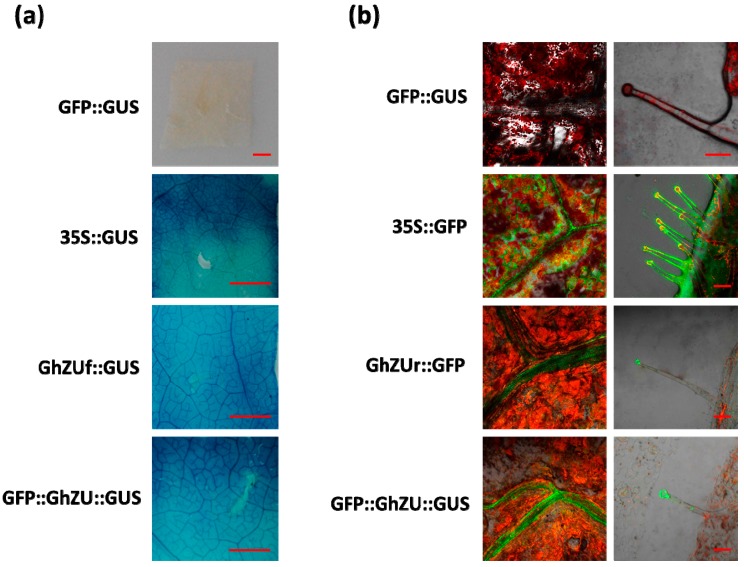
Agrobacterium strains carrying the transient expression vector were introduced into young leaves of tobacco plantlets (6–8 leaf stage) by the infiltration transient expression assays. Histochemical GUS assays and GFP detection were performed 3 days after infection. The histochemical GUS assay results showed an obvious blue color in the whole leaf that was infected with Agrobacterium containing the vectors GhZUf::GUS and GUS::GhZU::GFP (Figure 4a). The GFP fluorescence detection results revealed green fluorescence in the leaves infected by Agrobacterium containing the vectors GhZUr::GFP and GUS::GhZU::GFP, whereas the gfp gene driven by GhZUr was preferentially expressed in leaf trichomes and veins, and no fluorescence signal was detected in other leaf sites (Figure 4b and Figure S3).
Figure 4.
Transient expression of the gus gene and gfp gene in tobacco leaves, using the epidermis infection method. (a) Schematic diagram of the promoter-reporter gene constructs GFP::GUS, 35S::GUS, GhZUf::GUS, and GFP::GhZU::GUS, used for the transient assay in the N. benthamiana leaf using the pCambia1305 vector. (b) Schematic diagram of the promoter-reporter gene constructs GFP::GUS, 35S::GFP, GhZUr::GFP, and GFP::GhZU::GUS, used for the transient assay in the N. benthamiana leaf using the pCambia1305 vector. Below each construct, a representative assay of transient gus expression detected histochemically, transient gfp expression detected based on fluorescence imaging via the Agrobacterium infiltration assay in N. benthamiana leaf, and the respective promoter, no promoter, 35S promoter, Ghuhrf1 promoter (GhZUf), and Ghrack1 promoter (GhZUr) activities are shown.
No detectable histochemical GUS staining was visualized in infected leaves from the untransformed control tobacco plants; however, strong histochemical GUS staining under CaMV35S was observed (Figure 4a). No detectable GFP fluorescence was visualized in infected leaves from the untransformed control tobacco plants, but strong GFP fluorescence under CaMV35S was observed (Figure 4b and Figure S3).
2.5. Detection and the Copy Number Analysis of Transgenic Arabidopsis thaliana
In order to obtain transgenic positive plants, we used the gus gene as the target gene and designed primers to identify them by PCR amplification. Finally, 246 transgenic positive plants were screened out from 320 individuals (Figure S4).
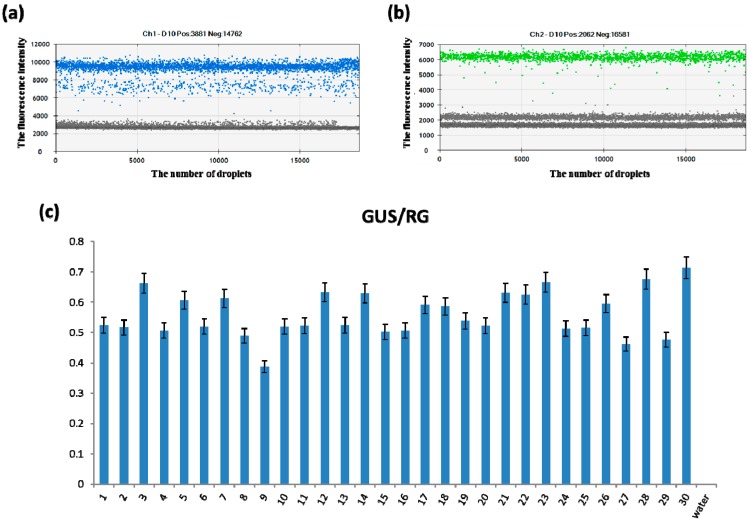
According to the Arabidopsis single-copy gene RG (AT1G03400.1) and the target gene gus, we designed a primer probe for a droplet digital PCR. The amplification results showed that the designed primer probe had high specificity, and the system could clearly distinguish between positive and negative microdrops (Figure 5a,b). The number of microdrops generated in the experiment exceeded 13,000, thus meeting the requirements of the Poisson distribution, and the relative standard deviation (RSD) value of the number of droplets formed by three replicates was less than 0.25, meeting the requirements of EU (European Union) nucleic acid molecular testing. The above results show that the droplet digital PCR system was stable and repeatable, and that the data was reliable [21,22,23,24].
Figure 5.
Detection of the copy number of transgenic Arabidopsis thaliana using the droplet digital PCR method. (a) Internal reference gene; (b) gus gene; (c) thirty selected single-copy gus gene individuals. The gray signal in the map represents the micro-droplets that had not been amplified by PCR, and the system was considered a negative signal; the blue signal shows the FAM fluorescence signal, and the green signal shows the HEX fluorescence signal representing the PCR amplification. The system was considered a positive signal.
Arabidopsis thaliana is a diploid plant, and the RG base exists in the form of a single copy. In the T0 generation of the transgenic Arabidopsis, the target gene gus is hemizygous. Theoretically, the content of the gene in the genome should be 1/2 RG to demonstrate that the transferred gus gene is a single-copy gene. Subsequently, pure and single-copy gus transgenic Arabidopsis plants were obtained through selfing. Using an established droplet digital PCR system to calculate the copy number of the target gene, 162 single-copy gus individuals were selected from 246 positive seedlings, and 30 individuals were selected for the subsequent experiments (Figure 5c).
2.6. Function Analysis of the Bidirectional Promoter GhZU in Transgenic Arabidopsis thaliana
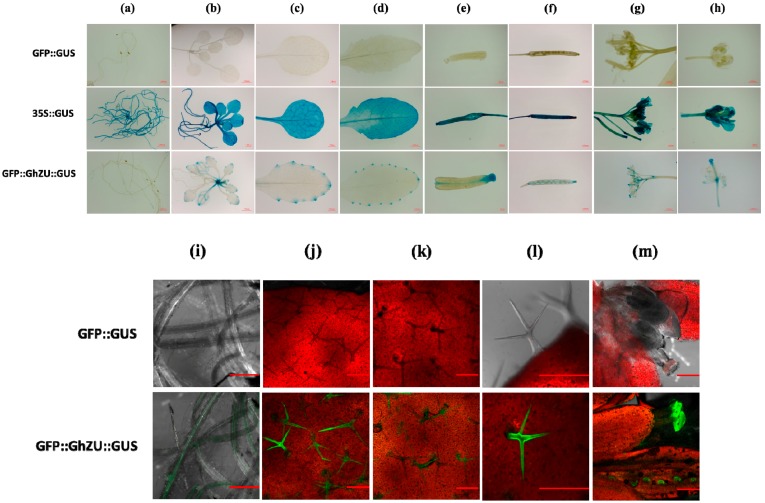
To evaluate the tissue-specific nature of the bidirectional promoter, independent transgenic Arabidopsis lines were generated for the constructs 1305.1-GFP::GUS, 1305.1-35S::GUS, and 1305.1-GFP::GhZU::GUS. In the GFP::GhZU::GUS plants, during the growth and development of Arabidopsis, strong histochemical GUS staining under GhZUf was observed in the leaf tip, apical meristem, stigma, and petiole base regions (Figure 6a–h). In contrast, strong GFP expression of GhZUr was detected in the young leaf trichomes and old leaf trichomes, and weak GFP expression was detected in the whole roots and lateral roots (Figure 6i–m, Figure S3). In the reproductive tissues of Arabidopsis, GFP expression under GhZUr showed weak expression in the anther, stigma, and developing silique, whereas GUS expression under GhZUf was detected in the floral bud, stigma, and developing silique (Figure 6h).
Figure 6.
Localization of GUS and GFP in vegetative and reproductive tissues of GFP::GUS, 35S::GUS, and GFP::GhZU::GUS transgenic Arabidopsis plants. (a–h) Histochemical GUS localization data of GFP::GUS (up), 35S::GUS (middle), and GFP::GhZU::GUS (down) in vegetative and reproductive tissues of Arabidopsis plants. (a) Root region; (b) two-week-old Arabidopsis seedlings; (c) young leaf; (d) old leaf; (e) developing silique; (f) mature silique; (g) inflorescence; and (h) flower. Histochemical GUS images of GFP::GUS (up), 35S::GUS (middle), and GFP::GhZU::GUS (down) are shown. (i–m) Confocal laser-scanning microscopic analysis of gfp expression under GFP::GUS and GFP::GhZU::GUS in Arabidopsis plants. (i) Root, (j) young leaf trichomes, (k) old leaf trichomes, (l) trichomes, and (m) flower. Green fluorescence images of GFP::GUS (up) and GFP::GhZU::GUS (down) are shown.
No detectable histochemical GUS staining was visualized in various tissues from the untransformed control Arabidopsis plants, including the root, leaf, flower, and immature and mature silique. However, strong histochemical GUS staining under CaMV35S was observed in all tissues during Arabidopsis growth and development (Figure 6a–h). Also, no detectable GFP fluorescence was observed in the vegetative tissues from the untransformed control Arabidopsis plants (Figure 6i–m, Figure S3).
The tissue-specific expression pattern of the bidirectional promoter GhZU was confirmed with the reporter genes (gus and gfp). According to the expression of the gus gene driven by GhZUr, GUS was mainly expressed in sites of vigorous plant growth, such as the leaf tip, apical meristem, and petiole base regions. GFP was predominantly expressed in young and vigorously growing plant tissues, such as young leaf trichomes. Therefore, the intergenic region GhZU was indeed found to be a bidirectional promoter, and was able to drive the reporter genes’ expression and in a similar manner according to its orientation.
2.7. Relative Expression Level of the gfp and gus Genes in Various Arabidopsis Tissues
The transcript abundance of the reporter genes gfp and gus were measured to better understand the regulation of the expression of these two tissue-specific genes (Ghrack1 and Ghuhrf1) under the bidirectional promoter GhZU. The relative transcript abundance of these two reporter genes (gfp and gus) in various tissues (root, young leaf, older leaf, flower, developing silique, and mature silique) were assayed by quantitative real-time PCR (qRT-PCR), using gene-specific primers to evaluate the tissue-specific expression of the bidirectional promoter in both orientations.
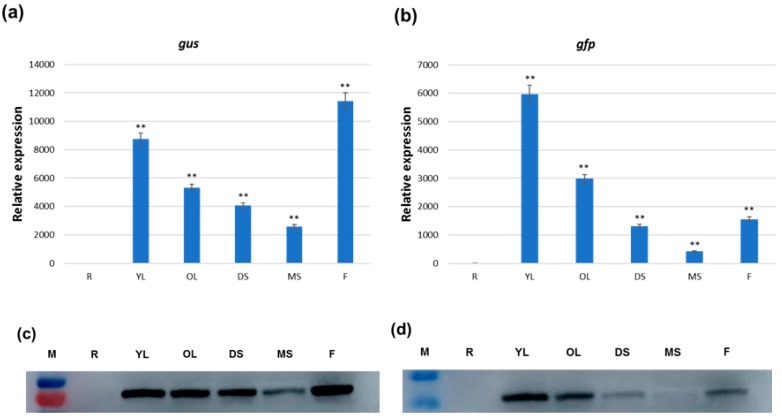
The expression level of the gus gene was in the following order: maximum expression in the flower tissue, followed by that of the young leaf, older leaf, developing silique, and mature silique; however, there was almost no expression in the root tissue (Figure 7a). The gfp transcript abundance was highest in the young leaf, followed by the older leaf, developing silique, flower tissue, and mature silique, whereas the lowest expression was detected in the root tissues (Figure 7b). These data indicate that this bidirectional promoter directed gene expression in an orientation-dependent manner during the Arabidopsis’ development and growth.
Figure 7.
GUS and GFP expression in various tissues of transgenic Arabidopsis plants generated for the constructs GFP::GhZU::GUS. (a,b) Relative expression (transcript) of two reporter genes, that is, gus (a) and gfp (b), detected in various Arabidopsis tissues by qRT-PCR. The data represent the relative expression of the gus and gfp transcripts ± SD of three biological replicates of each tissue (n = 3). Asterisks and double asterisks indicate significant deviations from the root at p < 0.05 and p < 0.01, respectively, using Student’s t-test for comparisons between the root and other tissues separately for both genes. (c,d) Relative expression (protein) of two reporter genes, that is, gus (c) and gfp (d), detected in various Arabidopsis tissues by protein hybridization. R (root), YL (young leaf), OL (old leaf), DS (developing silique), MS (mature silique), and F (inflorescence).
2.8. Detection of the Expression of the gus and gfp Genes in Various Tissues of Arabidopsis thaliana by Western Blot
The relative protein abundance of these two reporter genes in various tissues (root, young leaf, older leaf, flower, developing silique, and mature silique) were assayed by western blot using protein-specific antibodies to evaluate the tissue-specific expression of the bidirectional promoter in both orientations.
The western blot analysis using an anti-GUS antibody revealed strong GUS expression in the young leaves, older leaves, developing silique, and floral tissues, and moderate expression in the mature silique. However, no detectable GUS protein was visualized in the root (Figure 7c). Using an anti-GFP antibody revealed that GFP expression was highest in the young leaves, and had moderate expression in the old leaves. Whereas the lowest expression was detected in the developing silique and floral tissues, and GFP protein expression was not detected in the root (Figure 7d). The western blot results differed from the results obtained in the fluorescence quantitative analysis, potentially because the total protein from the plants degraded during extraction, or the protein expression was too weak and the sensitivity was too low, making it impossible to detect.
3. Discussion
With the rapid development of genome-sequencing technology today, more and more species have been completed genome sequencing. Functional studies of intergenic regions have attracted the most attention of researchers [11], and the role of noncoding DNA in phenotypic evolution has been reported many times in the literature [25]. The orientation of flanking genes may influence the evolution of intergenic regions, in which cis-regulatory elements are likely to be located [26,27]. The head-to-head clustering of genes in which two adjacent genes are separated by a short intergenic distance have been determined by the direction of gene expression in different transcription configurations, and are also prevalent and conserved in many eukaryotes, including yeasts, plants, invertebrates, and vertebrates [28]. With the availability of the complete genome sequences of several organisms, the functionality of intergenic regions has attracted increasing attention. Compared with unidirectional promoters, bidirectional promoters have some specific structural characteristics. Generally, the length of a bidirectional promoter is largely within 1000 bp in humans, but often longer than 1000 bp in plants [5,10]. The bidirectional promoter that drives rice chymotrypsin protease inhibitor genes (OCPI1 and OCPI2) is 1126 bp [29]. Also, an intergenic region (1258 bp) shared by At4g35985 and At4g35987 in Arabidopsis thaliana is a tissue-specific and stress-inducible bidirectional promoter analyzed in transgenic Arabidopsis and tobacco plants [11]. High C+G content is also an important feature of bidirectional promoters. Yang et al. found that 70.8% of human bidirectional promoters have a G+C content exceeding 60%, and that CpG-islands are present in 90% of bidirectional promoters [5,30]. The C+G content in bidirectional promoters in rice, Arabidopsis, and Poplar are 55%, 37%, and 48%, respectively, which are significantly higher than its content in randomly selected promoters [15]. The G+C content of bidirectional promoters are 55%, 48.2%, 31%, and 34% in sorghum, rice, soybean, and Arabidopsis genomes, respectively, while those in randomly selected promoters are 46.5%, 44.3%, 29.8%, and 32.1%, respectively. The G+C content of bidirectional promoters are generally higher than those in randomly selected promoters. This trend is consistent with that of bidirectional promoters in maize [18]. There is a growing interest in function analysis of intergenic regions, which would not only help us get a better understanding of divergent transcription, but also help us turn the information into a new tool for the manipulation of genomes [31]. In this study, the region shared by two adjacent genes Ghrack1 and Ghuhrf1 on chromosome D09 of Gossypium hirsutum was found to function as a promoter in both orientations. The intergenic region GhZU between the TSS of Ghrack1 and Ghuhrf1 was found to have a 38.7% C+G content, three CpG islands, and no TATA-box motif, which is why we conducted a critical analysis of this intergenic sequence.
We cloned the 1429 bp intergenic region sequence between the ATGs of the head-to-head gene pair. The intergenic region sequence was submitted to PLACE and PlantCARE to predict the putative cis-elements involved in the regulation of gene expression. Potential regulatory elements were identified within GhZU, including core elements such as the CAAT-Box and GC-Box, but no TATA-Box. Some cis-elements have been known to be involved in the growth and development of cotton fiber, such as MYB2AT, L1BOXATPDF1, and MYB2CONSENSUSAT; therefore, we linked the intergenic region sequence to the reporter genes in both orientations. We transformed the expression vectors into tobacco and Arabidopsis thaliana, where for tobacco, we demonstrated that the intergenic sequences had bidirectional promoter activity, and for transgenic Arabidopsis thaliana, the gus gene was mainly expressed in regions and tissues with more vigorous growth, and weak or no expression was observed in the mature tissues. Consistent with the Ghuhrf1 gene expression in cotton, this gene was mainly expressed during the initial stage of fiber progenitor cells, and the expression gradually decreased over time. Therefore, based on the expression of the gus gene in Arabidopsis thaliana and the Ghuhrf1 gene in cotton, the GhZUf promoter was shown to be a tissue- and period-specific promoter, whereas the gfp gene was mainly expressed in the epidermal hairs at the beginning of growth and the newly developed silique, but was not expressed in the mature tissues. Consistent with the expression of the Ghrack1 gene in cotton, this gene was mainly expressed during the early stage of fiber development, followed by a gradual decline with the growth and development of the fiber until the fiber matured. Many researchers have found that the development mechanism of Arabidopsis epidermis hair is similar to that of cotton fiber [32]. Therefore, based on the gfp gene expression in Arabidopsis thaliana and the Ghrack1 gene expression in cotton, the GhZUr promoter was determined to be a dominant fiber-expression promoter. In summary, the GhZU promoter is an orientation-dependent bidirectional promoter in cotton and Arabidopsis, and has certain dependencies in both directions of initiation; however, some differences exist in time and space. The expression patterns of GhZUf and GhZUr should be confirmed in future studies through its stable expression in transgenic cotton.
To improve plants by using molecular biology and genetic engineering methods, transferring multiple genes concomitantly is often necessary. Gene fusion technology or gene stacking technology is commonly used to recombine or superimpose two or more functional genes or to use multiple transgenes to transfer multiple functional genes into the same plant, which is usually completed by using a high-expression efficiency promoter [33]. Previous studies have shown that the presence of a homologous sequence between two promoters in the plant may cause “co-inhibition” of gene expression, leading to partial gene silencing [4]. Compared with unidirectional promoters, bidirectional promoters improve the efficiency of biotechnological improvements by regulating many genes, which is why bidirectional promoters have attracted considerable attention. The bidirectional promoters analyzed in this study received attention due to their similar tissue-specific expression patterns and variation in the expressional magnitude in both orientations. In one direction (GhZUf), the promoter is very active in the tip of the leaf and apical meristem regions, while in the other direction (GhZUr), the promoter is strongly expressed in young tissues, such as young leaf epidermal hairs. To increase the quantity and quality of fibers, several genes relatived with fiber growth and development can be simultaneously expressed in two directions to obtain more high-quality fibers. Therefore, the bidirectional promoters shared regulatory elements of expression in the opposite direction, and the genes driven by bidirectional promoters could be better expressed in the host cell under selective pressure than by other synthetic promoters. The bidirectional promoters could become the preferred promoters for transgene breeding in the future through the use of multigene co-transformation, which is invaluable in the field of genetic engineering research and its applications.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Nicotiana benthamiana was kindly provided by Professor Luo’s laboratory from the College of Life Science of Southwest University. Tobacco and cotton were grown in a greenhouse under a 16 h light/8 h dark cycle at 28–30 °C. Wild-type Arabidopsis thaliana (Col-0) and seeds of transgenic plants were surface-sterilized with 75% (v/v) ethanol for 8 min, followed by 1 min with 95% (v/v) ethanol. The sterilized Arabidopsis seeds were plated on containing 1/2 Murashige and Skoog medium. Seeds were stratified at 4 °C for 2 days, and the plates were subsequently transferred to a plant growth incubator under a 16 h light/8 h dark cycle at 23 °C.
4.2. Transcript Analysis by Semiquantitative PCR and Quantitative Real-Time PCR (qRT-PCR) in Upland Cotton
The total RNA was extracted from the root, leaf, anther, stigma, and fiber tissues at different periods (0, 5, 7, 14, 21, 26, and 28 dpa) from upland cotton K312, and cDNA was generated using a primescriptTM RT reagent kit and gDNA eraser (TaKaRa, Dalian, China) following the manufacturer’s instructions. For quantitative measurements of the Ghrack1-specific transcript, forward (q-Ghrack1-F) and reverse (q-Ghrack1-R) primers were used. For the quantitative measurements of the Ghuhrf1-specific transcript, forward (q-Ghuhrf1-F) and reverse (q-Ghuhrf1-R) primers were also used. Sad1 was used as a control with the primers q-sad-F and q-sad-R [34]. Quantification of the Ghrack1 and Ghuhrf1 transcript levels by semiquantitative PCR was also performed. Each reaction was performed in triplicate on a T-100 thermal cycler (BIO-RAD, CA, USA) using the following conditions: denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 30 s. The relative transcript abundance was assessed by semiquantitative PCR, following a published protocol [35]. PCR was performed in four replicates and repeated in three biological samples. Quantitative real-time transcription PCR (qRT-PCR) was carried out to confirm the sequencing data identifying the DEGs. Using the first cDNA strand as a template, qRT-PCR was performed on an ABI7500 Real-Time System (Applied Biosystems, USA) using a SYBR premix Ex Taq kit (TaKaRa, Dalian, China). For the qRT-PCR analysis, the sad1 gene was used as an internal control, and the relative quantification method was used to assess the fold changes in the target genes. Three technical replicates were performed using one biological sample. PCR was performed for 30 s at 95 °C, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. All primers are listed in Table S1.
4.3 Precise Identification of Transcription Start Sites in Ghrack1 and Ghuhrf1 by 5′-Rapid Amplification of cDNA Ends (5'-RACE)
Total RNA was extracted from the leaves of upland cotton K312 using an RNAprep pure plant kit (TIANGEN, Beijing, China), following the manufacturer’s instructions. Total RNA was treated with calf intestinalphosphatase (CIP) to remove the 5′ phosphate from partial transcripts. Dephosphory RNA was treated with tobacco acid pyrophosphatase (TAP), which removed the 5′ cap from capped mRNA and exposed the 5′ phosphate. The GeneRacer™ RNA Oligo (Thermo Fisher Scientific, MA, USA) was ligated to the TAP-treated mRNA with T4 RNA ligase. A cDNA template was generated by reverse transcription using SuperScript™ II RT and either the GeneRacer™ Oligo dT Primer or the gene-specific primer (GSP1 and GSP2). 5′ ends were PCR-amplified from these cDNA templates with a primer for the GeneRacer™ RNA Oligo (GeneRacer™ 5′ Primer) and gene-specific primer. Only cDNA containing the GeneRacer™ RNA Oligo sequence were amplified [36,37].
4.4. Cloning and Sequence Analysis of the Intergenic Region between Ghrack1 and Ghuhrf1
The primer pairs GhZU-F and GhZU-R were designed according to the DNA sequence between the rack1 and uhrf1 genes in the Gossypium hirsutum genome. The intergenic region between Ghrack1 and Ghuhrf1 was amplified using genome DNA from Gossypium hirsutum K312 as a template and GhZU-F/GhZU-R as primers. The PCR amplification with Phanta Max Super-Fidelity DNA Polymerase was performed according to the manufacturer’s instructions (Vazyme, Nanjing, China), under the following reaction conditions: 5 min at 95 °C, followed by 30 cycles of 30 s at 95 °C, 30 s at 58 °C, 2 min at 72 °C, and 10 min at 72 °C. The amplified DNA fragment was sequenced and analyzed using the software DNAman, PlantCARE [38], PLACE [39], and CpGPlot/CpGReport/Isochore.
4.5. Construction of Plant Expression Vectors
The target DNA fragments of the forward orientation promoter GhZUf and reverse orientation promoter GhZUr were cloned using the corresponding primers and template shown in Table S1. GhZUr-gfp was amplified by nested PCR, using previous PCR products of GhZUr and gfp as templates and GhZUr-F/gfp-R as primers. All target DNA fragments were ligated into the cloning vector pMD18-T (TaKaRa, Dalian, China) and sequenced.
The target DNA fragments of GhZUf, GhZUr, gus, and gfp were inserted between the XbaI, NcoI, and SphI sites of the plant expression vector pCambia1305.1 (Figure S5) using a homologous recombinant enzyme (TaKaRa, Dalian, China) to form the vector GhZUf::gus, GhZUr::gfp, gfp:GhZU::gus, and gfp::gus, respectively.
4.6. Transient Expression in Tobacco and Stable Expression in Arabidopsis
The final plant expression vectors were mobilized into the Agrobacterium tumefaciens strain GV3101 by using the freeze-thaw method. First, we transformed the vector into tobacco for the transient expression experiments. The transformed bacteria were grown on YEB medium containing 50 mg/L kanamycin, 100 mg/L rifampicin at 28 °C, and 220 rpm overnight. The cultures were diluted 1:100 with YEB and allowed to grow to an absorbance (measured at 600 nm) of <0.8. The young leaf epidermis nearest to the top of Nicotiana benthamiana at approximately the 6–8 leaf stage was used for infection with A. tumefaciens. The infection was stopped when at least two-thirds of the target leaf was consumed by A. tumefaciens. After the infection, the plants were cultured at a high humidity for 12 to 16 h in the dark, and then cultured under normal conditions for 3 days. Then, we transformed the vector into Arabidopsis for the stable expression experiments using a standard floral-dipping method [40]. The transformants were selected on MS medium supplemented with 50 mg/L kanamycin. Transgenesis was confirmed via PCR using GhZU-F/R. Transgene homozygotes selected from the T3 generation were used for the analysis of gfp and gus expression.
4.7. Detection and Copy Number Analysis of Transgenic Arabidopsis-Positive Plants
The gDNA was extracted from different plants using a DNAsecure plant kit (TIANGEN, Beijing), following the manufacturer’s instructions. These gDNA were used as templates and GUS-JC-F/GUS-JC-R as primers. The PCR amplification with Phanta Max Super-Fidelity DNA Polymerase was performed according to the manufacturer’s instructions (Vazyme, Nanjing, China) under the following reaction conditions: 5 min at 95 °C, followed by 30 cycles of 30 s at 95 °C, 30 s at 58 °C, 1 min at 72 °C, and 5 min at 72 °C.
The single-copy gene RG (AT1G03400) of Arabidopsis thaliana was selected as the internal reference gene, while the corresponding primers and probes were designated as RG-F/R, and the RG-probe fluorescent markers were designated as FAM (6-carboxy-fluorescein). The target gene probes and primers, including the GUS-F/R, GUS-probe, and fluorescently labeled HEX (hexachloro fluorescein), were designed using primer premier 5.0 software. A 20-μL droplet digital PCR probe reaction system was prepared, mixed well, and added to a microdroplet generator. Then, a 70-μL droplet of oil was added to the corresponding wells, which were covered with special pads and placed in a droplet generator. The oil droplets were transferred to a 96-well plate and placed with the red-labeled side of the membrane facing up, which was fixed and sealed with a heat sealer. The procedure consisted of an incubation at 180 °C for 10 s. Each reaction was performed in triplicate on a QX200 (BIO-RAD). The rate at which the PCR device raised and lowered the temperature was controlled within the range of ≤2.5 °C/s. The reaction conditions consisted of predenaturation at 94 °C for 10 min, denaturation at 94°C for 30 s, annealing at 62 °C for 60 s for 40 cycles, and incubation at 98 °C for 10 min. Then, the microdrop reader read the droplet signal and analyzed the experimental results using QuantaSoft software [41,42].
4.8. Histochemical GUS Assay and GFP Detection
The GUS assay was performed as described by Jefferson et al. [43]. Plant tissues or leaf sections were soaked in X-gluc solution, sealed closely to avoid evaporation, incubated overnight at 37 °C in the dark, and fixed in a formalin-isopropyl alcohol-glacial acetic acid (FAA) solution for 15 min. To remove chlorophyll, the fixed leaf sections were rinsed successively in 75%, 85%, 95%, and 100% ethanol.
For GFP detection, samples from various stages of plant development were placed on a slide glass and covered with a coverslip [11]. The laser-scanning confocal microscope Axio LSM 700 (Zeiss Co., Ltd., Jena, Germany) was used for the observations.
4.9. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) in Various Transgenic Arabidopsis Tissues
The total RNA was extracted from different plant tissues using an RNAprep pure plant kit (TIANGEN, Beijing, China) following the manufacturer’s instructions. For the quantitative measurements of the gfp transcript, the primer pair of q-gfp-F/R were used. For the quantitative measurements of the gus transcript using q-gus-F/R, the relative transcript abundance was assessed by qRT-PCR following a published protocol [35]. PCR was performed in four replicates and repeated in three biological samples. The transcript levels were measured following the comparative Ct method (Applied Biosystems bulletin, MA, USA). To normalize the amount of total RNA in all Arabidopsis samples, the actin gene-specific forward primer q-actin-F and reverse primer q-actin-R were used [44].
4.10. SDS-Polyacrylamide Gel Electrophoresis and Immunoblot Analysis
SDS-polyacrylamide gel electrophoresis was performed using 10% polyacrylamide gels, as previously described [45]. 40 micrograms of protein from young leaf, older leaf, flower, root, developing silique, and mature silique tissues from transgenic Arabidopsis plants were subjected to 10% SDS-polyacrylamide gel electrophoresis for the western blot. The Rubisco large subunit (LSU) was stained with Ponceau S as an internal control for loading uniformity.
For the determination of GFP expression in different tissues, a western blot analysis was performed using an anti-GFP tag mouse monoclonal antibody (Plant Specific) (1:3000–5000) from Beijing Emarbio Science & Technology Co., Ltd., as well as a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5000–10,000), and detected using a chemiluminescent reagent (abm, Vancouver, BC, Canada) following a published protocol [46].
For the determination of GUS expression in different tissues, a western blot analysis was conducted using an anti-GUS tag rabbit polyclonal antibody from Agrisera antibodies (Agrisera, Vannas, Swedish) (1:10,000) and a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:5000–10,000), and detected using a chemiluminescent reagent, following a published protocol.
Acknowledgments
This work was supported by grants from the Major Project of China on New Varieties of GMO Cultivation (2016zx08005-003).
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/11/3291/s1.
Author Contributions
Conception and design: J.Y., X.W. and Z.W. Writing the manuscript: J.Y. and X.W. Conceptualization and preparation of figures and tables: J.Y., X.W., A.H. and Z.W. All authors participated in the design and critical reviewing of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.James C. Global Status of Commercialized Biotech/GM Crops, 2017. Volume 36. International Service for the Acquisition of Agri-Biotech Applications; New York, NY, USA: 2018. pp. 1–8. [Google Scholar]
- 2.Kumari M., Rai A.K., Devanna B.N., Singh P.K., Kapoor R., Rajashekara H., Prakash G., Sharma V., Sharma T.R. Co-transformation mediated stacking of blast resistance genes Pi54 and Pi54rh in rice provides broad spectrum resistance against Magnaporthe oryzae. Plant Cell Rep. 2017;36:1747–1755. doi: 10.1007/s00299-017-2189-x. [DOI] [PubMed] [Google Scholar]
- 3.Shockey J., Mason C., Gilbert M., Cao H., Li X., Cahoon E., Dyer J. Development and analysis of a highly flexible multi-gene expression system for metabolic engineering in Arabidopsis seeds and other plant tissues. Plant Mol. Biol. 2015;89:113–126. doi: 10.1007/s11103-015-0355-5. [DOI] [PubMed] [Google Scholar]
- 4.Flavell R.B. Inactivation of gene expression in plants as a consequence of specific sequence duplication. Proc. Natl. Acad. Sci. USA. 1994;9:3490–3496. doi: 10.1073/pnas.91.9.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trinklein N.D., Aldred S.F., Hartman S.J., Schroeder D.I., Otillar R.P., Myers R.M. An abundance of bidirectional promoters in the human genome. Genome Res. 2004;14:62–66. doi: 10.1101/gr.1982804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermsen R., ten Wolde P.R., Teichmann S. Chance and necessity in chromosomal gene distributions. Trends Genet. 2008;24:216–219. doi: 10.1016/j.tig.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Wei W., Pelechano V., Jarvelin A.I., Steinmetz L.M. Functional consequences of bidirectional promoters. Trends Genet. 2011;27:267–276. doi: 10.1016/j.tig.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orekhova A.S., Rubtsov P.M. Bidirectional promoters in the transcription of mammalian genomes. Biochemistry (Biokhimiia) 2013;78:335–341. doi: 10.1134/S0006297913040020. [DOI] [PubMed] [Google Scholar]
- 9.Wakano C., Byun J.S., Di L.J., Gardner K. The dual lives of bidirectional promoters. Biochim. Biophys. Acta. 2012;1819:688–693. doi: 10.1016/j.bbagrm.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitra A., Han J.G., Zhang Z.Y.J., Mitra A. The intergenic region of Arabidopsis thaliana cab1 and cab2 divergent genes functions as a bidirectional promoter. Planta. 2009;229:1015–1022. doi: 10.1007/s00425-008-0859-1. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee J., Sahoo D.K., Dey N., Houtz R.L., Maiti I.B. An Intergenic Region Shared by At4g35985 and At4g35987 in Arabidopsis thaliana Is a Tissue Specific and Stress Inducible Bidirectional Promoter Analyzed in Transgenic Arabidopsis and Tobacco Plants. PLoS ONE. 2013;8:e79622. doi: 10.1371/journal.pone.0079622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang R., Yan Y., Zhu M., Yang M., Zhou F., Chen H., Lin Y. Isolation and Functional Characterization of Bidirectional Promoters in Rice. Front. Plant Sci. 2016;7:766. doi: 10.3389/fpls.2016.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Wan L., Li D., Zhu L., Qian M., Deng M. Searching for bidirectional promoters in Arabidopsis thaliana. BMC Bioinform. 2009;10(Suppl. 1):S29. doi: 10.1186/1471-2105-10-S1-S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krom N., Ramakrishna W. Comparative analysis of divergent and convergent gene pairs and their expression patterns in rice, Arabidopsis, and Populus. Plant Physiol. 2008;147:1763–1773. doi: 10.1104/pp.108.122416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhadi S.R., Krom N., Ramakrishna W. Genome-wide comparative analysis of putative bidirectional promoters from rice, Arabidopsis and Populus. Gene. 2009;429:65–73. doi: 10.1016/j.gene.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 16.Shwarz Z., Jolly S.O., Steinmetz A.A. Overlapping divergent genes in maize chloroplast chromosome and in vitro transcription of the gene for tRNAHIS. Proc. Natl. Acad. Sci. USA. 1981;78:3423–3428. doi: 10.1073/pnas.78.6.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin Y., Kim M.J., Paek K.H. The CaTin1 (Capsicum annuum TMV-induced clone 1) and CaTin1-2 genes are linked head-to-head and share a bidirectional promoter. Plant Cell Physiol. 2003;44:549–554. doi: 10.1093/pcp/pcg069. [DOI] [PubMed] [Google Scholar]
- 18.Liu X.Q., Zhou X.J., Li Y., Tian J., Zhang Q.X., Li S.Z., Wang L., Zhao J., Chen R.M., Fan Y.L. Identification and functional characterization of bidirectional gene pairs and their intergenic regions in maize. BMC Genom. 2014;15:338. doi: 10.1186/1471-2164-15-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra R.C., Grover A. Intergenic Sequence between Arabidopsis Caseinolytic Protease B-Cytoplasmic/Heat Shock Protein100 and Choline Kinase Genes Functions as a Heat-Inducible Bidirectional Promoter. Plant Physiol. 2014;166:1646–1658. doi: 10.1104/pp.114.250787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu S.J., Yue Q.J., Zhang W. Structural and functional analysis of an asymmetric bidirectional promoter in Arabidopsis thaliana. J. Integr. Plant Biol. 2015;57:162–170. doi: 10.1111/jipb.12308. [DOI] [PubMed] [Google Scholar]
- 21.Collier R., Dasgupta K., Xing Y.P., Hernandez B.T., Shao M., Rohozinski D., Kovak E., Lin J., de Oliveira M.L.P., Stover E., et al. Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. 2017;90:1014–1025. doi: 10.1111/tpj.13517. [DOI] [PubMed] [Google Scholar]
- 22.Sun Y., Joyce P.A. Application of droplet digital PCR to determine copy number of endogenous genes and transgenes in sugarcane. Plant Cell Rep. 2017;36:1775–1783. doi: 10.1007/s00299-017-2193-1. [DOI] [PubMed] [Google Scholar]
- 23.Xu X., Peng C., Wang X., Chen X., Wang Q., Xu J. Comparison of droplet digital PCR with quantitative real-time PCR for determination of zygosity in transgenic maize. Transgenic Res. 2016;25:855–864. doi: 10.1007/s11248-016-9982-0. [DOI] [PubMed] [Google Scholar]
- 24.Glowacka K., Kromdijk J., Leonelli L., Niyogi K.K., Clemente T.E., Long S.P. An evaluation of new and established methods to determine T-DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 2016;39:908–917. doi: 10.1111/pce.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wray G.A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 26.He F., Chen W.H., Collins S., Acquisti C., Goebel U., Ramos-Onsins S., Lercher M.J., de Meaux J. Assessing the Influence of Adjacent Gene Orientation on the Evolution of Gene Upstream Regions in Arabidopsis thaliana. Genetics. 2010;185:695–701. doi: 10.1534/genetics.110.114629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue M., Long Y., Zhao Z., Huang G., Huang K., Zhang T., Jiang Y., Yuan Q., Pei X. Isolation and Characterization of a Green-Tissue Promoter from Common Wild Rice (Oryza rufipogon Griff.) Int. J. Mol. Sci. 2018;19:2009. doi: 10.3390/ijms19072009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo Y.H., Li W.H. Gene clustering pattern, promoter architecture, and gene expression stability in eukaryotic genomes. Proc. Natl. Acad. Sci. USA. 2011;108:3306–3311. doi: 10.1073/pnas.1100210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh A., Sahi C., Grover A. Chymotrypsin protease inhibitor gene family in rice: Genomic organization and evidence for the presence of a bidirectional promoter shared between two chymotrypsin protease inhibitor genes. Gene. 2009;428:9–19. doi: 10.1016/j.gene.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 30.Yang M.Q., Elnitski L. Diversity of core promoter elements comprising human bidirectional promoters. BMC Genom. 2008;9(Suppl. 2):S3. doi: 10.1186/1471-2164-9-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venter M. Synthetic promoters: Genetic control through cis engineering. Trends Plant Sci. 2007;3:118–124. doi: 10.1016/j.tplants.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Hao J., Tu L., Hu H., Tan J., Deng F., Tang W., Nie Y., Zhang X. GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 2012;63:6267–6281. doi: 10.1093/jxb/ers278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X.Q., Yang W.Z., Mu B.N., Li S.Z., Li Y., Zhou X.J., Zhang C.Y., Fan Y.L., Chen R.M. Engineering of ‘Purple Embryo Maize’ with a multigene expression system derived from a bidirectional promoter and self-cleaving 2A peptides. Plant Biotechnol. J. 2018;16:1107–1109. doi: 10.1111/pbi.12883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang L., Chen J., Huang C., Liu Y., Jia S., Pan L., Zhang D. Validation of a cotton-specific gene, Sad1, used as an endogenous reference gene in qualitative and real-time quantitative PCR detection of transgenic cottons. Plant Cell Rep. 2005;24:237–245. doi: 10.1007/s00299-005-0929-9. [DOI] [PubMed] [Google Scholar]
- 35.Suttipanta N., Pattanaik S., Kulshrestha M., Patra B., Singh S.K., Yuan L. The Transcription Factor CrWRKY1 Positively Regulates the Terpenoid Indole Alkaloid Biosynthesis in Catharanthus roseus. Plant Physiol. 2011;157:2081–2093. doi: 10.1104/pp.111.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matteau D., Rodrigue S. Precise Identification of Genome-Wide Transcription Start Sites in Bacteria by 5′-Rapid Amplification of cDNA Ends (5′-RACE) Methods Mol. Biol. 2015;1334:143–159. doi: 10.1007/978-1-4939-2877-4_9. [DOI] [PubMed] [Google Scholar]
- 37.Fateh R., Zaini F., Kordbacheh P., Falahati M., Rezaie S., Ghazvini R.D., Borhani N., Safara M., Fattahi A., Kanani A., et al. Identification and Sequencing of Candida krusei Aconitate Hydratase Gene Using Rapid Amplification of cDNA Ends Method and Phylogenetic Analysis. Jundishapur J. Microbiol. 2015;8:e25218. doi: 10.5812/jjm.25218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lescot M., Dehais P., Thijs G., Marchal K., Moreau Y., Van de Peer Y., Rouze P., Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higo K., Ugawa Y., Iwamoto M., Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Gao F., Pfeifer E., Farah H., Karampini E., Dua D., Kamal N., Cane P., Tobal K., Sethi T., Spicer J., et al. Microdroplet digital PCR: Detection and quantitation of biomarkers in archived tissue and serial plasma samples in patients with lung cancer. J. Thorac. Oncol. 2015;10:212–217. doi: 10.1097/JTO.0000000000000349. [DOI] [PubMed] [Google Scholar]
- 42.Zhu P., Wang C., Huang K., Luo Y., Xu W. A Novel Pretreatment-Free Duplex Chamber Digital PCR Detection System for the Absolute Quantitation of GMO Samples. Int. J. Mol. Sci. 2016;17:402. doi: 10.3390/ijms17030402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jefferson R.A., Kavanagh T.A., Bevan M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;13:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Remans T., Smeets K., Opdenakker K., Mathijsen D., Vangronsveld J., Cuypers A. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta. 2008;227:1343–1349. doi: 10.1007/s00425-008-0706-4. [DOI] [PubMed] [Google Scholar]
- 45.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Sahoo D.K., Stork J., DeBolt S., Maiti I.B. Manipulating cellulose biosynthesis by expression of mutant Arabidopsis proM24::CESA3(ixr1-2) gene in transgenic tobacco. Plant Biotechnol. J. 2013;11:362–372. doi: 10.1111/pbi.12024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.