Abstract
Roots are important plant ground organs, which absorb water and nutrients to control plant growth and development. Phytohormones have been known to play a crucial role in the regulation of root growth, such as auxin and ethylene, which are central regulators of this process. Recent findings have revealed that root development and elongation regulated by ethylene are auxin dependent through alterations of auxin biosynthesis, transport and signaling. In this review, we focus on the recent advances in the study of auxin and auxin–ethylene crosstalk in plant root development, demonstrating that auxin and ethylene act synergistically to control primary root and root hair growth, but function antagonistically in lateral root formation. Moreover, ethylene modulates auxin biosynthesis, transport and signaling to fine-tune root growth and development. Thus, this review steps up the understanding of the regulation of auxin and ethylene in root growth.
Keywords: auxin, ethylene, phytohormones, root development, crosstalk
1. Introduction
Roots are important plant ground organs, which are crucial for plant survival and performs a wide range of functions, such as absorbing water and nutrients, supporting the plant body and interactions with soil microbiota. Generally, the root system consists of two principal root types: the primary root (PR), which is initiated during embryo development [1] and secondary roots, which form post-embryonically. These secondary roots encompass both lateral roots (LR), which develop as branches of the primary root and adventitious roots (AR), which develop on non-root tissue such as the hypocotyl, stems and leaves [2]. The root architecture of monocots and dicots is highly distinct. The dicotyledonous plant has a tap-root system with a central primary root and lateral roots, whereas monocotyledonous plants, such as rice (Oryza sativa) and maize (Zea mays), have fibrous root systems composed of a primary root, lateral roots and crown roots (also known as adventitious roots) [3].
In higher plants, root growth is maintained by coordinating cell proliferation and differentiation [4,5]. Phytohormones have been known to play a crucial role in the regulation of root growth [6,7,8,9]. Recent studies in Arabidopsis root have shown that different hormones control organ growth by regulating specific growth processes such as cell proliferation, differentiation or expansion in distinct tissues [9,10,11]. Plant hormones such as auxin and ethylene have been shown to be involved in root growth through a range of complex interactions [8].
Indole-3-acetic acid (IAA), the main auxin in plants, regulates almost every aspect of plant growth and development [12]. Genetic and biochemical studies indicated that tryptophan (Trp) is the main precursor for IAA in plants [13]. There are three pathways for IAA biosynthesis from Trp in plants: the indole-3-pyruvic acid (IPyA) pathway, the indole-3-acetamide pathway and the indole-3-acetaldoxime pathway [14]. The IPyA pathway has been proposed as the most important pathway to produce auxin in plant [15,16,17]. Once auxin is produced, it is perceived by TRANSPORT INHIBITOR RESPONSE 1 (TIR1) and AUXIN SIGNALING F-BOX PROTEIN (AFB). IAA directly interacts with the F-box protein TIR1 and promotes the degradation of the Aux/IAA transcriptional repressors to activate diverse auxin responsive genes [18,19]. Recent studies have shown that auxin biosynthesis, transport and auxin-dependent signaling processes all affect root development [13,20].
Ethylene is a simple and very important gaseous phytohormone that modulates multiple plant growth and development processes [21]. Ethylene is synthesized from methionine via a simple linear pathway, in which 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) and ACC oxidases (ACO) function as key enzymes [22]. Ethylene is sensed by a family of endoplasmic reticulum-located receptors, which are negative regulators of the signaling pathway [23,24]. In the absence of ethylene, the active receptors recruit a Raf-like protein kinase, CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1), to phosphorylate the C-terminal domain of ETHYLENE INSENSITIVE 2 (EIN2), thus causing the proteasome degradation of EIN2 by F-box proteins and repression of the downstream ethylene response [25,26,27]. In the presence of ethylene, ethylene binding to the receptors inhibits the interaction with CTR1, resulting in that CTR1 cannot phosphorylate EIN2. Unphosphorylated EIN2 is cleaved by unknown proteases and the EIN2 C-terminus translocate into the nucleus [25,28,29] or into the P-body [30,31]. In the P-body, the EIN2 C-terminus mediates translational repression of EIN3-BINDING F-BOX 1 (EBF1) and EBF2 [30,31,32]. In the nucleus, the EIN2 C-terminus transduces signals to the transcription factors EIN3 and EIN3-LIKE1 (EIL1), which are sufficient and necessary for activation of many ethylene-response genes. These changes ultimately cause different physiological responses [33,34,35]. Due to the pivotal regulation of ethylene and auxin in root growth, in which the ethylene elevates auxin accumulation and to trigger TIR1/AFB2-mediated signal transduction [36,37,38,39], here in this review we will focus our interest on the recent advances of ethylene, auxin and their crosstalk during root development, providing a different angle for analyzing the mechanisms of plant root development.
2. The Coordination of Ethylene and Auxin in Primary Root Growth
The primary root, initiated during embryo development, develops shortly after germination and is a fundamental part of the root system that absorbs mineral nutrients and provides mechanical support for shoot growth [1]. Plant root is characterized by a series of developmental zones: the meristematic zone (MZ), transition zone (TZ), elongation zone (EZ) and growth terminating zone (GTZ) [40,41]. In the center of the root tip there is a quiescent center (QC), which is mitotically inactive and functions as organizer of the root stem cell niche. [42].
Auxin is known to exert an inhibitory role on primary root growth. An auxin gradient, established by local auxin biosynthesis and transport, is important for primary root growth. Auxin synthesized in roots via the IPyA pathway is crucial in normal root elongation and root gravitropic responses [43]. Disruption of YUC genes and/or TAA genes can cause moderate to very severe root defects in Arabidopsis. For example, the taa1 tar1 tar2 triple mutant in Arabidopsis fails to make root meristem during embryogenesis [44]; similar phenotypes were observed in yuc1 yuc4 yuc10 yuc11 quadruple mutant as well [45]. But yuc3 yuc5 yuc7 yuc8 yuc9 quintuple mutant (yucQ) displays very short primary roots and agravitropic root growth. Histological analysis showed that yucQ had much smaller meristems, with enlarged cells [43]. In addition, overexpressing any member of the YUC family in Arabidopsis leads to primary root growth inhibition [15,43,46], indicating that basal levels of endogenous auxin are required to maintain normal root growth. In rice, disruption of the FISH BONE gene, an orthologue of TAA1, displays pleotropic phenotypes including agravitropic roots, long primary roots, few crown roots and a lack of lateral roots [47]. Overexpression of OsYUC genes enhances the development of crown roots, shortened primary roots and over proliferation of root hairs [48,49]. Knock-down OsYUC1 expression results in the inhibition of root formation and elongation [48]. In woodland strawberry (Fragaria vesca L.), silencing of YUCCA6 affected various post-embryonic organ developmental steps including root formation [50]. All these results evidence that auxin synthesized by the IPyA pathway plays important roles in primary root development.
Inhibition of root growth is also one of the characteristic effects of ethylene or its precursor ACC. In Arabidopsis, exogenous application of ethylene or ACC inhibits the primary root elongation [51]. Mutants of ETHYLENE OVERPRODUCER (eto1) and ctr1, with an enhanced ethylene biosynthesis or signaling, respectively, exhibit short primary roots [52,53,54]. In contrast, the inhibitory effect of ethylene on primary root growth is abolished in ethylene insensitivity mutants, such as ETHYLENE RESISTANT 1 (etr1), ein2, ein3 eil1 or in the presence of ethylene inhibitors, such as the biosynthesis inhibitor AVG [51] and pyrazinamide (PZA) [55], the perception inhibitor silver nitrate (AgNO3) [51] and 1-methylcyclopropene (1-MCP) [56]. In rice, ethylene inhibits primary root elongation [39,57,58]. Loss-of-function mutation of rice ethylene receptors shows significant shorter primary roots and a moderately enhanced ethylene response [6,59,60]. By screening rice ethylene response mutants, MHZ7/OsEIN2 and MHZ6/OsEIL1 have been identified [57,61]. Mutation of OsEIN2/OsEIL1 leads to completely insensitive to ethylene in primary root growth, OsEIN2/OsEIL1-overexpression rice seedlings showed very short and extremely twisted primary roots [57,61]. MHZ3, positively regulates the ethylene signaling by interacting with the OsEIN2 Nramp-like domain and regulating the stability of the OsEIN2 protein [62]. Loss-of-function mutation of MHZ3 leads to ethylene insensitivity in etiolated rice seedlings, whereas MHZ3 overexpression lines exhibited slightly but significantly longer coleoptiles and shorter primary roots when grown in the air [62]. In other monocotyledonous plants, such as maize, wheat, sorghum and B. distachyon, ethylene treatment also inhibits the primary root growth [58].
Ethylene affects root growth at two different levels. First, ethylene inhibits cell proliferation in the root apical meristem [11]. Second, ethylene inhibits cell expansion in the root elongation zone [51,63]. The reports of ethylene effects on cell division in roots are conflicting. Ruzicka et al. [51] showed that ethylene affects root growth primarily by regulating the elongation of cells that leave the root meristem but without impact on the root meristem size. Recent studies showed that ethylene negatively regulates root meristem size through inhibiting cell proliferation in the root meristem [11]. Through a targeted expression approach to map the tissue sites of ethylene growth regulation found that the epidermis is the main site of ethylene action controlling plant growth in both roots and shoots [10]. Taken together, ethylene inhibits primary root growth by inhibiting cell proliferation in meristem zone and cell elongation in elongation zone.
The auxin–ethylene crosstalk during root growth has been proposed based on several studies [36,37]. In Arabidopsis, ethylene application promotes the expression of IAA biosynthetic genes and IAA levels [44,51,64]. In ethylene-treated seedlings, an overall increase of auxin response at the root tip was observed and this is also reflected in direct auxin measurements [36,51]. By screening for mutants that display ethylene defects only in roots, some WEAK ETHYLENE INSENSITIVE (WEI) genes were identified, such as WEI2/ASA1, WEI7/ASB1 and WEI8/TAA1 [44,64]. WEI2/ASA1 and WEI7/ASB1 encode subunits of anthranilate synthase, a rate-limiting enzyme in Trp biosynthesis. Upregulation of WEI2/ASA1 and WEI7/ASB1 by ethylene results in the accumulation of auxin in the tip of primary root, whereas loss-of-function mutations in these genes prevent the ethylene-induced auxin increase [64]. WEI8/TAA1 encodes a long-anticipated tryptophan aminotransferase, a key enzyme in the IPyA pathway, catalyzes the conversion Trp to IPA. Analysis of WEI8/TAA1 and its paralogues revealed a link between local auxin production and tissue-specific ethylene effects [44]. The same phenotypes were also observed in simultaneously inactivated YUC mutant roots, such as yucQ. YUC catalyzes the conversion IPA to IAA, a rate-limiting step in the IPyA pathway [17]. These mutants underscore the link between ethylene signaling and auxin biosynthesis. However, it is not clear how ethylene signal is transmitted to auxin. Recent studies have shown that several transcription factors in the ethylene signaling pathway, such as EIN3, ETHYLENE RESPONSE FACTOR 1 (ERF1) and PHYTOCHROME INTERACTING FACTOR 4 (PIF4), function as crosstalk nodes between ethylene and auxin in root growth [65,66,67,68]. Particularly, PIF4 affects auxin-mediated growth by directly controlling the expression of TAA1 and YUC genes [65,66]. EIN3 directly binding to the YUC9 promoter to regulate auxin accumulation in root transition zone and root growth inhibition [67]. ERF1 binds the promoter of ASA1 in order to regulate auxin biosynthesis and ethylene-induced root growth inhibition [68]. Another interaction between ethylene and auxin biosynthesis was discovered in a chemical genetic strategy, using l-kynurenine, a small molecule that represses nuclear accumulation of the EIN3 transcription factor and TAA/TAR was identified as a target for l-kyn, to decrease ethylene-induced auxin biosynthesis and ethylene responses in roots [69]. Using yucasin [5-(4-chlorophenyl)-4H-1,2,4-triazole-3-thiol], an inhibitor of YUC activity, suppresses ethylene-inhibited root growth [39,70,71]. Recent studies in rice showed that exogenous application of yucasin largely recovered the short and coiled primary root phenotype of OsEIN2 and OsEIL1 overexpression transgenic lines and OsEIL1 directly activates the expression of OsYUC8 to modulate auxin biosynthesis and ethylene-inhibited primary root elongation [39].
In addition to auxin biosynthesis mutants, mutants related to auxin transport, perception and signaling also display abnormal responses to ethylene [37,51,64,72]. In Arabidopsis, mutants of auxin receptor tir1 and auxin response AUXIN RESISTANT axr2-1 and axr3-1, mutants in IAA7 and IAA17, respectively, were shown an ethylene-resistance root phenotype [37,51]. In rice, soil-surface rooting 1 (SOR1), which is a RING finger E3 ubiquitin ligase, controls root-specific ethylene responses by modulating Aux/IAA protein stability [38]. In conclusion, ethylene-regulated primary root growth requires auxin biosynthesis, perception, transport, or signaling.
3. The Integration of Ethylene and Auxin in Lateral Root Growth
Lateral roots are an essential component of root system architecture that maximizes the ability of the root system to acquire nutrients, therefore enabling the plant to adapt readily to the changes of the environment cues [73,74]. Lateral root development is initiated from asymmetric divisions of the pericycle founder cell of primary root and the whole process of development has been well established [75,76].
It is well known that the phytohormone auxin is involved in every stage of lateral root formation [75,77]. Endogenous auxin biosynthesis, polar auxin transport and auxin-dependent signaling processes all affect lateral root formation [78,79,80,81]. In Arabidopsis roots, gain-of-function yuc mutant (yuc1D), superroot1 (sur1) and sur2 showed an increase in endogenous auxin levels and lateral root number [46,82]. The mutants with defects in auxin transport, such as auxin resistant 1 (aux1), like aux1 3 (lax3), pin-formed (pin) multiple mutants, gnom and auxin resistant 4 (axr4), have reduced lateral root number or disturbed lateral root primodium [82]. The rice osaux1 mutant has a reduced number of lateral roots, accompanied by decreased endogenous auxin levels [79,83,84]. Furthermore, an elevated auxin level causes degradation of Aux/IAAs transcriptional regulators and releases ARFs [18,19], which promote lateral root proliferation [82,85]. A series of Arabidopsis gain-of-function Aux/IAA mutants, such as iaa1, iaa3, iaa14/solitaryroot 1 (slr1), iaa18, iaa19/massugu 2 (msg2), iaa28 [82,86] and osiaa11, osiaa13 and osiaa23 from rice, showed aberrant lateral roots, reduced lateral root number, or even complete absence of lateral root formation [87,88,89]. Accordingly, ARF mutants, such as arf7 arf19 double mutant, showed complete absence of lateral root formation [82].
Ethylene also plays an important role during the initiation and growth of lateral roots. For reviews of ethylene effects on lateral roots development, we refer to [90]. Treatment with ethylene or ACC reduces lateral root initiation in both Arabidopsis and tomato [91,92]. Similarly, enhanced ethylene synthesis (in eto1 mutant) or signaling (in ctr1 mutant) reduced lateral root formation [91]. By contrast, ethylene-insensitive mutants (etr1 and ein2), form an increased number of lateral roots [91], suggesting that ethylene exerts an inhibitory effect on lateral root formation.
Auxin and ethylene have antagonistic effects in lateral root development. Ethylene reduces DR5erv:GFP expression in the regions where lateral roots emerge, suggesting a locally reduced auxin responsiveness [93]. Mutants in auxin influx and efflux proteins, such as aux1, lax3, pin3 and pin7, were less sensitive to the inhibition of lateral root formation and stimulation of auxin transport following treatment with ACC [91,93]. By contrast, pin2 and abcb19 mutants exhibited normal responses to ACC [93]. Arabidopsis seedlings show a local depletion of PIN3 and PIN7 abundance in the region just below developing lateral root primordia, resulting in a local auxin accumulation at the lateral root primordia, thus promoting the formation of lateral roots. ACC treatment increased the abundance of transcripts PIN3 and PIN7, thus increasing rootward auxin transport, which suppresses the local auxin maxima at lateral root primordia, inhibiting their outgrowth [93,94].
4. The Role of Ethylene and Auxin in Adventitious Root Growth
Adventitious root, also known as crown root, constituting the major part of the monocotyledonous root system, emerges from any non-root tissue and is produced both during normal development and in response to stress conditions, such as flooding, nutrient deprivation and wounding during the growing period [95].
Similar to other development processes, adventitious root formation is highly dependent on auxin action [2]. Overexpression YUCCA genes in rice cause massive proliferation of crown roots. On the other hand, disruption of TAA1 greatly reduces crown root development [47,49]. Many genes essential for crown root development in rice are involved in the auxin-signaling pathway. For example, CROWNLESS ROOT 1 (CRL1)/ADVENTITIOUS ROOT LESS 1 (ARL1) encodes an ASYMMETRIC LEAVES 2 (AS2)/LATERAL ORGAN BOUNDARIES (LOB) transcription factor, which is transcriptionally regulated by the AUXIN RESPONSE FACTOR 16 (ARF16), acts as a positive regulator for crown root formation [96,97]. CRL4/OsGNOM1 plays an important role in crown root emergence by its influence on the polar localization of the auxin efflux carrier PIN1 [98]. The crl5 mutant produced fewer crown roots and displayed impaired initiation of crown root primordia. Its expression can also be induced by exogenous auxin treatment and may be a direct target of an ARF [99]. CRL6 influences crown root formation by regulating primordial initiation and development. It was shown that the expression of OsIAA genes were down-regulated in crl6, linking CRL6 to auxin regulatory network [100]. The WUSCHEL-related Homeobox (WOX) gene, WOX11, is involved in the activation of crown root emergence and growth. Its expression is induced by auxin and functions downstream IPyA pathway [3,49,101,102,103]. These studies show that the auxin is essential for crown root development in rice.
Ethylene is another important phytohormone in root development through interacting with auxin. However, ethylene function in crown root development remains unknown. In rice, OsERF3, which encodes an AP2/ETHYLENE-RESPONSIVE FACTOR (ERF) protein, negatively regulates ethylene biosynthesis and act as a WOX11-interacting partner involved in rice crown root development [3,104]. Moreover, the expression of OsERF3 is induced by auxin and ACC treatment, suggesting a potential relationship between ethylene and auxin in crown root development. Further research should focus on the ethylene effect on crown root development and illuminating the underlying molecular mechanism.
5. The Contribution of Ethylene and Auxin in Root Hair Growth
Plant root hairs are unicellular extensions of root epidermal cells that increase the root surface to enhance water and nutrient uptake and improve soil anchorage. Multiple phytohormones, including auxin and ethylene, play vital roles in regulating cell development in the root epidermis [105,106]. Constitutive activation of ethylene signaling or exogenous application of ethylene or ACC promotes root hair elongation. By contrast, the root hairs of ethylene-insensitive mutants and seedlings treated with inhibitor of ethylene synthesis or signaling are significantly shorter [105,107,108]. Root hair elongation is enhanced by exogenous auxin treatment [51]. Similarly, enhanced auxin synthesis (in sur1 and yuc1D mutants) promoted root hair elongation [13,46]. Moreover, ethylene-signaling mutants and auxin-signaling mutants develop fewer and shorter root hairs [8]. These data strongly indicate that auxin and ethylene are both required for root hair elongation. Further studies found that auxin is able to rescue root hair defects in the ein2-1 mutant and mutant aux1 on an eto1 background suppressed the long-root-hair phenotype of eto1 mutant [109]. Recent studies demonstrated that several ARFs, which are central transcriptional regulators of auxin signaling, bind the ROOT HAIR DEFECTIVE 6-LIKE 4 (RSL4) promoter and directly activate its expression [110], thus providing the molecular link between auxin signaling and transcriptional control of root hair development [105]. RSL4 also involved in ethylene-promoted root hair growth, ethylene-activated transcription factor EIN3 physically interacts with ROOT HAIR DEFECTIVE 6 (RHD6), a well-documented positive regulator of hair cells and that the two factors directly co-activate RSL4 to promote root hair elongation [107]. However, whether ARFs and EIN3 act synergistically or competitively to activate RSL4 in root hair development is unclear.
6. Conclusions and Future Perspectives
Over the years, the crosstalk between ethylene and auxin in Arabidopsis root development has been elucidated. However, their interaction in other plants is still unclear. In this review, we summarize the recent advances in the study of auxin–ethylene crosstalk in plant root development (Figure 1), step up our understanding of the mechanisms of plant root development.
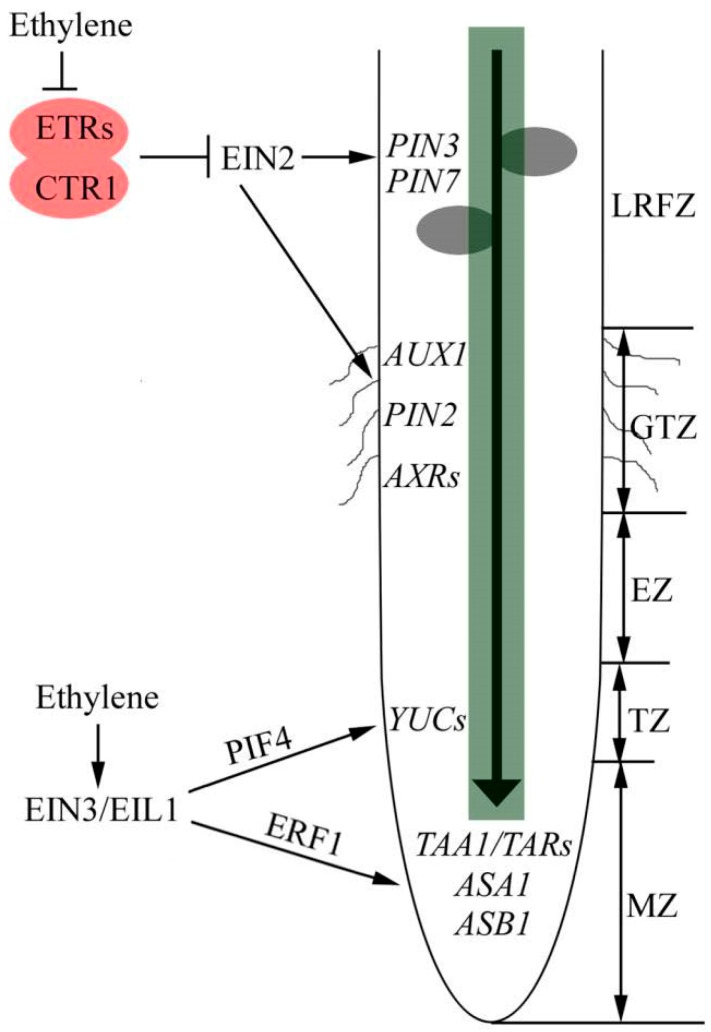
Figure 1.
Ethylene directs auxin to control root growth. The growth zones in root apex, including meristematic zone (MZ), transition zone (TZ), elongation zone (EZ) and growth terminating zone (GTZ). Ethylene promotes auxin biosynthesis in MZ and TZ through ASA1, ASB1, TAA1/TARs and YUCs, leading to inhibition of primary root elongation. ERF1 and PIF4 function as crosstalk nodes between ethylene and auxin in this process. In GTZ, ethylene promotes root hair initiation through modulation of the auxin levels mediated by auxin transporters. In the lateral root-forming zone (LRFZ), ethylene increases rootward auxin transport by increasing transcription and translation of PIN3 and PIN7 in the central cylinder, which prevent the localized accumulation of auxin needed to drive lateral root formation. Arrows indicates positive regulation, T sharp symbol indicates negative regulation, and waves in GTZ zone represent root hairs.
Plant root systems adapt rapidly to environmental changes and enable the plant to survive adverse conditions. In response to environmental cues, root growth is adapted through the modulation of endogenous auxin levels [111]. However, the underlying mechanisms of plants perceiving environmental changes and translating the cues into adaptive responses involved in the integrative regulation of auxin and ethylene in this process is largely unclear. Moreover, root growth is maintained by coordinating cell proliferation and differentiation [4,5]. Recent studies showed that epidermis is a key cell type required for ethylene-mediated growth inhibition [10]. Whether ethylene is required in multiple cell types for crosstalk with auxin or whether its effect is direct or indirect remains unknown. Cell type-specific promoters allow scientists to target the expression of a desired effector protein to particular cell types and reveal the necessity of the ethylene and auxin in a given cell type.
Ethylene and auxin are known to interact in the regulation of root growth. Several factors function as crosstalk nodes between ethylene and auxin have been identified in Arabidopsis, such as EIN3, ERF1 and PIF4 [65,66,67,68]. However, information in crops is limited. Moreover, hormonal crosstalk is not linear and should be tackled in a multidimensional space, sparking that the spatial and temporal relationships between the two hormones and the interdependent relationships with other hormones should be focused. The use of new chemicals known to target specific hormonal pathway components and the advent of high-throughput nucleotide sequencing techniques and CRISPR-CAS technology allowing generating specific mutants will easy to elucidate the underlying mechanisms of plant root development.
Author Contributions
H.Q. wrote the manuscript, designed the figures and edited, R.H. created the review’s outline and edited the manuscript.
Funding
This work was funded by [the Major Special Foundation of Transgenic Plants in China] grant number [2018ZX0800913B], [the National Natural Science Foundation of China] grant number [31871551 and 31801445], [the China Postdoctoral Science Foundation Grant] grant number [2017M620964 and 2018T110161] and [the Agricultural Sciences and Technology Innovation Mission] grant number [CAAS-XTCX2016].
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Zheng H., Pan X., Deng Y., Wu H., Liu P., Li X. AtOPR3 specifically inhibits primary root growth in Arabidopsis under phosphate deficiency. Sci. Rep. 2016;6:24778. doi: 10.1038/srep24778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellini C., Pacurar D.I., Perrone I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014;65:639–666. doi: 10.1146/annurev-arplant-050213-035645. [DOI] [PubMed] [Google Scholar]
- 3.Zhao Y., Cheng S.F., Song Y.L., Huang Y.L., Zhou S.L., Liu X.Y., Zhou D.X. The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell. 2015;27:2469–2483. doi: 10.1105/tpc.15.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji H.T., Wang S.F., Li K.X., Szakonyi D., Koncz C., Li X. PRL1 modulates root stem cell niche activity and meristem size through WOX5 and PLTs in Arabidopsis. Plant J. 2015;81:399–412. doi: 10.1111/tpj.12733. [DOI] [PubMed] [Google Scholar]
- 5.Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. A genetic framework for the control of cell division and differentiation in the root meristem. Science. 2008;322:1380–1384. doi: 10.1126/science.1164147. [DOI] [PubMed] [Google Scholar]
- 6.Ma B., Yin C.C., He S.J., Lu X., Zhang W.K., Lu T.G., Chen S.Y., Zhang J.S. Ethylene-induced inhibition of root growth requires abscisic acid function in rice (Oryza sativa L.) seedlings. PLoS Genet. 2014;10:e1004701. doi: 10.1371/journal.pgen.1004701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv B., Tian H., Zhang F., Liu J., Lu S., Bai M., Li C., Ding Z. Brassinosteroids regulate root growth by controlling reactive oxygen species homeostasis and dual effect on ethylene synthesis in Arabidopsis. PLoS Genet. 2018;14:e1007144. doi: 10.1371/journal.pgen.1007144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y., Vandenbussche F., van der Straeten D. Regulation of seedling growth by ethylene and the ethylene-auxin crosstalk. Planta. 2017;245:467–489. doi: 10.1007/s00425-017-2651-6. [DOI] [PubMed] [Google Scholar]
- 9.Li J.T., Zhao Y., Chu H.W., Wang L.K., Fu Y.R., Liu P., Upadhyaya N., Chen C.L., Mou T.M., Feng Y.Q., et al. SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation. PLoS Genet. 2015;11:e1005464. doi: 10.1371/journal.pgen.1005464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaseva I.I., Qudeimat E., Potuschak T., Du Y., Genschik P., Vandenbussche F., van der Straeten D. The plant hormone ethylene restricts Arabidopsis growth via the epidermis. Proc. Natl. Acad. Sci. USA. 2018;115:E4130–E4139. doi: 10.1073/pnas.1717649115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Street I.H., Aman S., Zubo Y., Ramzan A., Wang X., Shakeel S.N., Kieber J.J., Schaller G.E. Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol. 2015;169:338–350. doi: 10.1104/pp.15.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanneste S., Friml J. Auxin: A trigger for change in plant development. Cell. 2009;136:1005–1016. doi: 10.1016/j.cell.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y. Auxin biosynthesis: The Arabidopsis Book. BioOne. 2014;12:e0173. doi: 10.1199/tab.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mashiguchi K., Tanaka K., Sakai T., Sugawara S., Kawaide H., Natsume M., Hanada A., Yaeno T., Shirasu K., Yao H., et al. The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011;108:18512–18517. doi: 10.1073/pnas.1108434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stepanova A.N., Yun J., Robles L.M., Novak O., He W., Guo H., Ljung K., Alonso J.M. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23:3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Won C., Shen X., Mashiguchi K., Zheng Z., Dai X., Cheng Y., Kasahara H., Kamiya Y., Chory J., Zhao Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dharmasiri N., Dharmasiri S., Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 19.Tan X., Calderon-Villalobos L.I.A., Sharon M., Zheng C.X., Robinson C.V., Estelle M., Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 20.Olatunji D., Geelen D., Verstraeten I. Control of Endogenous Auxin Levels in Plant Root Development. Int. J. Mol. Sci. 2017;18:2587. doi: 10.3390/ijms18122587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bleecker A.B., Kende H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000;16:1–18. doi: 10.1146/annurev.cellbio.16.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Yang S.F. Hoffman, N.E. Ethylene biosynthesis and its regulation in higher-plants. Annu. Rev. Plant Physiol. 1984;35:155–189. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- 23.Hua J., Meyerowitz E.M. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/S0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- 24.Hua J., Sakai H., Nourizadeh S., Chen Q.G., Bleecker A.B., Ecker J.R., Meyerowitz E.M. EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ju C., Yoon G.M., Shemansky J.M., Lin D.Y., Ying Z.I., Chang J., Garrett W.M., Kessenbrock M., Groth G., Tucker M.L., et al. CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2012;109:19486–19491. doi: 10.1073/pnas.1214848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao H., Chang K.N., Yazaki J., Ecker J.R. Interplay between ethylene, ETP1/ETP2 F-box proteins and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009;23:512–521. doi: 10.1101/gad.1765709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salehin M., Estelle M. Ethylene prunes translation. Cell. 2015;163:543–544. doi: 10.1016/j.cell.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 28.Qiao H., Shen Z., Huang S.S., Schmitz R.J., Urich M.A., Briggs S.P., Ecker J.R. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338:390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wen X., Zhang C.L., Ji Y.S., Zhao Q., He W.R., An F.Y., Jiang L.W., Guo H.W. Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 2012;22:1613–1616. doi: 10.1038/cr.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merchante C., Brumos J., Yun J., Hu Q., Spencer K.R., Enriquez P., Binder B.M., Heber S., Stepanova A.N., Alonso J.M. Gene-specific translation regulation mediated by the hormone-signaling molecule EIN2. Cell. 2015;163:684–697. doi: 10.1016/j.cell.2015.09.036. [DOI] [PubMed] [Google Scholar]
- 31.Li W., Ma M., Feng Y., Li H., Wang Y., Ma Y., Li M., An F., Guo H. EIN2-directed translational regulation of ethylene signaling in Arabidopsis. Cell. 2015;163:670–683. doi: 10.1016/j.cell.2015.09.037. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Y., Zhu Z. Relaying the ethylene signal: New roles for EIN2. Trends Plant Sci. 2016;21:2–4. doi: 10.1016/j.tplants.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Chao Q.M., Rothenberg M., Solano R., Roman G., Terzaghi W., Ecker J.R. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/S0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 34.Chang K.N., Zhong S., Weirauch M.T., Hon G., Pelizzola M., Li H., Huang S.S., Schmitz R.J., Urich M.A., Kuo D., et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. Elife. 2013;2:e00675. doi: 10.7554/eLife.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller M., Munne-Bosch S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015;169:32–41. doi: 10.1104/pp.15.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swarup R., Perry P., Hagenbeek D., van der Straeten D., Beemster G.T., Sandberg G., Bhalerao R., Ljung K., Bennett M.J. Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell. 2007;19:2186–2196. doi: 10.1105/tpc.107.052100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell. 2007;19:2169–2185. doi: 10.1105/tpc.107.052068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H., Ma B., Zhou Y., He S.J., Tang S.Y., Lu X., Xie Q., Chen S.Y., Zhang J.S. E3 ubiquitin ligase SOR1 regulates ethylene response in rice root by modulating stability of Aux/IAA protein. Proc. Natl. Acad. Sci. USA. 2018;115:4513–4518. doi: 10.1073/pnas.1719387115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin H., Zhang Z., Wang J., Chen X., Wei P., Huang R. The activation of OsEIL1 on YUC8 transcription and auxin biosynthesis is required for ethylene-inhibited root elongation in rice early seedling development. PLoS Genet. 2017;13:e1006955. doi: 10.1371/journal.pgen.1006955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jaillais Y., Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 2010;17:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verbelen J.P., de Cnodder T., Le J., Vissenberg K., Baluska F. The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: Meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signal. Behav. 2006;1:296–304. doi: 10.4161/psb.1.6.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perilli S., di Mambro R., Sabatini S. Growth and development of the root apical meristem. Curr. Opin. Plant Biol. 2012;15:17–23. doi: 10.1016/j.pbi.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 43.Chen Q., Dai X., De-Paoli H., Cheng Y., Takebayashi Y., Kasahara H., Kamiya Y., Zhao Y. Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol. 2014;55:1072–1079. doi: 10.1093/pcp/pcu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., DoleZal K., Schlereth A., Jurgens G., Alonso J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 45.Cheng Y.F., Dai X.H., Zhao Y.D. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell. 2007;19:2430–2439. doi: 10.1105/tpc.107.053009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 47.Yoshikawa T., Ito M., Sumikura T., Nakayama A., Nishimura T., Kitano H., Yamaguchi I., Koshiba T., Hibara K.I., Nagato Y., et al. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 2014;78:927–936. doi: 10.1111/tpj.12517. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto Y., Kamiya N., Morinaka Y., Matsuoka M., Sazuka T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007;143:1362–1371. doi: 10.1104/pp.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang T., Li R., Xing J., Yan L., Wang R., Zhao Y. The YUCCA-Auxin-WOX11 module controls crown root development in rice. Front. Plant Sci. 2018;9:523. doi: 10.3389/fpls.2018.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu H., Xie W.F., Zhang L., Valpuesta V., Ye Z.W., Gao Q.H., Duan K. Auxin biosynthesis by the YUCCA6 flavin monooxygenase gene in woodland strawberry. J. Integr. Plant Biol. 2014;56:350–363. doi: 10.1111/jipb.12150. [DOI] [PubMed] [Google Scholar]
- 51.Ruzicka K., Ljung K., Vanneste S., Podhorska R., Beeckman T., Friml J., Benkova E. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell. 2007;19:2197–2212. doi: 10.1105/tpc.107.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kieber J.J., Rothenberg M., Roman G., Feldmann K.A., Ecker J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-B. [DOI] [PubMed] [Google Scholar]
- 53.Woeste K.E., Ye C., Kieber J.J. Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol. 1999;119:521–529. doi: 10.1104/pp.119.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogel J.P., Woeste K.E., Theologis A., Kieber J.J. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA. 1998;95:4766–4771. doi: 10.1073/pnas.95.8.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun X., Li Y., He W., Ji C., Xia P., Wang Y., Du S., Li H., Raikhel N., Xiao J., et al. Pyrazinamide and derivatives block ethylene biosynthesis by inhibiting ACC oxidase. Nat. Commun. 2017;8:15758. doi: 10.1038/ncomms15758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serek M., Tamari G., Sisler E.C., Borochov A. Inhibition of Ethylene-Induced Cellular Senescence Symptoms by 1-Methylcyclopropene, a New Inhibitor of Ethylene Action. Physiol. Plant. 1995;94:229–232. doi: 10.1111/j.1399-3054.1995.tb05305.x. [DOI] [Google Scholar]
- 57.Ma B., He S.J., Duan K.X., Yin C.C., Chen H., Yang C., Xiong Q., Song Q.X., Lu X., Chen H.W., et al. Identification of rice ethylene-response mutants and characterization of MHZ7/OsEIN2 in distinct ethylene response and yield trait regulation. Mol. Plant. 2013;6:1830–1848. doi: 10.1093/mp/sst087. [DOI] [PubMed] [Google Scholar]
- 58.Yang C., Lu X., Ma B., Chen S.Y., Zhang J.S. Ethylene signaling in rice and Arabidopsis: Conserved and diverged aspects. Mol. Plant. 2015;8:495–505. doi: 10.1016/j.molp.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 59.Yin C.C., Ma B., Collinge D.P., Pogson B.J., He S.J., Xiong Q., Duan K.X., Chen H., Yang C., Lu X., et al. Ethylene responses in rice roots and coleoptiles are differentially regulated by a carotenoid isomerase-mediated abscisic acid pathway. Plant Cell. 2015;27:1061–1081. doi: 10.1105/tpc.15.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Q., Zhang W., Yin Z.M., Wen C.K. Rice CONSTITUTIVE TRIPLE-RESPONSE2 is involved in the ethylene-receptor signalling and regulation of various aspects of rice growth and development. J. Exp. Bot. 2013;64:4863–4875. doi: 10.1093/jxb/ert272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang C., Ma B., He S.J., Xiong Q., Duan K.X., Yin C.C., Chen H., Lu X., Chen S.Y., Zhang J.S. MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 regulate ethylene response of roots and coleoptiles and negatively affect salt tolerance in rice. Plant Physiol. 2015;169:148–165. doi: 10.1104/pp.15.00353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma B., Zhou Y., Chen H., He S.J., Huang Y.H., Zhao H., Lu X., Zhang W.K., Pang J.H., Chen S.Y., et al. Membrane protein MHZ3 stabilizes OsEIN2 in rice by interacting with its Nramp-like domain. Proc. Natl. Acad. Sci. USA. 2018;115:2520–2525. doi: 10.1073/pnas.1718377115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Alarcon M.V., Lloret P.G., Salguero J. Synergistic action of auxin and ethylene on root elongation inhibition is caused by a reduction of epidermal cell length. Plant Signal. Behav. 2014;9:e28361. doi: 10.4161/psb.28361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stepanova A.N., Hoyt J.M., Hamilton A.A., Alonso J.M. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franklin K.A., Lee S.H., Patel D., Kumar S.V., Spartz A.K., Gu C., Ye S.Q., Yu P., Breen G., Cohen J.D., et al. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. USA. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun J.Q., Qi L.L., Li Y.N., Chu J.F., Li C.Y. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu G.C., Gao S., Tian H.Y., Wu W.W., Robert H.S., Ding Z.J. Local transcriptional control of YUCCA regulates auxin promoted root-growth inhibition in response to aluminium stress in Arabidopsis. PLoS Genet. 2016;12:e1006360. doi: 10.1371/journal.pgen.1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mao J.L., Miao Z.Q., Wang Z., Yu L.H., Cai X.T., Xiang C.B. Arabidopsis ERF1 mediates cross-talk between ethylene and auxin biosynthesis during primary root elongation by regulating ASA1 expression. PLoS Genet. 2016;12:e1005760. doi: 10.1371/journal.pgen.1005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He W.R., Brumos J., Li H.J., Ji Y.S., Ke M., Gong X.Q., Zeng Q.L., Li W.Y., Zhang X.Y., An F.Y., et al. A small-molecule screen identifies l-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nishimura T., Hayashi K., Suzuki H., Gyohda A., Takaoka C., Sakaguchi Y., Matsumoto S., Kasahara H., Sakai T., Kato J., et al. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2014;77:352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- 71.Tsugafune S., Mashiguchi K., Fukui K., Takebayashi Y., Nishimura T., Sakai T., Shimada Y., Kasahara H., Koshiba T., Hayashi K.I. Yucasin DF, a potent and persistent inhibitor of auxin biosynthesis in plants. Sci. Rep. 2017;7:13992. doi: 10.1038/s41598-017-14332-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Veloccia A., Fattorini L., Della Rovere F., Sofo A., D’Angeli S., Betti C., Falasca G., Altamura M.M. Ethylene and auxin interaction in the control of adventitious rooting in Arabidopsis thaliana. J. Exp. Bot. 2016;67:6445–6458. doi: 10.1093/jxb/erw415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dash M., Yordanov Y.S., Georgieva T., Tschaplinski T.J., Yordanova E., Busov V. Poplar PtabZIP1-like enhances lateral root formation and biomass growth under drought stress. Plant J. 2017;89:692–705. doi: 10.1111/tpj.13413. [DOI] [PubMed] [Google Scholar]
- 74.Hao Y.J., Wei W., Song Q.X., Chen H.W., Zhang Y.Q., Wang F., Zou H.F., Lei G., Tian A.G., Zhang W.K., et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011;68:302–313. doi: 10.1111/j.1365-313X.2011.04687.x. [DOI] [PubMed] [Google Scholar]
- 75.Du Y., Scheres B. Lateral root formation and the multiple roles of auxin. J. Exp. Bot. 2018;69:155–167. doi: 10.1093/jxb/erx223. [DOI] [PubMed] [Google Scholar]
- 76.Chiatante D., Rost T., Bryant J., Scippa G.S. Regulatory networks controlling the development of the root system and the formation of lateral roots: A comparative analysis of the roles of pericycle and vascular cambium. Ann. Bot. 2018;5:697–710. doi: 10.1093/aob/mcy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fukaki H., Tasaka M. Hormone interactions during lateral root formation. Plant Mol. Biol. 2009;69:437–449. doi: 10.1007/s11103-008-9417-2. [DOI] [PubMed] [Google Scholar]
- 78.Cai X.T., Xu P., Zhao P.X., Liu R., Yu L.H., Xiang C.B. Arabidopsis ERF109 mediates cross-talk between jasmonic acid and auxin biosynthesis during lateral root formation. Nat. Commun. 2014;5:5833. doi: 10.1038/ncomms6833. [DOI] [PubMed] [Google Scholar]
- 79.Zhao H., Ma T., Wang X., Deng Y., Ma H., Zhang R., Zhao J. OsAUX1 controls lateral root initiation in rice (Oryza sativa L.) Plant Cell Environ. 2015;38:2208–2222. doi: 10.1111/pce.12467. [DOI] [PubMed] [Google Scholar]
- 80.Porco S., Larrieu A., Du Y.J., Gaudinier A., Goh T., Swarup K., Swarup R., Kuempers B., Bishopp A., Lavenus J., et al. Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development. 2016;143:3340–3349. doi: 10.1242/dev.136283. [DOI] [PubMed] [Google Scholar]
- 81.Zhang G., Xu N., Chen H., Wang G., Huang J. OsMADS25 regulates root system development via auxin signalling in rice. Plant J. 2018;95:1004–1022. doi: 10.1111/tpj.14007. [DOI] [PubMed] [Google Scholar]
- 82.Peret B., de Rybel B., Casimiro I., Benkova E., Swarup R., Laplaze L., Beeckman T., Bennett M.J. Arabidopsis lateral root development: An emerging story. Trends Plant Sci. 2009;14:399–408. doi: 10.1016/j.tplants.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 83.Giri J., Bhosale R., Huang G., Pandey B.K., Parker H., Zappala S., Yang J., Dievart A., Bureau C., Ljung K., et al. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat. Commun. 2018;9:1408. doi: 10.1038/s41467-018-03850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu C.L., Sun C.D., Shen C.J., Wang S.K., Liu F., Liu Y., Chen Y.L., Li C.Y., Qian Q., Aryal B., et al. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.) Plant J. 2015;83:818–830. doi: 10.1111/tpj.12929. [DOI] [PubMed] [Google Scholar]
- 85.Jing H., Yang X., Zhang J., Liu X., Zheng H., Dong G., Nian J., Feng J., Xia B., Qian Q., et al. Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 2015;6:7395. doi: 10.1038/ncomms8395. [DOI] [PubMed] [Google Scholar]
- 86.Lavenus J., Goh T., Roberts I., Guyomarc’h S., Lucas M., de Smet I., Fukaki H., Beeckman T., Bennett M., Laplaze L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013;18:450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 87.Ni J., Wang G.H., Zhu Z.X., Zhang H.H., Wu Y.R., Wu P. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 2011;68:433–442. doi: 10.1111/j.1365-313X.2011.04698.x. [DOI] [PubMed] [Google Scholar]
- 88.Kitomi Y., Inahashi H., Takehisa H., Sato Y., Inukai Y. OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 2012;190:116–122. doi: 10.1016/j.plantsci.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 89.Zhu Z.X., Liu Y., Liu S.J., Mao C.Z., Wu Y.R., Wu P. A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol. Plant. 2012;5:154–161. doi: 10.1093/mp/ssr074. [DOI] [PubMed] [Google Scholar]
- 90.Muday G.K., Rahman A., Binder B.M. Auxin and ethylene: Collaborators or competitors? Trends Plant Sci. 2012;17:181–195. doi: 10.1016/j.tplants.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 91.Negi S., Ivanchenko M.G., Muday G.K. Ethylene regulates lateral root formation and auxin transport in Arabidopsis thaliana. Plant J. 2008;55:175–187. doi: 10.1111/j.1365-313X.2008.03495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Negi S., Sukumar P., Liu X., Cohen J.D., Muday G.K. Genetic dissection of the role of ethylene in regulating auxin-dependent lateral and adventitious root formation in tomato. Plant J. 2010;61:3–15. doi: 10.1111/j.1365-313X.2009.04027.x. [DOI] [PubMed] [Google Scholar]
- 93.Lewis D.R., Negi S., Sukumar P., Muday G.K. Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development. 2011;138:3485–3495. doi: 10.1242/dev.065102. [DOI] [PubMed] [Google Scholar]
- 94.Aloni R. Role of hormones in controlling vascular differentiation and the mechanism of lateral root initiation. Planta. 2013;238:819–830. doi: 10.1007/s00425-013-1927-8. [DOI] [PubMed] [Google Scholar]
- 95.Steffens B., Rasmussen A. The physiology of adventitious roots. Plant Physiol. 2016;170:603–617. doi: 10.1104/pp.15.01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Shibata Y., Gomi K., Umemura I., Hasegawa Y., Ashikari M., Kitano H., Matsuoka M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 2005;17:1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Coudert Y., Le V.A., Adam H., Bes M., Vignols F., Jouannic S., Guiderdoni E., Gantet P. Identification of CROWN ROOTLESS1-regulated genes in rice reveals specific and conserved elements of postembryonic root formation. New Phytol. 2015;206:243–254. doi: 10.1111/nph.13196. [DOI] [PubMed] [Google Scholar]
- 98.Liu S.P., Wang J.R., Wang L., Wang X.F., Xue Y.H., Wu P., Shou H.X. Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 2009;19:1110–1119. doi: 10.1038/cr.2009.70. [DOI] [PubMed] [Google Scholar]
- 99.Kitomi Y., Ito H., Hobo T., Aya K., Kitano H., Inukai Y. The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 2011;67:472–484. doi: 10.1111/j.1365-313X.2011.04610.x. [DOI] [PubMed] [Google Scholar]
- 100.Wang Y.H., Wang D., Gan T., Liu L.L., Long W.H., Wang Y.L., Niu M., Li X.H., Zheng M., Jiang L., et al. CRL6, a member of the CHD protein family, is required for crown root development in rice. Plant Physiol. Biochem. 2016;105:185–194. doi: 10.1016/j.plaphy.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 101.Zhao Y., Hu Y.F., Dai M.Q., Huang L.M., Zhou D.X. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell. 2009;21:736–748. doi: 10.1105/tpc.108.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhou S., Jiang W., Long F., Cheng S., Yang W., Zhao Y., Zhou D.X. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell. 2017;29:1088–1104. doi: 10.1105/tpc.16.00908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cheng S., Zhou D.X., Zhao Y. WUSCHEL-related homeobox gene WOX11 increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 2016;11:e1130198. doi: 10.1080/15592324.2015.1130198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang H., Zhang J., Quan R., Pan X., Wan L., Huang R. EAR motif mutation of rice OsERF3 alters the regulation of ethylene biosynthesis and drought tolerance. Planta. 2013;237:1443–1451. doi: 10.1007/s00425-013-1852-x. [DOI] [PubMed] [Google Scholar]
- 105.Zhang S., Huang L., Yan A., Liu Y., Liu B., Yu C., Zhang A., Schiefelbein J., Gan Y. Multiple phytohormones promote root hair elongation by regulating a similar set of genes in the root epidermis in Arabidopsis. J. Exp. Bot. 2016;67:6363–6372. doi: 10.1093/jxb/erw400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang T., Li C.X., Wu Z.H., Jia Y.C., Wang H., Sun S.Y., Mao C.Z., Wang X.L. Abscisic acid regulates auxin homeostasis in rice root tips to promote root hair elongation. Front. Plant Sci. 2017;8:1121. doi: 10.3389/fpls.2017.01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Feng Y., Xu P., Li B.S., Li P.P., Wen X., An F.Y., Gong Y., Xin Y., Zhu Z.Q., Wang Y.C., et al. Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2017;114:13834–13839. doi: 10.1073/pnas.1711723115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Song L., Yu H., Dong J., Che X., Jiao Y., Liu D. The molecular mechanism of ethylene-mediated root hair development induced by phosphate starvation. PLoS Genet. 2016;12:e1006194. doi: 10.1371/journal.pgen.1006194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Strader L.C., Chen G.L., Bartel B. Ethylene directs auxin to control root cell expansion. Plant J. 2010;64:874–884. doi: 10.1111/j.1365-313X.2010.04373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mangano S., Denita-Juarez S.P., Choi H.S., Marzol E., Hwang Y., Ranocha P., Velasquez S.M., Borassi C., Barberini M.L., Aptekmann A.A., et al. Molecular link between auxin and ROS-mediated polar growth. Proc. Natl. Acad. Sci. USA. 2017;114:5289–5294. doi: 10.1073/pnas.1701536114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kramer E.M., Ackelsberg E.M. Auxin metabolism rates and implications for plant development. Front. Plant Sci. 2015;6:150. doi: 10.3389/fpls.2015.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]