Abstract
DNA methylation and histone post-translational modifications have been described as epigenetic regulation mechanisms involved in developmental transitions in plants, including seasonal changes in fruit trees. In species like almond (Prunus dulcis (Mill.) D.A: Webb), prolonged exposure to cold temperatures is required for dormancy release and flowering. Aiming to identify genomic regions with differential methylation states in response to chill accumulation, we carried out Illumina reduced-representation genome sequencing on bisulfite-treated DNA from floral buds. To do this, we analyzed almond genotypes with different chilling requirements and flowering times both before and after dormancy release for two consecutive years. The study was performed using epi-Genotyping by Sequencing (epi-GBS). A total of 7317 fragments were sequenced and the samples compared. Out of these fragments, 677 were identified as differentially methylated between the almond genotypes. Mapping these fragments using the Prunus persica (L.) Batsch v.2 genome as reference provided information about coding regions linked to early and late flowering methylation markers. Additionally, the methylation state of ten gene-coding sequences was found to be linked to the dormancy release process.
Keywords: Prunus, flowering, bisulfite sequencing, genomics, epigenetics, breeding
1. Introduction
The almond tree (Prunus dulcis (Mill.) D.A. Webb), like the rest of the Prunus species, is a deciduous fruit tree that undergoes a cyclical process of flowering, sprouting, development, and winter rest, called dormancy. The dormancy state protects the plant from potential damage from cold weather during the winter [1,2]. The dormancy period is overcome when the tree accumulates sufficient chilling hours (the chilling requirement). After dormancy release, the tree is able to sprout and flower under favorable climatic conditions [3,4]. The study of molecular mechanisms leading to dormancy release and flowering is of great interest for almond breeding programs aiming to adapt new cultivars to specific growing areas [5,6]. The dormancy release process involves sensing environmental cues (such as temperature), signal transduction, and gene expression regulation to establish a suitable response according to the stimuli received [7,8]. Transcription reprogramming leading to dormancy release may thus be mediated by epigenetic mechanisms [9,10].
Epigenetics are chemical modifications affecting DNA or structural proteins (histones) within the chromatin. Two types of epigenetic modifications have been described: DNA methylation (5′ Methylated Cytosine, 5mC) in plants and histone Post-Translational Modifications (PTMs), which include the acetylation and methylation of histones [11,12].
Epigenetic changes are part of the transcriptional regulation machinery of genomes. The dynamic but heritable character of such modifications makes them interesting regulators mediating adaptive responses to environmental changes, such as seasonal cycles, and in the long term, climate change [13]. DNA methylation is associated with cell status stability and regulation of expression. DNA methylation occurs in three sequence contexts: CG and CHG, which are found in promoter and coding regions, and CHH (where H = A, C or T), found in non-coding regions and transposable elements (TEs) [14].
Several works have described the role of epigenetics in the regulation of dormancy in deciduous plant species. Santamaría et al. [15], for instance, described a methylation decrease concomitant with H4 deacetylation and the progress of dormancy release in Castanea sativa Mill. In peach (Prunus persica), de la Fuente et al. [16] identified a genome-wide pattern of the PTM Trymethylation of Histone 3 on Lis (K) residue 27 (H3K27me3) during bud dormancy release, and Lloret et al. [17] found a relationship between gene expression, PTMs, and sorbitol synthesis during bud dormancy progression and release. Rothkegel et al. [18] showed that DNA methylation is one of the mechanisms participating in the regulation of MADS-box (MCM1-AGAMOUS-DEFICIENS-SRF) genes controlling bud dormancy in sweet cherry (Prunus avium L.). In apple (Malus domestica (Suckow) Borkh.), genome methylation patterns have been linked to chilling acquisition during dormancy [19]. In the case of almond, preliminary results from the transcriptome sequencing of non-dormant and dormant flower buds showed differential expression in a DNA methyltransferase gene and in the S-ADENOSYL METHIONINE SYNTHETASE gene responsible for the synthesis of the molecule SAM (S-adenosyl methionine), which donates the methyl group to the DNA molecule [20]. In addition, DNA methylation phenomena have also been associated with floral self-incompatibility [21] and with bud falling phenomena [22] in this species.
Genome-wide analysis of DNA methylation can be done by bisulfite sequencing, which uses Next Generation Sequencing (NGS) to analyze digested and bisulfite-treated DNA samples. The epi-Genotyping by Sequencing (epiGBS) technique was developed to represent a small part of the genome for cost-effective exploration and comparative analysis of DNA methylation and genetic variation in hundreds of de novo samples. Furthermore, this method makes it possible to genotype samples without a prior reference genome [23].
The objective of this work was to analyze the DNA methylation status of almond flower buds using epi-GBS for the first time. For this purpose, we evaluated dormant and non-dormant flower buds from two almond genotypes with different chilling requirements and flowering times using the epi-GBS protocol.
2. Results
2.1. Evaluation of the Quality of the Epi-GBS Analysis
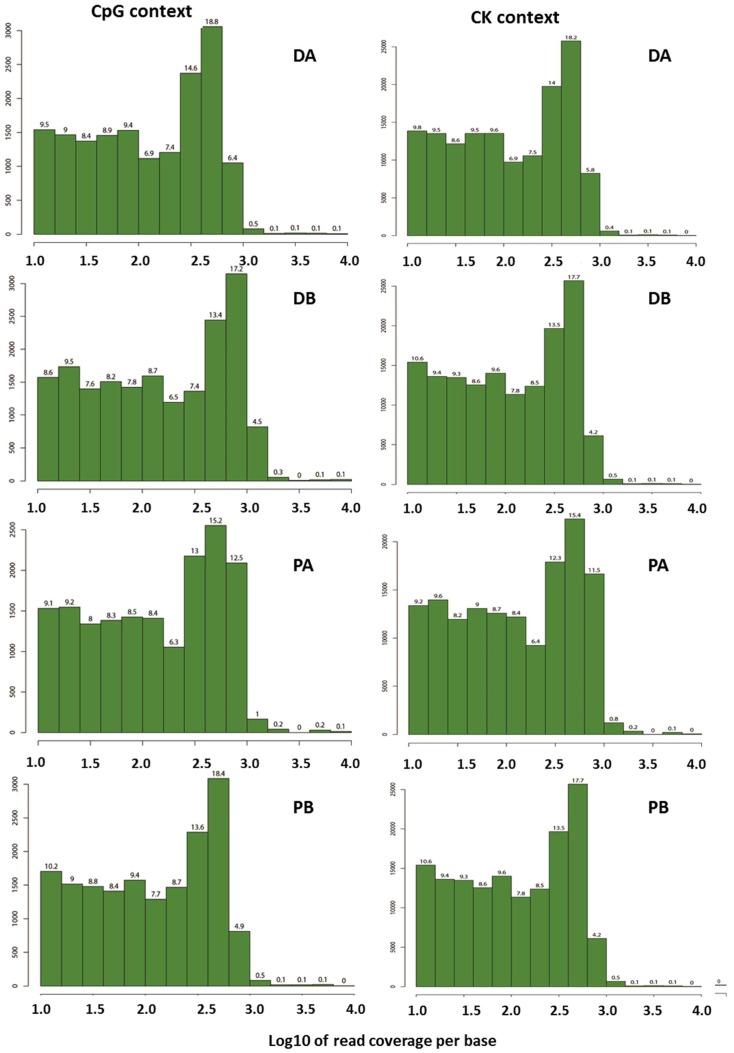
We sequenced 9518 fragments (about a 1244 kb size) and identified 7317 methylated or unmethylated fragments. Furthermore, we were able to reconstruct the original sequence of 4377 fragments. The total length of the “mock genome” obtained by merging the reconstructed fragments was 662,458 bp. Regarding the quality of this epi-GBS analysis, the absence of a secondary peak towards the right of the read coverage histograms shows that the data do not suffer from PCR duplication bias in either year (Figure 1; data from the flower buds sampled in 2015–2016). These read coverage results show the uniformity of the reads and correct PCR amplification (good quality) around the whole genome in both contexts and seasons of study.
Figure 1.
Read coverage of the samples tested in the CpG and CK (including CHG and CHH) contexts during the 2015–2016 season. “D” = ‘Desmayo Largueta’, “P” = ‘Penta’, “A” = dormant bud stage, “B” = non-dormant bud stage.
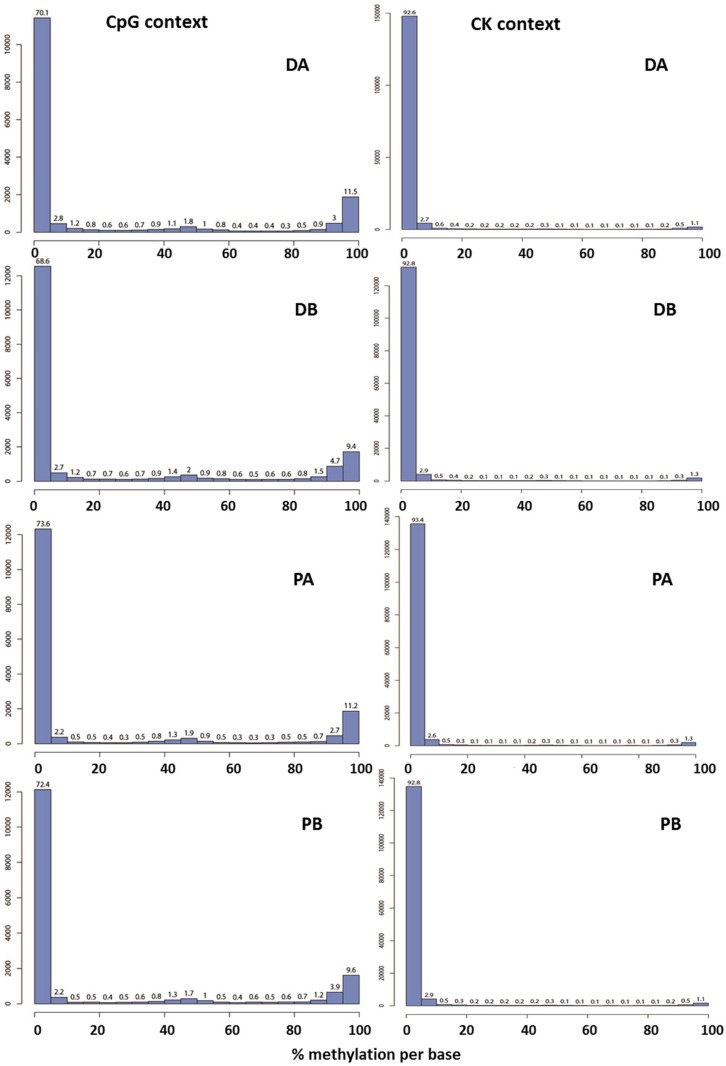
The histograms of CpG methylation showed that roughly 70% to 75% of the cytosine positions in the CpG context of the mock genome were unmethylated, around 10% of the positions were completely methylated and the remaining positions were partially methylated to varying degrees in both seasons of study (Figure 2; data from 2015–2016 season).
Figure 2.
Percentage of DNA methylation of the samples tested in the CpG and CK (including CHG and CHH) contexts during the 2015–2016 season. “D” = ‘Desmayo Largueta’, “P” = ‘Penta’, “A” = dormant bud stage, “B” = non-dormant bud stage.
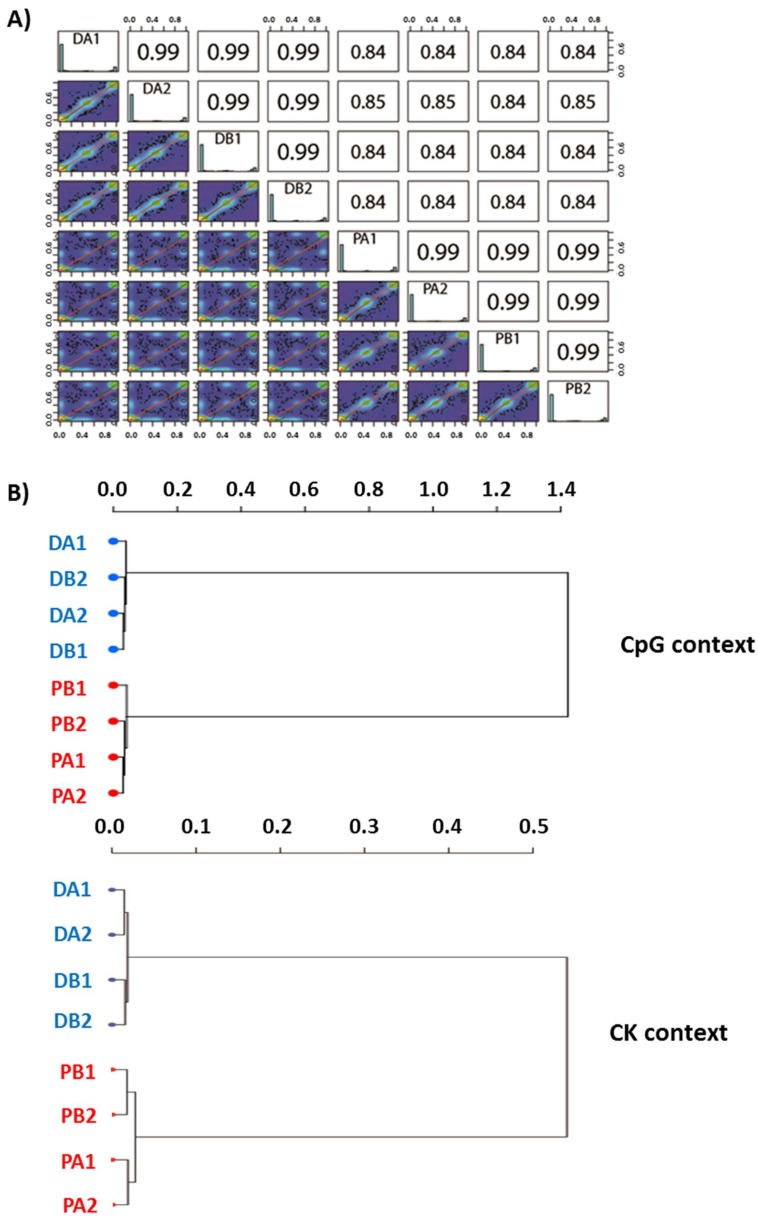
Furthermore, the correlation analyses clearly show that samples of the same variety cluster together independently of the developmental stage. The Pearson’s correlation coefficient was constantly 0.99 in comparisons within each variety and in the range of 0.84 to 0.85 in comparisons between samples of different varieties (Figure 3A). These results were also corroborated by clustering analysis, in which samples belonging to the same variety were close together while the two varieties were separated by long branches (Figure 3B). The DNA methylation pattern is generally variety-dependent rather than dormancy-dependent.
Figure 3.
(A) Almond correlations and (B) clustering analysis of the methylated fragments in both CpG and CK contexts. “D” = ‘Desmayo Largueta’, “P” = ‘Penta’, “A” = dormant bud stage, “B”=non-dormant bud stage. “1” samples are from the 2015–2016 season and “2” samples are from the 2016–2017 season.
2.2. Differentyally Methylated Genes Detected
Quantitative analysis showed that 7317 different fragments were methylated in at least one sample: 5109 ‘Cs’ were methylated in ‘Desmayo Largueta’ A samples; 5089 ‘Cs’ were methylated in ‘Desmayo Largueta’ B samples; 4955 ‘Cs’ were methylated in ‘Penta’ A samples; and 5003 ‘Cs’ were methylated in ‘Penta’ B samples.
Table 1 shows that the number of differentially methylated fragments (DMFs) detected was variable depending on the comparison performed. Furthermore, a total of 677 DMFs were found between ‘Desmayo Largueta’ and ‘Penta’ genotype samples in all stages analyzed. However, when comparing dormancy state samples (A and B), 23 DMFs were found between ‘Desmayo Largueta’ stage A and ‘Desmayo Largueta’ stage B samples and 48 DMFs were found between ‘Penta’ stage A samples and ‘Penta’ stage B samples. Of those DMFs, ten were common between ‘Desmayo Largueta’ and ‘Penta’ in the A to B stage comparison. The DMFs were divided into hypermethylated or hypomethylated categories using ‘Penta’ or ‘stage B’ samples as the reference. The fragment sequences are included in Table S1.
Table 1.
Number of differentially methylated fragments (DMFs) detected by epi-Genotyping by Sequencing (epi-GBS) according to sample comparisons.
| Differentially Methylated Fragments | ||
|---|---|---|
| D–P genotype comparison | ||
| Hypo (<) | Hyper (>) | Stages |
| 307 | 370 | A and B |
| 677 DMFs | ||
| A–B stage comparison | ||
| Hypo (<) | Hyper (>) | Genotype |
| 3 | 20 | D |
| 21 | 27 | P |
| 3 | 7 | D and P |
| 10 DMFs | ||
“D” = ‘Desmayo Largueta’, “P” = ‘Penta’; “A” = dormant bud stage, “B” = non-dormant bud stage.
More than 99% of the identified DMFs were mapped on the Prunus persica v2.1 genome (Table S2), and those located between 2 kb upstream and 1 kb downstream from the gene coding sequences were selected for subsequent annotation analysis. The number of differentially methylated genes (DMGs) thus identified is shown in Table 2 and Table 3.
Table 2.
The number of differentially methylated genes (DMGs) identified from sequenced fragments mapping onto the Prunus persica genome (v2.1).
| Differentially Methylated Genes | |||
|---|---|---|---|
| Methylation State | Gene Position | Gene Hits | Genes Identified |
| Hypermethylated | Upstream | 36 | |
| Inside | 291 | ||
| Downstream | 134 | ||
| Total | 461 | 423 | |
| Hypomethylated | Upstream | 19 | |
| Inside | 201 | ||
| Downstream | 80 | ||
| Total | 300 | 281 | |
| Equally-methylated | Upstream | 6 | |
| Inside | 41 | ||
| Downstream | 8 | ||
| Total | 55 | 27 | |
| Total DMGs | 816 | 731 | |
The methylation state refers to the number of 5′ Methylated Cytosines (5mCs) in ‘Desmayo Largueta’ samples compared to ‘Penta’ samples. The category “equally methylated” refers to genes whose number of 5mCs was the same between samples but in which the 5mCs were located in different fragment positions. The gene position is based on gene orientation with respect to the fragment-mapping region (“upstream”, “inside”, and “downstream”).
Table 3.
The number of differentially methylated genes between the A and B dormancy states of flower buds, identified from sequenced fragments mapped to the P. persica genome (v2.1).
| Methylation State | Gene Position | Differentially Methylated Genes | ||
|---|---|---|---|---|
| ‘Desmayo Largueta’ | ‘Penta‘ | Common | ||
| Hyper-methylated | Upstream | 7 | 3 | 1 |
| Inside | 5 | 2 | 2 | |
| Downstream | 5 | 9 | 4 | |
| Total | 17 | 14 | 7 | |
| Hypo-methylated | Upstream | 1 | - | - |
| Total | 18 | 14 | 7 | |
The methylation state refers to the number of 5mCs in stage A (dormant buds) samples with respect to the number found in stage B (non-dormant buds) samples. The gene position is based on gene orientation with respect to the fragment-mapping region.
DMGs were classified according to their position with respect to the fragment-mapping region. The most frequently mapped fragments were those within gene regions (“inside” DMGs) followed by the 5′regulatory regions (“downstream” DMGs) and, finally, in the 3′ regions (“upstream” DMGs).
Data shown in Table 2 indicate that DMGs were found as hypermethylated in ‘Desmayo Largueta’ samples (in both the A and B stages) to a greater extent than in ‘Penta’ samples (423).
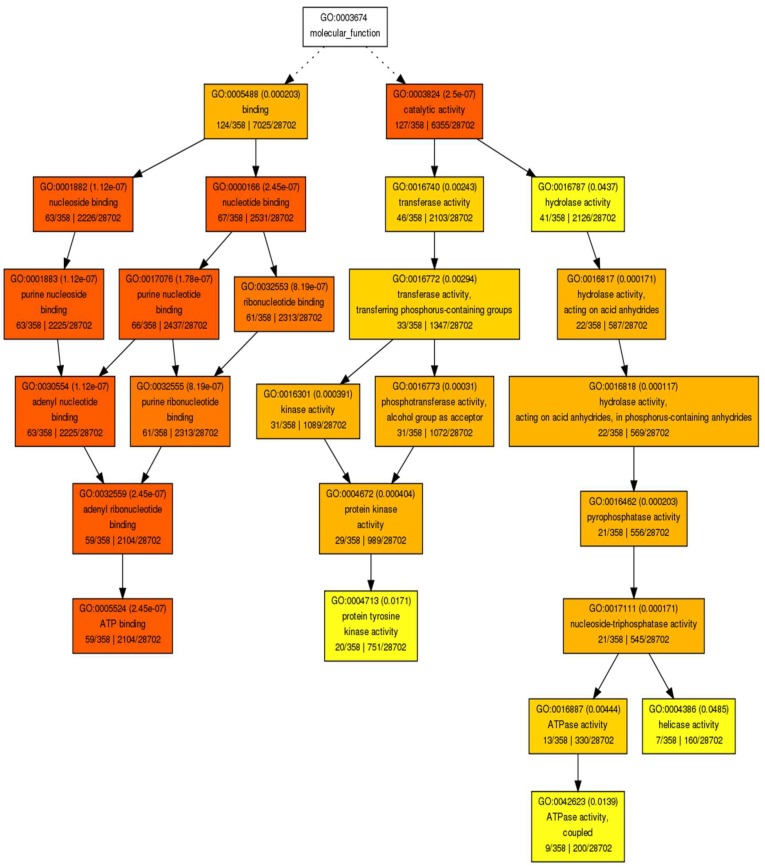
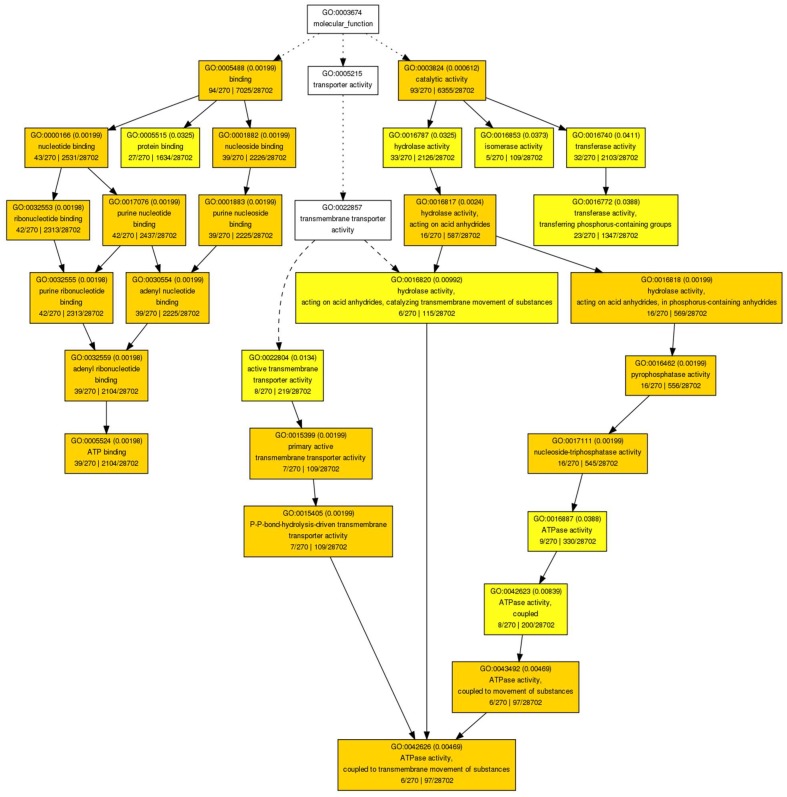
We found enriched hypermethylated genes in ‘Desmayo Largueta’ flower bud samples in the following processes related to primary metabolism in the “biological function” GO (Gene Ontology) category: amino-acid and carbohydrate synthesis and protein phosphorylation (Figure S1). ATP binding and protein kinase and phosphatase activity were the two main “molecular function” GO terms found (Figure 4).
Figure 4.
GO terms of the “Molecular function” category represented in genes identified as hypermethylated in ‘Desmayo Largueta’ flower buds in both the “A” = dormant bud stage and “B” = non dormant bud stage.
In a gene-level analysis, the following candidate genes appeared as hypermethylated in ‘Desmayo Largueta’ flower bud samples: genes related to transcription regulation, including transcription factors (Prupe.1G395600, Prupe.5G088700, Prupe.6G343100); genes linked to RNA-mediated silencing (Prupe.7G221200); genes linked to chromatin remodelling (Prupe.8G221300, and LATE ELONGATED HYPOCOTYL (LHY), encoded by Prupe.2G200400); and, especially, genes involved in the auxin response (Prupe.1G000200, Prupe.1G067400, Prupe.7G048400) and AUXIN RESPONSE FACTOR (ARF) signal transduction (Prupe.3G010900, Prupe.5G217700, Prupe.7G228800). We also identified DNA repair proteins, such as those encoded by Prupe.1G510000, Prupe.2G013900, Prupe.3G029600, Prupe.3G16000 and Prupe.5G066100. Finally, proteins participating in oxidoreduction processes, such as LATE EMBRYOGENESIS ABUNDANT (LEA) proteins encoded by Prupe.4G026900 and Prupe.4G02700, also appeared as hypermethylated in ‘Desmayo Largueta’ flower bud samples (Table S3).
Regarding the hypomethylated genes in the ‘Desmayo Largueta’ samples, we found cellular protein localization within the “biological function” GO category (Figure S2). ATP-coupled transmembrane transport and ATP-binding activity, on the other hand, appeared in the “molecular function” GO category (Figure 5).
Figure 5.
“Molecular function” GO terms represented in genes identified as hypomethylated in ‘Desmayo Largueta’ flower buds in both the “A” = dormant bud stage and “B” = non dormant bud stage.
We were able to identify a wide range of DNA-binding proteins encoded by the hypomethylated genes in the ‘Desmayo Largueta’ samples: histone methyltransferases (encoded by Prupe.1G050800 and Prupe.7G271600); the transcription factor NUCLEAR FACTOR-Y (NF-Y) (encoded by Prupe.2G47600); DNA topoisomerases (encoded by Prupe.1G173400 and Prupe.1G173500); and FAR-RED IMPAIRED RESPONSE 1 (FAR1) (Prupe.1G196400). Interestingly, a single gene coding for a HYDROPHOBIC SEED PROTEIN (HSP) also appeared as hypomethylated in the ‘Desmayo Largueta’ samples.
2.3. Differentyally Methylated Genes Related to Bud Dormancy
More identified DMGs were found as hypermethylated in stage A samples (dormant buds) than in stage B samples (non-dormant buds) (Table 3). Furthermore, just one hypomethylated gene could be functionally annotated, and it was mapped in the 3’ regulatory region of the gene Prupe.4G277200, which encodes for a REGULATION OF CHROMOSOME CONDENSATION (RCC1) protein (Table 4).
Table 4.
Hypermethylated genes identified in ‘Desmayo Largueta’ (D) and ‘Penta’ (P) stage A flower buds compared to those found in stage B flower buds (Chromosome, Fragment ID, Prupe.ID, Functional annotation).
| FragmentID | Comparison | Chromosome | Gene Position | Prupe.Gene Code |
|---|---|---|---|---|
| fragment1289 | AB | Pp01 | Downstream | Prupe.1G287200 |
| fragment1294 | AB | Pp01 | Upstream | Prupe.1G125600 |
| fragment3735 | PAPB | Pp01 | Downstream | Prupe.1G099900 |
| fragment32 | AB | Pp03 | Downstream | Prupe.3G130700 |
| fragment341 | AB | Pp06 | Downstream | Prupe.6G307900 |
| fragment797 | AB | Pp02 | Inside | Prupe.2G074400 |
| fragment1708 | DADB | Pp02 | Inside | Prupe.2G019300 |
| fragment4206 | PAPB | Pp02 | Inside | Prupe.2G019300 |
| fragment341 | DADB | Pp02 | Downstream | Prupe.2G031100 |
| fragment255 | DADB | Pp02 | Upstream | Prupe.2G053300 |
| fragment92 | DADB | Pp02 | Inside | Prupe.2G057100 |
| fragment92 | DADB | Pp02 | Inside | Prupe.2G057800 |
| fragment797 | DADB | Pp02 | Upstream | Prupe.2G074300 |
| fragment3707 | PAPB | Pp04 | Downstream | Prupe.4G186400 |
| fragment4154 | PAPB | Pp04 | Downstream | Prupe.4G253800 |
| fragment238 | AB | Pp04 | Downstream | Prupe.4G270800 |
| fragment3299 | PAPB | Pp06 | Downstream | Prupe.6G307900 |
| fragment1289 | DADB | Pp02 | Inside | Prupe.2G146000 |
| fragment341 | DADB | Pp03 | Upstream | Prupe.3G026400 |
| fragment797 | DADB | Pp03 | Downstream | Prupe.3G130700 |
| fragment2263 | DADB | Pp05 | Inside | Prupe.5G036900 |
| fragment157 | PAPB | Pp06 | Upstream | Prupe.6G014500 |
| fragment1289 | DADB | Pp05 | Downstream | Prupe.5G038500 |
| fragment483 | PAPB | Pp02 | Upstream | Prupe.2G039500 |
| fragment507 | DADB | Pp06 | Inside | Prupe.6G097800 |
| fragment1708 | DADB | Pp06 | Upstream | Prupe.6G331300 |
| fragment2727 | DADB | Pp06 | Upstream | Prupe.6G331300 |
| fragment849 | PAPB | Pp01 | Downstream | Prupe.1G105700 |
Common stage A hypermethylated genes coded for a MITOGEN-ACTIVATED PROTEIN (MAP)-kinase and a phosphatase (Prupe.4G270800 and Prupe.1G287200); an LEUCINE RICH REPEAT-TOLL INTERLEUKIN 1 RECEPTOR (LRR-TIR) apoptotic ATPase associated with disease resistance (Prupe.3G130700); a GDSL (Gly, Asp, Ser and Leu motif) lipase (Prupe.6G307900); an Nt-C2 family protein (Prupe.2G074400); and a Glycerophosphatidylinositol (GPI) anchor synthase (Prupe.2G019300). The Prupe.1G125600 gene was annotated, but its protein function is unknown (Table 4). Moreover, genes coding for VACUOLAR PROTEIN SORTING 1 (VPS1) proteins were detected as hypermethylated in stage A in both ‘Desmayo Largueta’ and ‘Penta’. VPS1 genes corresponded to Prupe.3G026400 in ‘Desmayo Largueta’ samples and Prupe.2G029500 in ‘Penta’ samples.
Three additional LRR-TIR apoptotic ATPases were identified as encoded by hypermethylated genes in ‘Desmayo Largueta’ stage A samples (Table 4). Other genes that were found coded for defense proteins, a phosphatase, a cornichon protein associated with cell polarity and a Cytochrome P-450 (CytP450) protein member. On the other hand, hypermethylated genes in ‘Penta’ stage A samples coded for the ASSEMBLY PROTEIN 180 (AP180) clathrin assembly protein, the Tryptophan-Aspartic acid-Sterile Alpha Motif domain containing protein (WDSAM1) ubiquitination protein, lipases, a glycosyl hydrolase and a FCF2 rRNA processing protein recently described in yeast (Table 4).
3. Discussion
Using our epiGBS variant as a first approach is a less expensive technique than complete GBS with highly accurate results. Furthermore, without the sequenced genome of the species, it is easier to perform bioinformatic analysis with well-defined fragments obtained by epiGBS. In this work, a conversion with bisulfite and a subsequent sequencing were performed to evaluate the 5mC variants of the samples analyzed. Subsequently, using bioinformatic analysis, these differentially methylated regions were mapped in the reference genome.
Applying epi-GBS to ‘Desmayo Largueta’ and ‘Penta’ flower bud gDNA samples provided data about methylation (5mC) variants depending on the genotype and dormancy state of the flower buds. Quality evaluation of the analysis showed that more than 90% of all cytosine positions were completely unmethylated and that only 1.0% to 1.3% of the positions were completely methylated, with higher methylation in the CpG context. These results agree with previous results in different plant species indicating that CG methylation is the typical genomic region for DNA methylation with less methylation abundance in the CHG and CHH contexts [23]. As a result of the specificity of the methylome of plants with respect to that of animals, we adapted bisulfite conversion methods to allow for correct analysis in plants for all cytosine contexts [24].
DNA methylomes have now been analyzed in many plants species, including Arabidopsis, rice, maize, and tomato. DNA methylation results in these species indicate that the distribution of methylation marks across the genomes is generally conserved, although variations can be observed between species depending on several factors, including transposon abundance and genome size [25]. In agreement with our results, polymorphism (5mC variants) can be observed by comparing different genotypes or even accessions within the same species. In addition, recent results evaluating methylomes from 1,227 different accessions of Arabidopsis distributed worldwide have shown important polymorphisms between accessions [26].
It is interesting to note the high degree of differential methylation that seems to be fixed between the two almond genotypes analyzed. This fact is of practical importance in cultivar improvement for developing epigenetic markers based on methylation variants and taking into account the high flexibility of methylation patterns in relation to external signals in order to identify markers based on methylation polymorphisms. In contrast to standard sequencing, bisulfite sequencing makes it possible to obtain information that conditions the phenotype. As a consequence, knowing the methylation state might help us understand the genetic determinism of important agronomic traits more deeply. Although the methylation patterns are highly variable in response to different external factors, the markers that we have detected in our almond genotypes are conserved in different stages of development and in different years and can therefore be considered as stable and conserved epigenetic marks.
In this study, data showed important differences between genotypes, which displayed different phenotypes in terms of breeding traits (chilling requirements for dormancy release, flowering and ripening times, almond production and almond characteristics) (Table 1 and Table 2). It is remarkable that more hypermethylated than hypomethylated fragments were identified in stage A (dormant flower buds) almond samples in both genotypes (Table 3). This is concordant with the general decrease in 5mC during dormancy progression in C. sativa [15]. On the other hand, the most frequently mapped fragments were within the gene regions (“inside” DMGs), followed by the 5′regulatory regions (“downstream” DMGs) and, in last place, in the 3′ regions (“upstream” DMGs). According to Vining et al. [27], 5mC in promoters and gene body parts is related to a repressed state of chromatin, a condition that inhibits the accessibility of the transcriptional machinery.
Among the genes found as hypermethylated in ‘Desmayo Largueta’ with respect to ‘Penta’ flower buds (Table S3), ARF transcription factors were highly represented. Its known that the expression of genes like ARFs involved in the auxin response are subjected to epigenetic regulation [9,28], and ARF transcriptional regulation is required for developmental processes like germination [29]. Accordingly, Zhang et al. [30] observed a flowering delay in Arabidopsis when ARF6 and ARF8 were repressed. Nonetheless, the reason underlying the hypermethylated state of genes participating in the auxin response pathway in both dormant and non-dormant flower buds of the early flowering genotype ‘Desmayo Largueta’ has yet to be unraveled.
Another hymermethylated gene in ‘Desmayo Largueta’ flower bud samples was a member of the LEA family (Table S3). LEA proteins are involved in osmoprotection, which is activated in response to low temperatures [31]. When hypermethylated, this gene showed a repressed state of expression, although in low chilling requirement cultivars like ‘Desmayo Largueta’, osmoprotection would not be so necessary or may be regulated in a different way. The LEA gene family has been characterized by Du et al. [32] in Prunus mume (Siebold) Siebold & Zucc., and differential expression has been identified during bud dormancy in this species [33].
The LHY protein, on the other hand, is a well-described flowering time regulator in response to the photoperiod [34,35], and the gene network controlling this trait has been studied [36]. ‘Desmayo Largueta’ is a low-chill cultivar whose dormancy period takes place under short photoperiod conditions such as the experimental conditions of this work. It would be interesting to study LHY behavior during dormancy progression in different almond cultivars.
The methylation variants observed may be associated with evolutionary changes related to each genotype’s features [37]. It will be interesting to distinguish which variants are related to traits of agronomic interest in order to explore adaptive mechanisms to the environment [38]. Recently, for instance, Garg et al. [39] identified conserved methylation polymorphisms distributed throughout rice varieties with different responses to drought resistance.
We found other hypermethylated genes in dormant (A) flower buds with respect to dormancy released (B) flower buds, including a MAP kinase (MAPK) and a phosphatase (Prupe.1G287200). MAP kinases and phosphatases have been found to participate in the initial response to cold induced by an increase in Ca2+ [31]. Furthermore, MAPK3 has been shown to be a central regulator of seed dormancy in barley [40]. Regarding the other genes hypermethylated in the A state, LRR-TIR apoptotic ATPases may be activated in a type of programmed cell death (PCD) called developmental cell death (DCD), leading to a differentiation of cells after dormancy release, as occurs in floral morphogenesis or in the pollen tube [41,42]. Nt-C2 and VSP1 proteins, on the other hand, are involved in vesicular trafficking from the cell membrane, and this process has been linked to cell wall differentiation and appears to be important in the dormancy release process [43,44]. Finally, GPI anchoring (a post-translational modification of proteins consisting of glycosylation) proteins are involved in intercellular signaling, as occurs in flowering transition as shown in Populus genus by Rinne et al. [45].
4. Materials and Methods
4.1. Plant Material and Experimental Design
We used flower buds from ‘Desmayo Largueta’, a traditional almond cultivar with very low chilling requirements and an extra-early flowering time, and ‘Penta’, a cultivar released from the CEBAS-CSIC Almond Breeding Program (Murcia, South-East Spain) with high chilling requirements and an extra-late flowering time. The plant material consisted of flower buds at stages A (dormancy phase) and B (after dormancy release) that were referenced to the phenological stages described by Felipe [46] (Figure 6). Dormancy release evaluation was performed by the forcing method according to Prudencio et al. [6]. Almond flower buds were picked from the experimental field of CEBAS-CSIC during two seasons of study: 2015–2016 and 2016–2017.
Figure 6.
Plant material from ‘Desmayo Largueta’ and ‘Penta’ almond cultivars. Flower buds in the dormant state (A) and after dormancy release (B) according to Felipe [41]. Scale bars represent 1 mm in each case.
4.2. Epi-GBS Protocol
Every sample (‘Desmayo Largueta’ stage A, ‘Penta’ stage A, ‘Desmayo Largueta’ stage B, and ‘Penta’ stage B from the first and second season of study) consisted of a pool of ten flower buds. Genomic DNA was extracted from each sample following the method described by Doyle and Doyle [47]. The DNA samples were quantified using Qubit (Thermo Fisher Scientific, Alcobendas, Spain) and diluted to 1 µg in 100 µL. A total of 20 µL was digested using PstI restriction enzyme. Adaptors consisting of barcoded oligos were ligated to every sample (Table S4). Non-phosphorylated hemimethylated adapters were used to reduce costs. Fragmented samples (libraries generated by restriction) were pooled and purified and subsequently subjected to nick translation with C-dNTPs (Zymo Research, Irvine, CA, USA) and 7.5 µL of DNA PolI (NEB, Ipswich, MA, USA) in NEB buffer 2. An EZ DNA Methylation-Lightning kit (Zymo Research) was used for bisulfite treatment, and fragments were selected by size with a Thermo Scientific Size Selection kit (Thermo Fisher Scientific). Libraries were amplified using the Kapa HiFi HotStart Uracil+ ReadyMix (Roche, Barcelona, Spain) and purified with Magjet NGS Cleanup (Thermo Fisher Scientific). Paired-end Illumina 2500 reads (2 × 100 bases) were generated by Macrogen (Seoul, Korea) [23].
4.3. Bioinformatic Analysis of DNA Methylation
The process_radtags program of the Stacks 1.48 pipeline [48]. Checking the integrity of the restriction site was disabled with the “-disable_rad_check” option and quality filtering with the default settings was disabled with the exception the rad_check. This was necessary because the bisulfite treatment changes unmethylated cytosines in the recognition sequence of PstI, and, as a result, checking the restriction cut site would filter out all fragments. The ustacks program of the pipeline was used to align the fragments into perfectly matching stacks. The default settings were used with the exception of -M, which was set to 4 in order to increase the maximum distance (in nucleotides) between stacks. Finally, cstacks was used to build a catalog of consensus loci. A custom C program was used for the reconstruction of the original sequences of the fragments by comparing the reads with origins in the “Watson” and “Crick” strands of the genomic DNA. The reconstructed DNA fragments were merged by another custom C program to produce one continuous “mock genome”. Bismark_v0.19.0 [49] was used to align the original fragments to the mock genome and to extract the methylation information. The Bismark coverage reports were used as input for the methylKit R package [50]. A methyl kit was used to elaborate histograms of C-methylation and coverage and to assess sample similarity and correlation using the default settings. For the hierarchical clustering of the samples, dist was set to “correlation” and method to “ward”. Finally, we used the calculate DiffMeth function of a MethylKit to search for differentially methylated cytosines with the settings difference = 25, qvalue = 0.01. We looked for both hypermethylated and hypomethylated bases setting type = hyper and = hypo, respectively. The positions of the differentially methylated cytosines were extracted from the MethylKit files. Another custom made C-program was used to identify the original fragments where these differentially methylated cytosines were located.
4.4. Gene Finding and Annotation
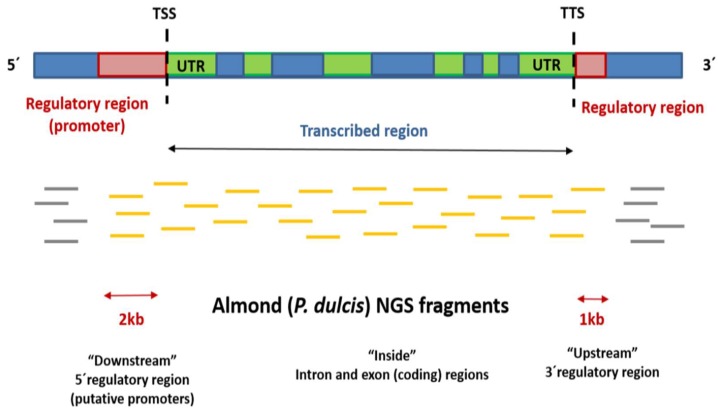
The sequence of each fragment was mapped against the P. persica reference genome (v2.0) [51] with Gmap [52]. Two different output files, in the gff3 and SAM format, were obtained. The gff3 ouput files were processed to extract the boundary coordinates (start and end positions) of each hit using command line tools. After that, the boundary coordinates were used by a second custom python script to retrieve three different categories of annotations based on gene locations on the P. persica reference genome: upstream and downstream genes (in a size window of 10,000 bp) and “inside genes” (fragments within gene sequence). Finally, SAM format files were processed using a custom python script to extract the alignment information (number of exons, percentage of coverage, percentage of identity, and amino acid changes). Functional annotation of genes selected by distance to the mapped fragment was carried out using AgriGO software using Singular Enrichment Analysis (SEA), and Fisher’s test [53] (Figure 7).
Figure 7.
Schematic representation of a plant gene and classification of almond (Prunus dulcis) DMFs mapped in the P. persica genome. DMFs mapping from 2 kb upstream TSS to 1 kb downstream TTS were selected for functional annotation (fragments colored in orange). DMFs were classified according to the gene position—downstream, inside, or upstream—with respect to the fragment-mapping region. Fragments mapped in intergenic regions (colored in grey) were discarded as putative gene regulatory regions. TSS: Transcriptional Start Site, UTR: Untranslated Transcription Region, TTS: Transcriptional Terminal Site. Exons and introns within the transcribed region are colored in blue and green, respectively.
5. Conclusions
In this study, we applied the epi-GBS protocol to almond (P. dulcis) DNA samples for the first time. The technical potential is evident in the discovery of epigenetic variants, based on 5mC, that are genotype-dependent. According to the results obtained, the DNA methylation (5mC) pattern is generally genotype-dependent rather than dormancy state-dependent. Comparative DNA methylation studies of both almond varieties released from breeding programs and traditional varieties will surely contribute to our knowledge of methylation variants and provide candidate epialleles linked to agronomic traits. Such polymorphisms can be screened in large populations using NGS to confirm the locus methylation state associated with a given character of interest. In spite of coverage limitation, we were able to identify genes whose DNA methylation state changed between the dormant and active state of the flower buds. This was possible in both the traditional early-flowering genotype ‘Desmayo Largueta’ and the extra-late-flowering genotype ‘Penta’ from the CEBAS-CSIC Almond Breeding Program. Furthermore, common genes arose from the analysis. In the future, it would be interesting to improve the technique coverage to obtain a greater representation of the genome. Ultimately, the results will be an essential complement to RNAseq experiments in bud dormancy progression.
Abbreviations
| 5mC | 5′ Methylated Cytosine |
| AP180 | ASSEMBLY PROTEIN 180 |
| ARF | AUXIN RESPONSE FACTOR |
| CytP450 | Cytochrome P-450 |
| DMF | Differentially Methylated Fragment |
| DMG | Differentially Methylated Gene |
| epiGBS | Epi-Genotyping By Sequencing |
| FAR1 | FAR-RED IMPAIRED RESPONSE 1 |
| GDSL | Gly, Asp, Ser, and Leu motif |
| GO | Gene Ontology |
| GPI | Glycerophosphatidylinositol |
| H3K27me3 | Trymethylation of Histone 3 on Lis (K) residue 27 |
| HSP | HYDROPHOBIC SEED PROTEIN |
| LEA | LATE EMBRYOGENESIS ABUNDANT |
| LHY | LATE ELONGATED HYPOCOTYL |
| LRR-TIR | LEUCINE RICH REPEAT-TOLL INTERLEUKIN 1 RECEPTOR |
| MADS | MCM1-AGAMOUS-DEFICIENS-SRF |
| MAPK | MITOGEN-ACTIVATED PROTEIN KINASE |
| NGS | Next Generation Sequencing |
| NF-Y | NUCLEAR FACTOR-Y |
| PTMs | Post-Translational Modifications |
| SAM | S-ADENOSYL METHIONINE |
| VPS1 | VACUOLAR PROTEIN SORTING 1 |
| WDSAM1 | Tryptophan-Aspartic acid-Sterile Alpha Motif 1 |
Supplementary Materials
The supplementary materials can be found at http://www.mdpi.com/1422-0067/19/11/3542/s1. Figure S1. GO terms of the “Biological Function” category represented in genes identified as hypermethylated in ‘Desmayo Largueta’ flower buds, in both the A and B dormancy states, compared to ‘Penta’ flower buds. Figure S2. GO terms of the “Biological Function” category represented in genes identified as hypomethylated in ‘Desmayo Largueta’ flower buds, in the both A and B dormancy states, compared to ‘Penta’ flower buds. Table S1. Total NGS fragments identified by Fragment ID. Table S2. Percentage of DMFs mapped in the Prunus persica genome (v2.1), Mapping region, Coverage, Percent identity, Number of exons, Amino acid changes, Comparison and Category. Table S3. Hypermethylated, Hypomethylated and Equally-methylated genes detected in ‘Desmayo Largueta’ flower buds with respect to ‘Penta’ flower buds in both the A and B stages (Chromosome, Fragment ID, Prupe.ID, Functional annotation). Gene list for GO annotation. Table S4. Sample barcodes. “D” = ‘Desmayo Largueta’, “P” = ‘Penta’, “A” = dormant bud stage; “B” = non-dormant bud stage. DNA samples with number “1” correspond to the 2015–2016 season, and those with number “2” correspond to the 2016–2017 season.
Author Contributions
A.S.P. and O.W. participated in the design and coordination of the study. A.S.P. and O.W. carried out the epiGBS protocol. A.S.P., O.W. and P.J.M.-G. carried out data analysis. A.S.P., O.W., P.J.M.-G., R.M.R., P.M.-G. and F.D. participated in the manuscript elaboration and discussion.
Funding
This study has been supported by Grants 19308/PI/14 and 19879/GERM/15 of the Seneca Foundation of the Region of Murcia (Spain) and the Almond Breeding project of the Spanish Ministry of Economy and Competiveness.
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Vitasse Y., Lenz A., Körner C. The interaction between freezing tolerance and phenology in temperate deciduous trees. Front. Plant Sci. 2014;5 doi: 10.3389/fpls.2014.00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beauvieux R., Wenden B., Dirlewanger E. Bud Dormancy in Perennial Fruit Tree Species: A Pivotal Role for Oxidative Cues. Front. Plant Sci. 2018;9:657. doi: 10.3389/fpls.2018.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang B.A., Early J.D., Martin G.C., Darnell R.L. Endo-, para-, and eco-dormancy physiological terminology and classification for dormancy research. HortScience. 1987;22:371–377. [Google Scholar]
- 4.Egea J., Ortega E., Martínez-Gómez P., Dicenta F. Chilling and heat requirements of almond cultivars for flowering. Environ. Exp. Bot. 2003;50:79–85. doi: 10.1016/S0098-8472(03)00002-9. [DOI] [Google Scholar]
- 5.Martínez-Gómez P., Prudencio A.S., Gradziel T.M., Dicenta F. The delay of flowering time in almond: A review of the combined effect of adaptation, mutation and breeding. Euphytica. 2017;213:197. doi: 10.1007/s10681-017-1974-5. [DOI] [Google Scholar]
- 6.Prudencio A.S., Martínez-Gómez P., Dicenta F. Evaluation of breaking dormancy, flowering and productivity of extra-late and ultra-late flowering almond cultivars during cold and warm seasons in South-East of Spain. Sci. Hort. 2018;235:39–46. doi: 10.1016/j.scienta.2018.02.073. [DOI] [Google Scholar]
- 7.Cooke J.E.K., Eriksson M.E., Juntilla O. The dynamic nature of bud dormancy in trees: Environmental control and molecular mechanisms. Plant Cell Environ. 2012;35:1707–1728. doi: 10.1111/j.1365-3040.2012.02552.x. [DOI] [PubMed] [Google Scholar]
- 8.Abbott A.G., Zhebentyayeva T., Barakat A., Liu Z. The genetic control of bud-break in trees. Adv. Bot. Res. 2015;74:201–228. [Google Scholar]
- 9.Yaish M.W., Colasanti J., Rothstein S.J. The role of epigenetics processes in controlling flowering time exposed to stress. J. Exp. Bot. 2011;62:3727–3735. doi: 10.1093/jxb/err177. [DOI] [PubMed] [Google Scholar]
- 10.Ríos G., Leida C., Conejero C., Badenes M.L. Epigenetic regulation of bud dormancy events in perennial plants. Front. Plant Sci. 2014;5:247. doi: 10.3389/fpls.2014.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saze H. Epigenetic memory transmission through mitosis and meiosis in plants. Semin. Cell Dev. Biol. 2008;19:527–536. doi: 10.1016/j.semcdb.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 12.Feng S., Jacobsen S.E. Epigenetic modifications in plants: An evolutionary perspective. Curr. Opin. Plant Biol. 2011;14:179–186. doi: 10.1016/j.pbi.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lämke J., Bäurle I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017;18:124. doi: 10.1186/s13059-017-1263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pascual J., Cañal M.J., Correia B., Escandon M., Hasbún R., Meijón M., Pinto G., Valledor L. Can Epigenetics Help Forest Plants to Adapt to Climate Change? In: Alvarez-Venegas R., De la Peña C., Casas-Mollano J.A., editors. Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications: Transcriptional Regulation and Chromatin Remodelling in Plants. Springer; Cham, Switzerland: 2014. pp. 125–146. [Google Scholar]
- 15.Santamaría M., Hasbún R., Valera M., Meijón M., Valledor L., Rodríguez J.L., Rodríguez R. Acetylated H4 histone and genomic DNA methylation patterns during bud set and bud burst in Castanea sativa. J. Plant Physiol. 2009;166:1360–1369. doi: 10.1016/j.jplph.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 16.de la Fuente L., Conesa A., Lloret A., Badenes M.L., Ríos G. Genome-wide changes in histone H3 lysine 27 trimethylation associated with bud dormancy release in peach. Tree Genet. Gen. 2015;11:45. doi: 10.1007/s11295-015-0869-7. [DOI] [Google Scholar]
- 17.Lloret A., Martínez-Fuentes A., Agustí M., Badenes M.L., Ríos G. Chromatin-associated regulation of sorbitol synthesis in flower buds of peach. Plant Mol. Biol. 2017;95:507–517. doi: 10.1007/s11103-017-0669-6. [DOI] [PubMed] [Google Scholar]
- 18.Rothkegel K., Sánchez E., Montes C., Greve M., Tapia S., Bravo S., Almeida A.M. DNA methylation and small interference RNAs participate in the regulation of MADS-box genes involved in dormancy in sweet cherry (Prunus avium L.) Tree Physiol. 2017;37:1739–1751. doi: 10.1093/treephys/tpx055. [DOI] [PubMed] [Google Scholar]
- 19.Kumar G., Rattan U.K., Singh A.K. Chilling-Mediated DNA Methylation Changes during Dormancy and Its Release Reveal the Importance of Epigenetic Regulation during Winter Dormancy in Apple (Malus × domestica Borkh.) PLoS ONE. 2016;11:e0149934. doi: 10.1371/journal.pone.0149934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prudencio A.S., Dicenta F., Martínez-Gómez P. Gene expression analysis of flower bud dormancy breaking in almond using RNA-Seq; Proceedings of the VII International Symposium on Almonds & Pistachios; Adelaida, Australia. 5–9 November 2017. [Google Scholar]
- 21.Fernández i Martí A., Gradziel T.M., Socias i Company R. Methylation of the Sf locus in almond is associated with S-RNase loss of function. Plant Mol. Biol. 2014;86:681–689. doi: 10.1007/s11103-014-0258-x. [DOI] [PubMed] [Google Scholar]
- 22.Fresnedo-Ramírez J., Chan H.M., Parfitt D.E., Crisosto C.H., Gradziel T.M. Genome-wide DNA-(de) methylation is associated with Noninfectious Bud-failure exhibition in Almond (Prunus dulcis [Mill.] D.A.Webb) Sci. Rep. 2017;7:42686. doi: 10.1038/srep42686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Gurp T.P., Wagemaker N.V., Wouters B., Vergeer P., Ouborg J.N., Werhoeben K.J.V. epiGBS: Reference-free reduced representation bisulfite sequencing. Nat. Meth. 2016;13:322–329. doi: 10.1038/nmeth.3763. [DOI] [PubMed] [Google Scholar]
- 24.Niederhuth C.E., Bewick A.J., Ji L. Widespread natural variation of DNA methylation within angiosperms. Gen. Biol. 2016;17:194. doi: 10.1186/s13059-016-1059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.How-Kit A., Emeline T., Deleuze J.F., Gallusci P. Locus-Specific DNA Methylation Analysis and Applications to Plants. In: Rajewsky N., Jurga S., Barciszewski J., editors. Plant Epigenetics. Springer; Berlin, Germany: 2017. pp. 303–328. [Google Scholar]
- 26.Kawakatsu T., Huang S.S., Jupe F. Epigenomic diversity in a global collection of Arabidopsis thaliana accesions. Cell. 2016;166:492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vining K.J., Pomraning K.R., Wilhelm L.J., Priest H.D., Pellegrini M., Mockler T.C. Dynamic DNA cytosine methylation in the Populus trichocarpa genome: Tissue-level variation and relationship to gene expression. BMC Gen. 2012;13:27. doi: 10.1186/1471-2164-13-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao W., Custard K.D., Brown R.C., Lemmon B.E., Harada J.J., Goldberg R.B., Fischer R.L. DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell. 2006;18:805–814. doi: 10.1105/tpc.105.038836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P.P., Montgomery T.A., Fahlgren N., Kasschau K.D., Nonogaki H., Carrington J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 30.Zhang G.-Z., Jin S.-H., Li P., Jiang X.-Y., Li Y.-J., Hou B.K. Ectopic expression of UGT84A2 delayed flowering by indole-3-butyric acid-mediated transcriptional repression of ARF6 and ARF8 genes in Arabidopsis. Plant Cell Rep. 2017;36:1995–2006. doi: 10.1007/s00299-017-2225-x. [DOI] [PubMed] [Google Scholar]
- 31.Bañuelos M.L.G., Moreno L.V., Winzerling J., Orozco J.A., Gardea A.A. Winter metabolism in deciduous trees: Mechanisms, genes and associated proteins. Rev. Fitotec. Mex. 2008;31:295–308. [Google Scholar]
- 32.Du D., Zhang Q., Cheng T., Pan H., Yang W., Sun L. Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol. Biol. Rep. 2013;40:1937–1946. doi: 10.1007/s11033-012-2250-3. [DOI] [PubMed] [Google Scholar]
- 33.Yamane H., Kashiwa Y., Kakehi E., Yonemori K., Mori H., Hayashi K., Iwamoto K., Tao R., Kataoka I. Differential expression of dehydrin in flower buds of two Japanese apricot cultivars requiring different chilling requirements for bud break. Tree Physiol. 2006;26:1559–1563. doi: 10.1093/treephys/26.12.1559. [DOI] [PubMed] [Google Scholar]
- 34.Fujiwara S., Oda A., Yoshida R., Niinuma K., Miyata K., Tomozoe Y., Tajima T., Nakagawa M., Hayashi K., Coupland G., et al. Circadian Clock Proteins LHY and CCA1 Regulate SVP Protein Accumulation to Control Flowering in Arabidopsis. Plant Cell. 2008;20:2960–2971. doi: 10.1105/tpc.108.061531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park B.S., Eo H.J., Jang I.-C., Kang H.-G., Song J.T., Seo H.S. Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem. Biophys. Res. Commun. 2010;398:242–246. doi: 10.1016/j.bbrc.2010.06.067. [DOI] [PubMed] [Google Scholar]
- 36.Park M.-J., Kwon Y.-J., Gil K.-E., Park C.-M. LATE ELONGATED HYPOCOTYL regulates photoperiodic flowering via the circadian clock in Arabidopsis. BMC Plant Biol. 2016;16:114. doi: 10.1186/s12870-016-0810-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Varriale A. DNA Methylation in Plants and Its Implications in development, Hybrid Vigour, and Evolution. In: Rajewsky N., Jurga S., Barciszewski J., editors. Plant Epigenetics. Springer; Berlin, Germany: 2017. pp. 263–280. [Google Scholar]
- 38.Viggiano L., de Pinto M.C. Dynamic DNA Methylation Patterns in Stress Response. In: Rajewsky N., Jurga S., Barciszewski J., editors. Plant Epigenetics. Springer; Berlin, Germany: 2017. pp. 281–302. [Google Scholar]
- 39.Garg R., Narayana Chevala V.V.S., Shankar R., Jain M. Divergent DNA methylation patterns associated with gene expression in rice cultivars with contrasting drought and salinity stress response. Sci. Rep. 2015;5:14922. doi: 10.1038/srep14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura S., Pourkheirandish M., Morishige H., Kubo Y., Nakamura M., Ichimura K., Seo S., Kanamori H., Wu J., Ando T., et al. Mitogen-Activated Protein Kinase Kinase 3 Regulates Seed Dormancy in Barley. Curr. Biol. 2016;26:775–781. doi: 10.1016/j.cub.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 41.Koonin E.V., Aravind L. Origin and evolution of eukaryotic apoptosis: The bacterial connection. Cell Death Differ. 2002;9:394. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- 42.Del Duca S., Serafini-Fracassini D., Cai G. Senescence and programmed cell death in plants: Polyamine action mediated by transglutaminase. Front. Plant Sci. 2014;5:120. doi: 10.3389/fpls.2014.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ebine K., Ueda T. Roles of membrane trafficking in plant cell wall dynamics. Front. Plant Sci. 2015;6:878. doi: 10.3389/fpls.2015.00878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S.J., Brandizzi F. The plant secretory pathway: An essential factory for building the plant cell wall. Plant Cell Physiol. 2014;55:687–693. doi: 10.1093/pcp/pct197. [DOI] [PubMed] [Google Scholar]
- 45.Rinne P.L.H., Welling A., Vahala J., Ripel L., Ruonala R., Kangasjärvi J., van der Schoot C. Chilling of Dormant Buds Hyperinduces FLOWERING LOCUS T and Recruits GA-Inducible 1,3-β-Glucanases to Reopen Signal Conduits and Release Dormancy in Populus. Plant Cell. 2011;23:130–146. doi: 10.1105/tpc.110.081307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Felipe A.J. Phenological states of almond; Proceedings of the Third GREMPA Colloquium; Bari, Italy. 3–7 October 1977; pp. 101–103. (In Italian) [Google Scholar]
- 47.Doyle J.J., Doyle M. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987;19:11–15. [Google Scholar]
- 48.Catchen J., Hohenlohe P.A., Bassham S., Amores A., Cresko W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013;22:3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krueger F., Andrews S.R. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akalin A., Kormaksson M., Li S., Garrett-Bakelman F.E., Figueroa M.E., Melnick A., Mason C.E. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13:R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verde I., Jenkins J., Dondini L., Micali S., Pagliarani G., Vendramin E. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Gen. 2017;18:225. doi: 10.1186/s12864-017-3606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu T.D., Watanabe C.K. GMAP: A genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 53.Tian T., Yue L., Hengyu Y., Qi Y., Xin Y., Zhou D., Wenying X., Zhen S. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017;45:W122–W129. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.