Abstract
Quorum sensing (QS) enables single-celled bacteria to communicate with chemical signals in order to synchronize group-level bacterial behavior. Pseudoalteromonas are marine bacteria found in versatile environments, of which QS regulation for their habitat adaptation is extremely fragmentary. To distinguish genes required for QS regulation in Pseudoalteromonas, comparative genomics was deployed to define the pan-genomics for twelve isolates and previously-sequenced genomes, of which acyl-homoserine lactone (AHL)-based QS traits were characterized. Additionally, transposon mutagenesis was used to identify the essential QS regulatory genes in the selected Pseudoalteromonas isolate. A remarkable feature showed that AHL-based colorization intensity of biosensors induced by Pseudoalteromonas most likely correlates with QS regulators genetic heterogeneity within the genus. This is supported by the relative expression levels of two of the main QS regulatory genes (luxO and rpoN) analyzed in representative Pseudoalteromonas isolates. Notably, comprehensive QS regulatory schema and the working model proposed in Pseudoalteromonas seem to phylogenetically include the network architectures derived from Escherichia coli, Pseudomonas, and Vibrio. Several associated genes were mapped by transposon mutagenesis. Among them, a right origin-binding protein-encoding gene (robp) was functionally identified as a positive QS regulatory gene. This gene lies on a genomic instable region and exists in the aforementioned bioinformatically recruited QS regulatory schema. The obtained data emphasize that the distinctly- and hierarchically-organized mechanisms probably target QS association in Pseudoalteromonas dynamic genomes, thus leading to bacterial ability to accommodate their adaption fitness and survival advantages.
Keywords: quorum sensing, Pseudoalteromonas, regulatory architecture, dissemination, horizontal transfer
1. Introduction
Newly characterized in 1995, the Pseudoalteromonas genus is ubiquitously present in marine environments, and is able to synthesize broad bioactive chemical molecules [1]. They are greatly versatile bacteria existing in diverse life habitats, as well as on animal and plant tissues, such as Pseudoallteromonas tunicate isolated from the sea lions, Pseudoalteromonas citrea isolated from sponges, and Pseudoalteromonas nigrifaciens isolated from mussels [2,3,4,5]. Broad life habitats suggest their versatile evolution in adaptation patterns. Pseudoalteromonas is a genus of ubiquitous marine bacteria typically recognized as model organism to explore the adaptation/fitness mechanisms under extreme conditions [6].
In fact, the genomes of several Pseudoalteromonas spp. have been sequenced, evidencing their genetic heterogeneity accounting for different adaptation capabilities [6,7]. Genomic analysis uncovered the genetic basis of Pseudoalteromonas flavipulchra JG1 in marine environment [1]. The potential fitness of Pseudoalteromonas haloplanktis TAC125 to cold environments was elucidated by genome sequencing [8]. The comparative genomics analysis on Pseudoalteromonas tunicata D2 has revealed the adaptation mechanism of its epiphytic life on algae surface [9]. Whole genome sequencing, together with comparative genomics analysis within Pseudoalteromonas sp. SM9913 and P. haloplanktis TAC125, have illustrated the adaption of Pseudoalteromonas to marine sediment environments as well [10].
Diverse studies have proven that bacteria use quorum sensing (QS) to coordinate the population behaviors through cellular information exchange for adaption/fitness to complex and versatile environments [11]. QS is the unique process by which bacteria produce, release, and detect specific chemical signal molecules, and successively alter the expression of specific genes to maintain population replication/fitness for survival benefit [12]. By QS-related strategy, Pseudomonas aeruginosa alters adaptation abilities in different external environments [13,14]. QS is very important for surface- and biofilm-living bacteria that often reach high densities of microbiota [15]. QS is involved not only in the production of virulence factor and biofilm formation, but also bioluminescence, sporulation, motility, and antibiotic production [11,12,13,14,15,16].
First discovered in marine Vibrio fischeri, QS has now been determined in both Gram-positive and Gram-negative bacteria [16]. However, very few reports have dealt with QS in Pseudoalteromonas. It was found that QS is probably involved in the violacein production in Pseudoalteromonas [17,18]. The putative promoter regions of gene clusters induced by population-dependent signaling molecules were mapped in violacein-producing Pseudoalteromonas sp. 520P1 [19]. QS-related swimming motility and swarming motility of Pseudoalteromonas sp. SM9913 provided a mechanistic insight into its multifaceted lifestyle in deep-sea sediment [20]. Biofilm formation in Pseudoalteromonas ulvae TC14 was found to be influenced by heterogenous QS regulatory molecules [21].
Previous studies on QS in Pseudoalteromonas were merely focused on the identification of signal molecules and their cognate genes, together with QS regulation on bacterial physiology [22]. Herein, we explored the correlation between environmental adaptation potential and QS architecture from the 12 sequenced Pseudoalteromonas isolates. We also reported transposon (mini-Tn10) mutagenesis to identify QS-related regulators in Pseudoalteromonas. Several QS regulatory genes were successfully mapped, and right origin-binding protein-encoding gene (robp) was demonstrated to serve as a positive QS regulatory gene that localizes on a genomic instable region. The obtained results provide a better understanding of QS regulation in Pseudoalteromonas, highlighting the importance of horizontal gene transfer in their adaptation capacities.
2. Results
2.1. Characterization of Quorum Sensing Related Colorization Derived from Pseudoalteromonas Isolates
Based on 16S rRNA gene sequencing, the taxonomy of total bacteria isolated from the Mangrove forest sediments in Ximen Island (28°20′33′′N, 121°10′28′′E) of Zhejiang Province was determined. 16S rRNA sequence comparison indicated that the 12 isolates belong to Pseudoalteromonas (Table 1). AHLs-related colorization was then tested by cross-feeding bioassay using A. tumefaciens A136 as a biosensor [23] on a LB-2216E plate. The results in Table 1 showed that 11 out of 12 isolates result in the colorization of A136, and different isolates exhibit distinguished colorization intensities, indicating diverse capabilities of AHLs production. T1lg65 yields the strongest colorization, whereas T1lg88 does not result in any visualizable colorization. Based on these scenarios, we speculate that genetic patterns and genomic discrepancy among different Pseudoalteromonas isolates probably influence their traits in producing QS signals. Data in Supplementary Figure S1 indicated that there is no direct correlation between the intensity of QS signals and biofilm formation or motility within these 12 isolates.
Table 1.
Colony morphology and cross-feeding bioassay for AHL-based quorum sensing phenomenon of Pseudoalteromonas*.
| Strain | Similarity (%) | 16S RNA Top-Hit Strain | Colony | Color | Size ** | Morphology | Colorization Intensity *** | |
|---|---|---|---|---|---|---|---|---|
| T1lg10 | 97.80 | P. tetraodonis IAM14160 | moist | white | big | round | + | |
| T1lg21 | 98.73 | P. shioyasakiensis SE3 | moist | white | big | round | +++ | |
| T1lg22 | 97.92 | P. shioyasakiensis SE3 | moist | white | big | round | + | |
| T1lg23Z | 99.43 | P. byunsanensis FR1199 | moist | purple | small | round | ++ | |
| T1lg23B | 99.22 | P. byunsanensis FR1199 | moist | white | small | round | ++ | |
| T1lg24 | 99.71 | P. spongiae UST010723-006 | moist | orange | quite small | round | + | |
| T1lg48 | 97.82 | P. aestuariivivensDB-2 | moist | white | big | round | +++ | |
| T1lg65 | 99.15 | P. elyakovii ATCC 700519 | moist | yellow | small | round | ++++ | |
| T1lg75 | 97.91 | P. robra ATCC 29570 | moist | white | big | round | + | |
| T1lg76 | 97.92 | P. tetraodonis IAM14160 | moist | white | big | round | ++ | |
| T1lg88 | 97.79 | P. aestuariivivens DB-2 | moist | white | big | round | - | |
| T1lg122 | 97.92 | P. aestuariivivens DB-2 | moist | white | big | round | ++ | |
* This table reports the main features of Pseudoalteromonasphenotypic characteristics assayed after 12 h culture. ** Relative size. *** +: colorization intensity from biosensor strain A136 which was used to sense the QS signal molecules of AHLs with acyl side chain length from C6 to C14.
2.2. Genomic Quality Control and Diversified Sequence Features
The genomes of the 12 Pseudoalteromonas isolates were then sequenced and analyzed. It was found that genomic size, protein-coding gene numbers, and G+C contents significantly vary in the 12 Pseudoalteromonas isolates (Table 2), consistent with a previous report [24] reflecting genomic heterogeneity.
Table 2.
Genomic characteristics of twelve Pseudoalteromonas isolates in this study.
| Strain | Contig | Length (bp) | Gene Number | (G+C)% | WGS Accession Number | NCBI BioProject | tRNA | rRNA | tmRNA | CDS * | Repeat Region | Pigmen-Tation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1lg10 | 57 | 3,430,516 | 3128 | 49.46 | PQBV00000000 | PRJNA430922 | 82 | 1 | 1 | 3039 | 1 | NO |
| T1lg21 | 90 | 4,664,131 | 4298 | 41.33 | PQBX00000000 | PRJNA430929 | 88 | 10 | 1 | 4199 | NO | |
| T1lg22 | 42 | 3,429,413 | 3155 | 49.43 | PQBY00000000 | PRJNA430930 | 82 | 5 | 1 | 3078 | NO | |
| T1lg23B | 55 | 4,584,947 | 4063 | 43.24 | PQBZ00000000 | PRJNA430934 | 85 | 6 | 1 | 3961 | NO | |
| T1lg23Z | 54 | 4,583,886 | 4039 | 43.23 | PQCA00000000 | PRJNA430936 | 86 | 8 | 1 | 3950 | YES | |
| T1lg24 | 40 | 4,692,206 | 4241 | 40.83 | PQCB00000000 | PRJNA430938 | 73 | 4 | 1 | 4163 | YES | |
| T1lg48 | 24 | 3,589,059 | 3293 | 49.89 | PQCC00000000 | PRJNA430939 | 82 | 6 | 1 | 3206 | NO | |
| T1lg65 | 3683 ** | 6,875,118 | 6145 | 46.49 | PQBW00000000 | PRJNA430940 | 143 | 29 | - | 6027 | 16 | YES |
| T1lg75 | 78 | 3,654,576 | 3317 | 50.13 | PQCD00000000 | PRJNA430941 | 95 | 7 | 2 | 3213 | NO | |
| T1lg76 | 131 | 3,378,987 | 3084 | 49.63 | PQCE00000000 | PRJNA430942 | 97 | 7 | 1 | 2981 | 1 | NO |
| T1lg88 | 42 | 3,400,396 | 3097 | 49.49 | PQCF00000000 | PRJNA430943 | 71 | 9 | 1 | 3019 | NO | |
| T1lg122 | 47 | 4,592,438 | 4184 | 41.4 | PQCG00000000 | PRJNA430944 | 91 | 7 | 1 | 4094 | NO |
* CDS: protein coding sequences. ** a number of sequencing replicates to acquire more quality of the T1lg65 genome.
The identified regulators listed in Supplementary Table S1 further show that the 12 Pseudoalteromonas isolates have significant genomic heterogeneity. Given that T1lg65 facilitates the strongest colorization of A136, T1lg48 moderate colorization, T1lg24 weak colorization, and T1lg88 no colorization (Table 1), the comparative genomics of these Pseudoalteromonas isolates was performed using Pseudoalteromonas sp. SM9913 as control. The correlation between genomic heterogeneity and QS trait was accordingly exploited. The Venn diagram in Supplementary Figure S2 indicated that the non-conserved and specific genes in Pseudoalteromonas isolates with different QS traits are significantly different. For example, 1400 genes in T1lg65 are unique, whereas 412 genes in T1lg88 are specific.
Additionally, regulatory protein family classification was also performed to further investigate genetic patterns and QS-associated genes. The COG categories within T1lg65 and T1lg88 with opposite QS traits were determined by using SM9913 as control (Supplementary Figure S3). T1lg65 and T1lg88 are different in genomic size. Additionally, the orthologous proteins pertaining to COG ‘T’ and ‘J’, related to signal transduction mechanisms, and translation, ribosomal structure, and biogenesis, respectively, are obviously different in amount. The number of non-conserved sequences of ‘T’ in T1lg65 is higher than that in both T1lg88 and SM9913. Meanwhile, the number of conserved sequences of ‘J’ is high in all three representatives. The number of non-conserved sequences of ‘J’ in T1lg65 is higher than that in both T1lg88 and SM9913. All these data suggest a correlation between genomic heterogeneity and QS trait.
2.3. Dissemination of Regulatory Factors within Specific Genetic Regions in Pseudoalteromonas
In order to determine the detailed QS system in Pseudoalteromonas, over 200 references were collected from ‘PubMed’ and ‘Google Scholar’ using the search terms ‘Quorum sensing’ and ‘Quorum sensing & Pseudoalteromonas’, and by capturing QS-related citations within these academic sources (Supplementary Table S1). All references were manually curated for details of QSs and/or their regulation targets. Overall, 67 QS regulators were collected from the following experimentally-validated information and KEGG-provided retrievals. Our studies also included the relevant data on regulation factors which have been well studied, such as the RetS/LadS regulatory system of Pseudomonas and LuxR-based sensing mechanisms in Vibrio. Then, the conserved domains were carefully curated using COG-based RPS-BLAST [25] and Pfam-based HMMER [26] programs to acquire their conserved protein domains; thereby, 27 assignments were provided to 67 regulatory elements (22/27 present in Pseudoalteromonas) (Figure 1). Lastly, the regulatory targets were collected and the activator/repressor information was accommodated in order to acquire and encompass a reservoir of the corresponding genetic contexts (Supplementary Table S1).
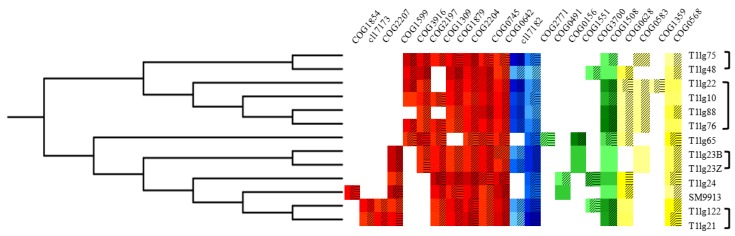
Figure 1.
Heatmap schema of quorum sensing regulator dissemination in selected Pseudoalteromonas genomes. The left panel of the heatmap is the evolutionary tree. The middle of the heatmap denotes the distribution of QS regulatory protein families in thirteen Pseudoalteromonas isolates. The top of the heatmap corresponds to the protein family IDs. Red: positive regulation; blue: both positive and negative regulation; green: negative regulation; yellow: unknown regulation direction. The color intensity represents the relative amount of the nucleotide base numbers of the genes containing cognate protein family (heavy: more similarity; moderate: intermediate similarity; light: less similarity). A range of BLAST hits were found to be classified into the same protein family. Notably, identified conserved and diverse genes are labeled with matching signatures; white: no hits; oblique stripes: relatively conservative; horizontal stripes: absolutely specific; others: absolutely conservative.
In accordance with Pseudoalteromonas phylogeny described using CVTree3, QS positive and negative regulator disseminations were found to exhibit comparable conservation (Figure 1). Interestingly, phylogenetic neighbors likely exhibit similar QS regulation patterns. Additionally, strains carrying stronger QS-based traits commonly exhibit similar QS regulatory architectures. The observation from T1lg65 and T1lg88 showed that QS-associated traits are related to regulator numbers and their disseminations. A variety of regulators were also found to localize on ‘genomic instable regions’ (shadow in blocks in Figure 1, indicating uniqueness in these strains). Lastly, Pseudoalteromonas heterogeneity fully indicated that regulators pertaining to different clades could simultaneously and/or sequentially influence QS-associated traits.
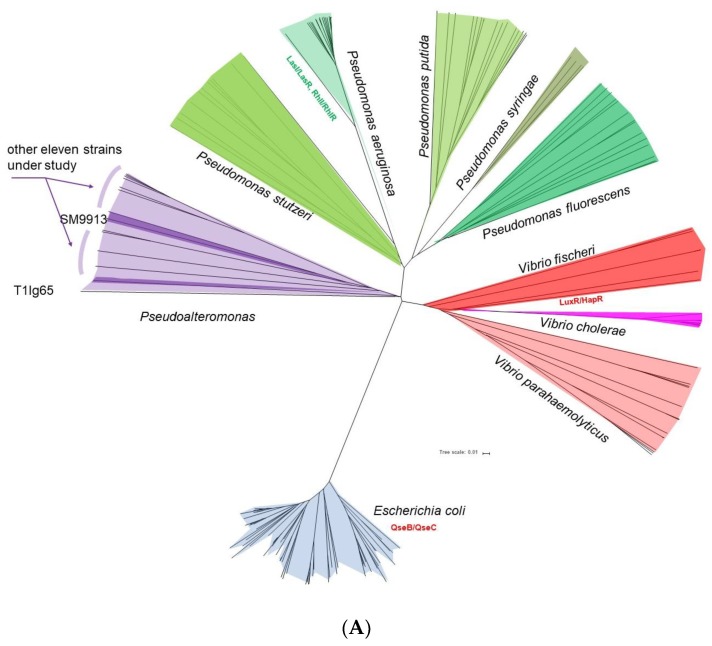
Besides the AHL-based colorization of A136 (Table 1), the inferred clade and candidate regulation factors in T1lg65 are obviously dissimilar to those from other isolates (Figure 1). Therefore, all identified QS genes of T1lg65 were reciprocally aligned with the other 12 Pseudoalteromonas genomic sequences. QS-related genes in T1lg65 were classified into three groups (Supplementary Table S2): absolutely conserved genes well-matched with other 12 genome sequences, non-conserved genes partially aligned, and completely unique genes unaligned with any other sequenced genomes (Supplementary Table S2). Similar results were obtained in T1lg88 as well (Supplementary Table S2). Moreover, the results denoted that most of QS related genes in T1lg65 are specific genes compared to other Pseudoalteromonas isolates, which is consistent with acquired adaption/survival fitness from horizontal genetic transfer events [27]. Phylogeny of the 12 Pseudoalteromonas strains in this work and representatives of Pseudomonas, Vibrio, and Escherichia coli implied that Pseudoalteromonas species likely emerge from the fascinating transition modulation form among a wide range of QS regulon representatives (Figure 2A). However, the T1lg65 clade is distinctly related to other isolates (Figure 2B).
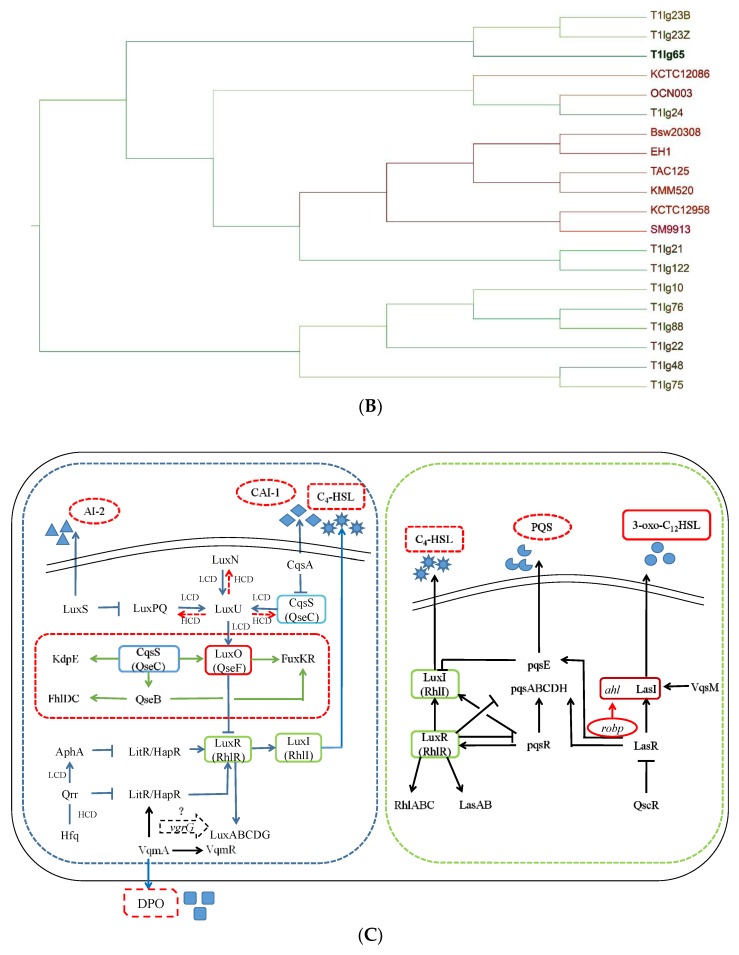
Figure 2.
Evolutionary relationship between bacterial representatives with well-studied quorum sensing regulon and quorum sensing working model proposed from Pseudoalteromonas. (A) Schematic presentation of evolutionary relationship among Pseudoaltermonas, Vibrio, Pseudomonas, and Escherichia coli based on phylogenetic analysis of concatenated protein-coding sequences. Representative QS regulon in each clade was indicated if applicable: LuxR/HapR in V.choleraeV52 and V. parahaemolyticus RIMD 2210633; QseB/QseC in E. coli O157:H7; LasI/LasR in P. aeruginosa PAO1, P. putida KT2440 and P. syringaepv. tomato strain DC3000. (B) Pseudoalteromonas phylogeny. Fully-sequenced Pseudoalteromonas strains in this phylogeny: Pseudoalteromonas phenolica KCTC 12086: chr1 (CP013187), chr2 (CP013188); Pseudoalteromonas sp. OCN003: chr1 (CP009888), chr2 (CP009889); Pseudoalteromonas sp. Bsw20308: chr1 (CP013138), chr2 (CP013139); Pseudoalteromonas aliena EH1: chr (CP019628); Pseudoalteromonas haloplanktis TAC125: chr1 (CR954246), chr2 (CR954247); Pseudoalteromonas translucida KMM 520:chr1 (CP011034), chr2 (CP011035); Pseudoalteromonas issachenkonii KCTC 12958: chr1 (CP013350); chr2 (CP013351); Pseudoalteromonas sp. SM9913: chr1 (CP001796); chr2 (CP001797). (C) Quorum sensing working model proposed from Pseudoalteromonas. The blue panel on the left in the working model schematics shows the QS system in Vibrio [11,12,33,34,35,36,37,38,39], whereas the red panel for that of E. coli [12,33]. The large light green dotted box presents the QS regulators in Pseudomonas aeruginosa [40,41,42,43]. Arrows indicate positive regulation (activation) and T-bars indicate negative regulation (inhibition). Red dotted arrows indicate positive regulation at high cell densities. Triangle represents signal molecule AI-2, diamond represents signal molecule CAI-1, and polygon represents signal molecule C4-HSL, together with 3/4 donut and droplet indicating signal molecule PQS and 3-oxo-C12-HSL, respectively (molecules with dotted line indicating non-AHL signals). Box represents signal molecule DPO (3,5-dimethylpyrazin-2-ol).
QS-related colorization in T1lg65, T1lg88, and T1lg24 was examined by using cross-feeding bioassay. The results in Supplementary Figure S4A showed that T1lg65 and T1lg88 respectively exhibit the strongest and the weakest QS-dependent colorization of A136, and T1lg24 has colorization comparable to that of T1lg65. Subsequently, two important regulator genes, luxO and rpoN, respectively considered as positive and negative regulator genes in Vibrio and Pseudomonas [28,29,30,31,32], were chosen for analysis of relative expression level in these three Pseudoalteromonas isolates. Supplementary Figure S4B and Supplementary Figure S4C showed that the relative expression of both luxO and rpoN in three isolates is obviously different. The relative expression levels of luxO are the highest, the lowest, and moderate in T1lg65, T1lg88, and T1lg24, respectively, consistent with the QS-based colorization traits (Supplementary Figure S4A). Most probably, LuxO positively regulates the QS trait in Pseudoalteromonas as well. In contrast, the overall expression levels of rpoN are the lowest, the highest, and moderate in T1lg65, T1lg88, and T1lg24, respectively, which is opposite to QS-based colorization (Supplementary Figure S4A). Most probably, RpoN also negatively regulates QS trait in Pseudoalteromonas. Our data collectively demonstrate that both LuxO and RpoN play similar roles in regulating the QS system in Pseudoalteromonas as those in Vibrio and Pseudomonas [30]. Moreover, our survey further reveals that the QS trait correlates with genetic heterogeneity in Pseudoalteromonas, which is implicated in their phylogenetics (Figure 1 and Figure 2A).
2.4. Complicated Intermediate Quorum Sensing Schema in Pseudoalteromonas
Comparative genomics was performed to acquire the genetic contexts of QS regulators, including their position in the regulatory interaction and conserved enzymatic domains of proteinaceous regulators. Several regulators were identified by BLAST search carrying the same functional conserved domains, such as LuxO:QseF [protein family ID: COG2204] and CqsS:QseC [protein family ID: COG0642] (Supplementary Table S1 and Figure 2C). The collected information was systemically curated by text mining and comparative genomics.
As a result, a QS working model in Pseudoalteromonas was proposed, and such a schematic module sufficiently reflects the complexity of QS regulation in Pseudoalteromonas (Figure 2C). The current status of prior knowledge and data mining on these elements reveals a complex bacterial lifestyle. The scheme likely consists of three independent and cross-linked QS regulation networks related to those found in E. coli, Pseudomonas, and Vibrio. Four characteristics were presented by this model: (1) there are more regulators than any other known QS networks; (2) a variety of chemical molecules can be sensed by receptor proteins that, in turn, reflects cell density; (3) sRNA-like and proteinaceous regulators are included, accounting for this complicated system; (4) surprisingly, such diversified systems seem to exhibit in considerably perfect order. Interestingly, the interconnected regulation patterns under high and low cell density were clearly presented. For example, LuxN positively regulates LuxU at low cell densities, and LuxU activates LuxQ at high cell densities. Therefore, LuxO-, LuxR-, and LuxI-like regulation factors are characterized as unique nodes of ‘intermediate’ regulation status for insight into Pseudoalteromonas adaptive cascades.
2.5. Expanding the Proposed Schema via Identification of robp Essentiality for QS-Dependent Phenomenon
To acquire essential genes accounting for the highly enhanced QS-related colorization, the Tn-seq-based pipelines were performed in strain T1lg65. In total, 14 mutants with null AHL-based colorization of A136 due to mini-Tn10 insertion were identified (Supplementary Table S3). Overall, these mutants have similar patterns of cell growth as the wild-type, e.g., slow growth in the initial stage and fast growth in the middle stage, followed by slower growth in the later stage (Supplementary Figure S5). On the other hand, the disruption of genes causes differences in cell growth, such as the ompA::Tn mutant giving a slower growth and a lower final cell density than WT, and the dmcp::Tn mutant yielding a higher cell density than WT from 5 h. In general, all mutant strains show slower growth in the first 4–5 h, compared to WT (Supplementary Figure S5). Also, the disruption of genes (Supplementary Table S3) causes changes in biofilm formation and motility (Supplementary Figure S5). Even if there are no statistically-significant differences in biofilm formation ability, there is a clear trend. All mutants have lower abilities to form biofilms. Additionally, five mutants, i.e., rhtA::Tn, marR::Tn, robp::Tn, dmcp::Tn and capB::Tn lose abilities in terms of swimming motility (Supplementary Figure S5). Among identified genes from 14 mutants (Supplementary Table S3), robp is considered as a candidate gene for positive regulation of QS signal formation in T1lg65 (Supplementary Figure S6), since partial robp sequence-encoded protein domain belongs to COG2207-like family.
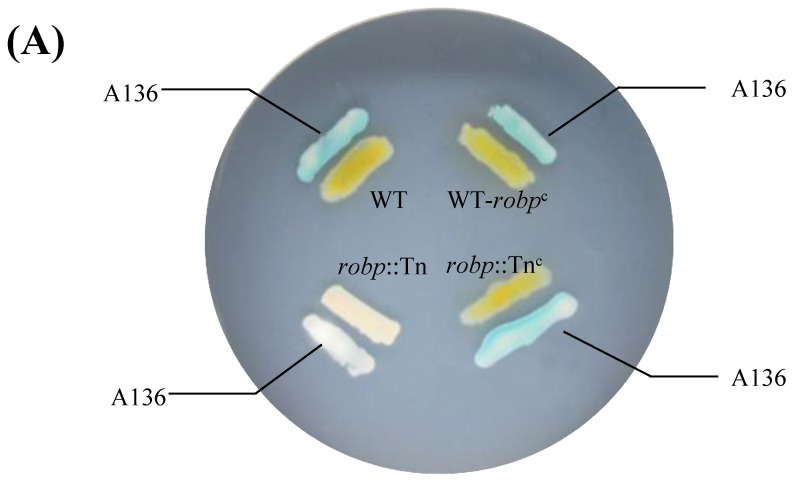
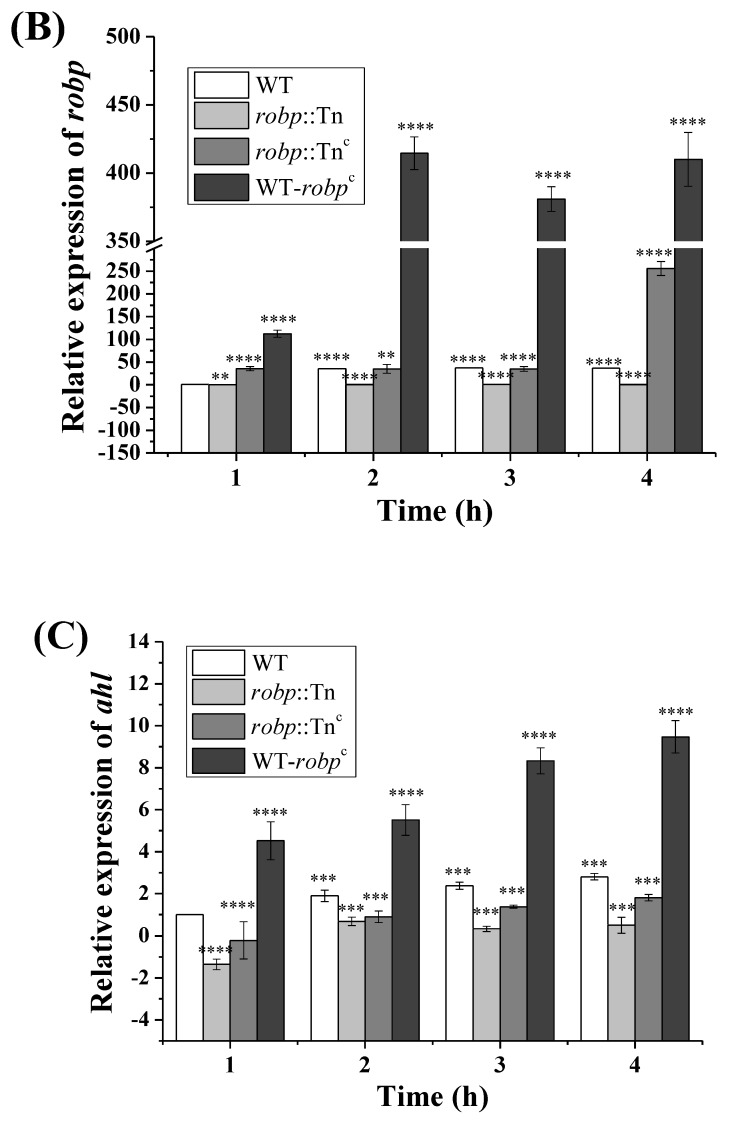
In order to demonstrate the robp role to QS trait in T1lg65, four strains were prepared: wild-type (WT), mutant (robp::Tn, disruption of robp gene in strain T1lg65 by Tn10), complon (robp::Tnc, complementation of robp in robp::Tn), and overexpressor (WT+robpc, overexpression of robp in strain T1lg65). The growth and QS phenotype in WT, robp::Tn, robp::Tnc and WT+robpc were accordingly compared. Figure 3A showed that the mutant (robp::Tn) yielded null colorization of A136. In contrast, the complon (robp::Tnc) generated almost completely restored colorization of A136, which is comparable to that from WT and overexpressor (WT+robpc). Our data further showed that the relative expression levels of both robp gene (Figure 3B) and ahl gene (Figure 3C) encoding acyl-homoserine lactone synthase (LasI in Figure 2) are overall high in WT+robpc, low in robp::Tn, and moderate in WT or robp::Tnc, indicating that robp is involved in positive regulation of QS signal formation in Pseudoalteromonas. Therefore, our bioinformatic analysis (Figure 1) and transposon mutagenesis mapping (Figure 3) congruously demonstrate that Robp serves as a positive regulator for QS system in Pseudoalteromonas.
Figure 3.
Growth profile and QS-related phenotype in wild-type, mutant, complicon and overexpressor. (A) Growth and QS-based colorization of biosensor A136; (B) Relative expression level of robp gene encoding right origin-binding protein; (C) Relative expression level of ahl gene encoding acyl homoserine lactone synthase. WT: wild-type (strain T1lg65); robp::Tn: mutant of strain T1lg65 with disrupted robp gene by Tn10; robp::Tnc: complon of robp::Tn with complementation of robp; WT+robpc: overexpressor of strain T1lg65 with overexpression of robp. All independent experiments were repeated in triplicate. Significant difference is respectively indicated by asterisks as follows: *** p < 0.001; **** p < 0.0001. Gene expression in WT at 1h was treated as 1 (internal reference).
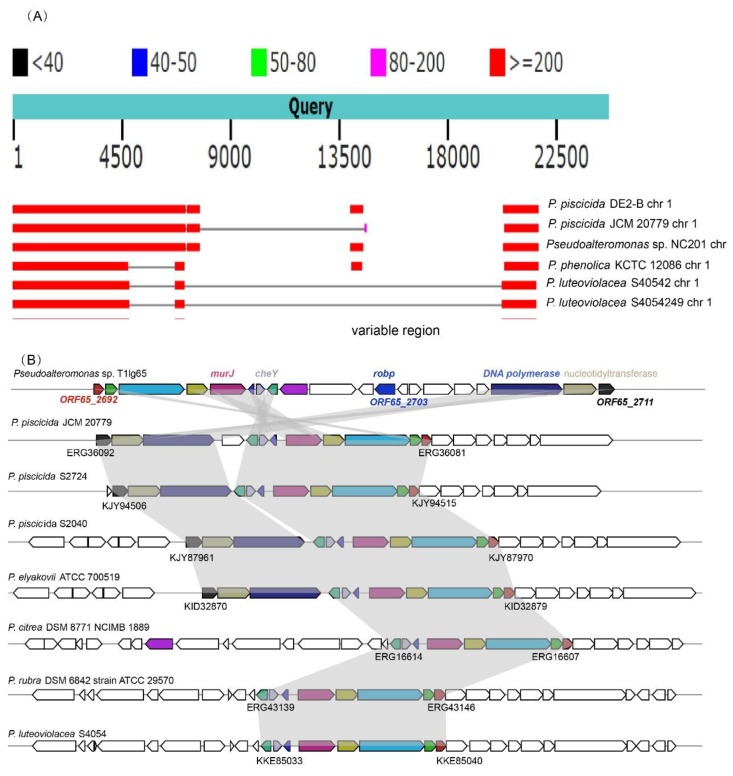
Intriguingly, our further analysis in Figure 4A showed that robp and its proximity localize on a large region of plasticity (labeled as ‘robp island’ in this work) [44] in Pseudoalteromonas. Of note, acquisition/loss of the ‘robp island’ needs to be further clarified, since no other homologs were detected in other our sequenced genomes. Furthermore, the intact ‘robp island’ is also absent in RefSeq-archived sequences (Figure 4B). Such findings greatly outline the efficacy and important roles of horizontally acquired robp facilitating AHL-based QS (Supplementary Figure S6). Supplementary Figure S7 further shows that the backbone of genomes between T1lg65 and ATCC 700519 is nearly identical, but their genomic sizes and genetic architecture are different. In addition, no ‘robp island’ was found in ATCC 700519, indicating individual characteristics. Interestingly, no identical homolog was found in other strains. However, a variety of homologs (>1000) with <60% identities were predicted. They commonly contain AraC-like regulatory domains. For instance, Robp has some similarity to VqsM [45] in P. aeruginosa PAO1. These two protein sequences both include COG2207- or AraC-like domains (Supplementary Table S1). However, another domain is obviously different (Robp: GryI; VqsM: Arabinose_bd). In addition, the position of AraC encoding fragment in their sequences are different as well. This finding expands a wider landscape of functionality for AraC family regulators on quorum sensing.
Figure 4.
Region of genomic plasticity in the context of robp. (A) NCBI-BLAST outputs against RefSeq-archived sequences as of Jan, 2018 suggest that robp and neighboring genes could constitute a candidate genetic region of variability in Pseudoalteromonas strains, but its up-/down-stream genomic surroundings are comparably conserved; (B) The synteny alignment scheme indicates that robp is located in a region of genomic plasticity, indicating robp and its vicinity acquired by isolate T1lg65 are probably resulted from a series of genomic transposition and rearrangement. Genes coding for DNA polymerase, nucleotidyltransferase and Chemotaxis Y (CheY) protein are found to exist immediately adjacent to the ‘robp island’. Schematic diagrams of robp and its vicinity are drawn to scale.
3. Discussion
Pseudoalteromonas species are commonly isolated from different marine environments, such as the Arctic, California, Antarctica, and China’s Chukchi Sea. The ORF number of previously-reported Pseudoalteromonas genomes ranges from 3612 to 5012, with an average G+C content of 41% (generally ranging from 38% to 47%, data from NCBI). It has been reported that the morphological traits of Pseudoalteromonas are closely correlated with production/category of QS signal molecule [1,20,22]. In this study, QS-related traits and genome sequences of the 12 Pseudoalteromonas isolates were characterized in order to explore the association between individual heterogeneity and population behavior. It was found that their morphological behavior is different (Table 1 and Supplementary Figure S1). In addition, the 12 Pseudoalteromonas isolates are also different in ORF number and G+C content (Table 2). The genomic scale emphasizes a potential relationship between genomic variation and ecological population behavior in Pseudoalteromonas.
The regulatory schema (Figure 1 and Figure 2) indicated that diverse and elaborate interplays exist in QS regulatory system of Pseudoalteromonas, including regulator number and dissemination conservation/diversity. The presence of QS genes in the Pseudoalteromonas isolates varies greatly. The copy number variation and transmission of QS-related genes mainly constitute the genetic description of such high complexity. Among the 12 Pseudoalteromonas isolates, T1lg65 and T1lg88 exhibit the strongest and null AHL-based colorization, respectively. One plausible explanation is that T1lg65 and T1lg88 harbor more and less QS-related specific genes, respectively (Supplementary Table S2). Therefore, QS in Pseudoalteromonas probably correlates with genomic heterogeneity.
Pseudoalteromonas has previously been phylogenetically related with Vibrio and Pseudomonas. Our comparative genomics analysis dissects that the QS-related scheme and working model in Pseudoalteromonas (Figure 1 and Figure 2) are more complicate than that in Vibrio, Pseudomonas, and Escherichia coli [14,33,46,47]. QS in Pseudoalteromonas is likely to be a transitional form comprised of that in Vibrio and Pseudomonas. Pseudoalteromonas are commonly considered to have special metabolic capacities [48,49]. Metabolic regulation in Pseudoalteromonas, as well as cell individual growth/replication, is related to cell density [9]. Pseudoalteromonas species seem to achieve elaborate cell physiology through a sophisticated QS regulation system, and thus, facilitate adaption advantages to various complex niches. Emerging studies on non-AHL signal molecules are required to provide insights into QS-related lifestyle and persistence/fitness of Pseudoalteromonas.
Diversified QS systems in Pseudoalteromonas (Figure 1 and Figure 2) could bridge the huge knowledge gap between Pseudoalteromonas ‘mobilome’ and physiology-associated traits. Sixteen repeat regions were found in the T1lg65 genome, and this finding was not observed in other selected strains. Repeat regions could be closely related to horizontal gene transfer and genetic duplication-driven overexpression. Our transposon mutagenesis has mapped 14 QS associated genes, including robp and the type-VI secretion system associated vgrG [50,51] (Supplementary Table S3). However, only robp was found to share similarity to the bioinformatically-recruited, COG2207-like family, with positive regulatory role in QS system of Pseudoalteromonas (Figure 4), while the statuses of the remaining thirteen genes (Supplementary Table S3) are yet to be clarified, pointing out that QS system in Pseudoalteromonas is more complicated than what was originally thought. The robp flanking genes localize on a large region of plasticity (‘robp island’: potential implications for the spread of robp) in T1lg65 (Figure 4A) and no other homologs were detected in other Pseudoalteromonas genomes (Figure 4B), emphasizing the efficacy and important roles of horizontally-acquired robp facilitating AHL-based QS (Supplementary Figure S6). The OmpA family proteins are closely related to diversified cell communication, such as peptidoglycan-binding function and bacterial protective efficacy against adversity [51,52]. In addition, a putative capB-encoded capsular protein responds for bacterial invasion ability in Francisella [53]. It is possible that they both also contribute to QS-driven changes. Therefore, the newly-identified QS-associated patterns involving genetic conservation and variation significantly extend the repository of diversified mobilome-bearing adaptive and auxiliary determinants in Pseudoalteromonas.
4. Methods
4.1. Strains, Plasmids and Media
Zobell 2216E medium (sea salt 30g/L, tryptone 5 g/L, yeast extract 1 g/L, pH 7.6~7.8, agar 1.5~2% for solid) was used to culture the 12Pseudoalteromonasisolates collected from the Mangrove forest sediments in Ximen Island (28°20′33′′N, 121°10′28′′E), Zhejiang Province, China. Unless otherwise stated, Pseudoalteromonas isolates were stored in our lab at Zhejiang University of Technology and grew at 28°C overnight. In addition of morphological traits (Table 1), the taxonomy of all Pseudoalteromonas isolates was characterized based on the comparison of 16S rRNA gene sequence between 27F:5′AGAGTTTGATCCTGGCTCAG3′ and 1492R: 5′GGTTACCTTGTTACGACTT3′ (Table 1). All Escherichia coli and plasmids used in this study were listed in Supplementary Table S3. Unless otherwise specified, E. Coli cells grew in Luria-Bertani (LB) medium (NaCl 10 g/L, tryptone 10 g/L, yeast extract 5 g/L, agar 1.5~2% for solid) at 37°C overnight. If necessary, antibiotics were added to final concentrations as below: ampicillin (Amp) 100 μg/mL, tetracycline (Tet) 40 μg/mL, gentamycin (Gm) 50 μg/mL and kanamycin (Km) 50 μg/mL.
4.2. Cross-Feeding Bioassay
AHL-based QS traits of Pseudoalteromonas isolates were characterized by cross-feeding bioassay [54,55]. Ensiferadhaerens X097 was used as positive control in cross-feeding bioassay because it can produce AHLs, resulting in a blue color of indicator/biosensor Agrobacterium tumefaciens A136 [56]. A136 carries a fused lacZ-traI. AHLs with acyl side chain length from C6 to C14 can induce the expression of lacZ in A136 and 5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside (X-gal) will be accordingly hydrolyzed to generate a blue color [54].
Of note, A136 grows well on the LB medium, but not on the Zobell 2216E medium, whereas Pseudoalteromonas isolates grow well on Zobell 2216E medium. Therefore, cross-feeding bioassay for QS trait of Pseudoalteromonas isolates was performed on solid LB-2216E medium (obtained by mixing equal amounts of LB and Zobell 2216E) [54,57]. In general, after cultivation at 30°C for 12 h, colorization intensity of A136 [55] was recorded to describe the AHL-driven QS trait of Pseudoalteromonas isolates.
4.3. Genomic Sequencing and Assembly/Annotation
Single colony was picked from 2216E plate, and then transferred into shaking flask for overnight cultivation. Next, genomic DNA was extracted using a bacterial genomic DNA extraction kit (GE, Shanghai, China) for sequencing (Illumina Hi-Seq 2500 system, Vazyme Biotech Co.,Ltd, Nanjin, China). Lower quality sequencing data were filtered for accurate assembly [58]. The reads with average 500-bp were assembled using Velvet short sequence assembly software based on the de-Bruijn algorithm [59].
PROKKA [60] was used for bacterial gene prediction. The protein sequences of predicted genes were compared with NR [61], Swiss/TrEMBL [62], COG [63], and GO [64] databases to obtain the annotated information. Genomic rRNA and tRNA were predicted by using RNAmmer-1.2 [65] and tRNAscan-SE [66], respectively.
4.4. Comparative Genomics and Pan-Genomic Characterization
All-against-all BLASTn-based alignment was performed to define the core genomes. Conserved and auxiliary genes were characterized by given cut-offs (BLASTn identity ≥ 25, E-value ≤ 0.01 and matching length ≥ 100 bp). The core proteome phylogeny of the 12 Pseudoalteromonas isolates and previously-sequenced Pseudoalteromonas sp. SM9913 were therefore calculated and inferred by using CVTree3 [67]. To further illustrate the gene pools of Pseudoalteromonas species, the VENN scheme was provided to describe the relationship of selected distantly-related Pseudoalteromonas isolates.
4.5. Quorum Sensing Regulatory and Functional Gene Classification
Manual literature mining for QS regulators and their targets were performed with a range of associated keywords. To obtain accurate and detailed regulatory repertoire, related life process information from collected literatures was extracted and collated. In summary, four categories of regulatory data were acquired: (1) regulation factors, (2) downstream binding target, (3) successive activation/repression, and (4) nucleotide and protein sequence if available.
4.6. Transposon Mutagenesis
To create a mutant library with altered AHL-based colorization, transposon mutagenesis [68,69,70] was applied in tetracycline-resistant Pseudoalteromonas sp. T1lg65 with the strongest AHL-based colorization of A136 among isolates (Table 1). First, strain T1lg65 was cultured in 2216E broth. Meanwhile, E. coli S17-1(λpir) carrying suicide vector pLOF/Km with mini-Tn10 was incubated in LB broth. After overnight growth, each culture was inoculated into fresh medium without any antibiotics, and reached an exponential growth phase. Second, 50 μL of T1lg65 receptor cells were spotted onto LB-2216E. After slight drying, another 50 μL of S17-1(λpir) donor cells were added onto T1lg65 cells. Negative controls with only S17-1(λpir) or T1lg65 were also prepared. After overnight conjugation, the cells were scraped and suspended in 1 mL of 2216E. After appropriate dilution, the cell mixture was spread onto 2216E with antibiotics for incubation. Finally, the target mutants with null AHL-based colorization of A136 were determined using cross-feeding bioassay [54,56].
4.7. Characterization of Transposon-Inserted Gene
Genomic DNA was prepared from Tn10-inserted mutant using a bacterial genomic DNA extraction kit (GE, USA). The Tn10-inserted DNA sequence in each mutant was characterized using high-efficiency thermal asymmetric interlaced PCR (hiTAIL-PCR) [70]. The primers for hiTAIL-PCR are listed in Supplementary Table S4. The first round PCR was conducted in a 20 μL system containing 20 ng DNA, 50 nM DTn10AP1, 1.0 μM of each LAD primer [70], 200 μM dNTPs, 2.0 μL of PCR buffer, and 2.5 U rTaq polymerases (TaKaRa, Dalian, China). Next, second round PCR was employed in a 50 μL system with 1.0 μL of appropriately-diluted PCR product from first round, 5.0 μL of PCR buffer, 50 nM DTn10AP2, 1.0 μM of each LAD primer, 200 μM dNTPs, and 2.5 U rTaq polymerases. Finally, third round PCR was performed in a 50 μL system with 1.0 μL of appropriately-diluted PCR product from second round, 5.0 μL of PCR buffer, 50 nM DTn10AP3, 1.0 μM of each LAD primer, 200 μM dNTPs, and 2.5U rTaq polymerases. The amplicons from the second and third rounds were evaluated on 1.5% agarose gel and the desired fragments were purified (Qiagen, MD, USA) for TA cloning (TaKaRa, Dalian, China). After sequencing in Sangon Biotech (Shanghai, China), the disrupted gene sequence in each mutant was obtained. After alignment with the relevant gene on our previously-collected genome, the transposon-inserted gene was accordingly determined.
4.8. Complementation and Overexpression of the Disrupted Gene
To complement the disrupted robp genein mutant of robp::Tn10 (Supplementary Table S3) with null AHL-based colorization of A136, the entire robp was amplified using paired primers (robpF:5′ATGGATCCTTACTCAATAGGCAAGTAAATATCGGT3′, with a BamH I restriction site; robpR:5′ATGGTACCATGTCAAAATATCAAAAGCGGTTTA3′, with a KpnI restriction site). Pfu DNA polymerase (Promega, Beijin, China) was used to amplify 867 bp of robp. Then, Taq DNA polymerase (TaKaRa, Dalian, China) was used to add 3’-adenine overhang. Next, the desired product was purified for TA cloning (TaKaRa, China). After KpnI/BamH I digestion, the gene was released, and subcloned into KpnI/BamH I digested pBBR1MCS-5 [70] to generate plasmid pBBR1MCS-5/robp for transformation into E. coli HB101. Finally, assisted by plasmid pBR2013 in E. coli DH5α [70], pBBR1MCS-5/robpin E. coli HB101was transferred into mutant strain robp::Tn10 and wild type strain T1lg65 for complementation and overexpression of robp gene, respectively, through tri-parental conjugation.
Tri-parental conjugation was performed as previously reported [70]. First, mutant or wild-type (receptor) E. coli HB101 with pBBR1MCS-5/robp (donor) and E. coli DH5α with pRK2013 (assistor) were separately cultured in 2216E with 40 μg/mL Tet and 50 μg/mL Km, LB with 50 μg/mL Gm and LB with 50 μg/mL Km, respectively. Next, 2 mL of each culture cells at exponential growth phase were pelleted after centrifugation at 5000 g for 5 min. After washing once, each cell pellet was gently resuspended in 0.5 mL of LB-2216E and mixed together. After centrifugation and removal of supernatant, the cell mixture was completely resuspended again in 0.2 mL of LB-2216E and dropped to sterilized microporous membrane (size diameter 1 cm; aperture 0.45 μm) on LB-2216E plate for overnight conjugation at 30°C. Finally, the microporous membrane was washed in 3 mL of 2216E to remove the cells. After appropriate dilution, 50 μL of cell mixtures were plated onto 2216E containing necessary antibiotics. After incubation at 30°C, the target colonies were molecularly confirmed.
4.9. Quantitative real-Time PCR for Detectionof Relative Gene Expression
The cultured Pseudoalteromonas cells were pelleted, and the total RNA was next prepared using RNAiso Plus kit (TaKaRa, Dalian, China). RNA integrity was determined according to OD260nm/OD280nm ratio and around 500 ng of RNA samples were amplified to cDNA using PrimeScriptTM RT Master Mix kit (TaKaRa, Dalian, China). After appropriate dilution, the reverse-transcribed cDNA was further amplified to target gene fragment using SYBR green Premix Ex TaqTM kit (TaKaRa, Dalian, China) [71]. All the primers (Supplementary Table S4) were designed according to the genome sequences of selected Pseudoalteromonas isolates. The PCR amplification was performed on CFX Connect Real-Time System (Bio-Rad, Hercules, CA) with a protocol of one cycle of denaturation at 95°C for 10 min and 40 cycles of denaturation at 95°C for 15 s and annealing/elongation at 60°C for 30 s. The 116 bp of 16S rRNA gene fragment (Supplementary Table S4) was used as internal control. The relative expression of target gene was calculated as previously reported [70].
4.10. Analysis of Data Significance
Unless otherwise specified, triplicate reactions per experiment were performed. All the data were presented as mean ± standard error and statistical significance was analyzed based on one-way analysis of variance followed by the Dunnett’s post hoc test using StatView 5.0 software. Significant difference is respectively indicated by asterisks as follows: * p < 0.05; ** p < 0.01; *** p < 0.001; **** p < 0.0001.
4.11. Availability of Data and Materials
The annotated draft genomes of Pseudoalteromonas isolates under this study have been submitted to WGS database under NCBI Accession Numbers listed in Table 2. The remaining data that support the findings of this study are available from the corresponding author upon request.
5. Conclusions
AHL-based colorization intensity of biosensors induced by Pseudoalteromonas most likely correlates with QS regulators genetic heterogeneity within the genus, providing a significantly better understanding of QS regulation in Pseudoalteromonas. Comprehensive QS regulatory schema and a working model proposed in Pseudoalteromonas seem to phylogenetically include the network architectures derived from Escherichia coli, Pseudomonas, and Vibrio, emphasizing that the distinctly- and hierarchically-organized mechanisms probably target QS regulation in bacterial dynamic genomes. Our studies improve understanding of the landscapes of QS regulation in Pseudoalteromonas and bacterial capabilities of accommodating their adaptation fitness and survival advantages.
Abbreviations
| QS | quorum sensing |
| AHL | acyl-homoserine lactone |
| COG | cluster of orthologous groups of proteins |
| robp | right origin-binding protein-encoding gene |
| Tn | transposon |
| LB | Luria-Bertani |
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/11/3636/s1.
Author Contributions
Conceptualization, Z.Y., J.Y., W.Z., J.Q. and J.L.; Data curation, Z.Y., Y.D., J.Y., J.Z., M.Z., M.D. and J.L.; Formal analysis, Y.D., D.Y., J.Z., M.Z., M.D. and J.L.; Funding acquisition, Z.Y. and W.Z.; Investigation, Z.Y., Y.D., D.Y., J.Z., M.Z., M.D. and J.L.; Methodology, Y.D.; Project administration, Z.Y.; Supervision, Z.Y. and J.Q.; Validation, Y.D., J.Y. and J.L.; Visualization, Z.Y.; Writing—original draft, Z.Y., Y.D. and J.L.; Writing—review & editing, Z.Y., Y.D., J.Y., W.Z., J.Q. and J.L.
Funding
This study was supported by the National Natural Science Foundation of China (31670114, 31670115, 31800118) and Natural Science Foundation of Zhejiang Province, China (LY16C010002) for which the authors are thankful.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Yu M., Tang K., Liu J., Shi X., Gulder T.A., Zhang X.H. Genome analysis of Pseudoalteromonas flavipulchra JG1 reveals various survival advantages in marine environment. BMC Genomics. 2013;14:707. doi: 10.1186/1471-2164-14-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmstrom C., James S., Neilan B.A., White D.C., Kjelleberg S. Pseudoalteromonas tunicata sp. nov., a bacterium that produces antifouling agents. Int. J. Syst. Bacteriol. 1998;48:1205–1212. doi: 10.1099/00207713-48-4-1205. [DOI] [PubMed] [Google Scholar]
- 3.Ivanova E.P., Kiprianova E.A., Mikhailov V.V., Levanova G.F., Garagulya A.D., Gorshkova N.M., Yumoto N., Yoshikawa S. Characterization and identification of marine Alteromonas nigrifaciens strains and emendation of the description. Int. J. Syst. Bacteriol. 1996;46:223–228. doi: 10.1099/00207713-46-1-223. [DOI] [PubMed] [Google Scholar]
- 4.Nam Y.D., Chang H.W., Park J.R., Kwon H.Y., Quan Z.X., Park Y.H., Lee J.S., Yoon J.H., Bae J.W. Pseudoalteromonas marina sp. nov., a marine bacterium isolated from tidal flats of the Yellow Sea, and reclassification of Pseudoalteromonas sagamiensis as Algicola sagamiensis comb. nov. Int. J. Syst. Evol. Microbiol. 2007;57:12–18. doi: 10.1099/ijs.0.64523-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhou M.Y., Chen X.L., Zhao H.L., Dang H.Y., Luan X.W., Zhang X.Y., He H.L., Zhou B.C., Zhang Y.Z. Diversity of both the cultivable protease-producing bacteria and their extracellular proteases in the sediments of the South China sea. Microb. Ecol. 2009;58:582–590. doi: 10.1007/s00248-009-9506-z. [DOI] [PubMed] [Google Scholar]
- 6.Bosi E., Fondi M., Orlandini V., Perrin E., Maida I., de Pascale D., Tutino M.L., Parrilli E., Lo Giudice A., Filloux A., et al. The pangenome of (Antarctic) Pseudoalteromonas bacteria: Evolutionary and functional insights. BMC Genomics. 2017;18:93. doi: 10.1186/s12864-016-3382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borchert E., Knobloch S., Dwyer E., Flynn S., Jackson S.A., Johannsson R., Marteinsson V.T., O’Gara F., Dobson A.D.W. Biotechnological potential of cold adapted Pseudoalteromonas spp. isolated from ‘Deep Sea’ sponges. Mar. Drugs. 2017;15:184. doi: 10.3390/md15060184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Medigue C., Krin E., Pascal G., Barbe V., Bernsel A., Bertin P.N., Cheung F., Cruveiller S., D’Amico S., Duilio A., et al. Coping with cold: The genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res. 2005;15:1325–1335. doi: 10.1101/gr.4126905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ballestriero F., Thomas T., Burke C., Egan S., Kjelleberg S. Identification of compounds with bioactivity against the nematode Caenorhabditis elegans by a screen based on the functional genomics of the marine bacterium Pseudoalteromonas tunicata D2. Appl. Environ. Microbiol. 2010;76:5710–5717. doi: 10.1128/AEM.00695-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin Q.L., Li Y., Zhang Y.J., Zhou Z.M., Zhang W.X., Chen X.L., Zhang X.Y., Zhou B.C., Wang L., Zhang Y.Z. Comparative genomics reveals a deep-sea sediment-adapted life style of Pseudoalteromonas sp. SM9913. ISME J. 2011;5:274–284. doi: 10.1038/ismej.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galloway W.R., Hodgkinson J.T., Bowden S.D., Welch M., Spring D.R. Quorum sensing in Gram-negative bacteria: Small-molecule modulation of AHL and AI-2 quorum sensing pathways. Chem. Rev. 2011;111:28–67. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- 12.Ng W.L., Bassler B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chugani S., Kim B.S., Phattarasukol S., Brittnacher M.J., Choi S.H., Harwood C.S., Greenberg E.P. Strain-dependent diversity in the Pseudomonas aeruginosa quorum-sensing regulon. Proc. Natl. Acad. Sci. USA. 2012;109:E2823–E2831. doi: 10.1073/pnas.1214128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feltner J.B., Wolter D.J., Pope C.E., Groleau M.C., Smalley N.E., Greenberg E.P., Mayer-Hamblett N., Burns J., Deziel E., Hoffman L.R., et al. LasR variant cystic fibrosis isolates reveal an adaptable quorum-sensing hierarchy in Pseudomonas aeruginosa. mBio. 2016;7:e1513–e1516. doi: 10.1128/mBio.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang H., Lovell C.R. Microbial surface colonization and biofilm development in marine environments. Microbiol. Mol. Biol. Rev. 2016;80:91–138. doi: 10.1128/MMBR.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobretsov S., Teplitski M., Paul V. Mini-review: Quorum sensing in the marine environment and its relationship to biofouling. Biofouling. 2009;25:413–427. doi: 10.1080/08927010902853516. [DOI] [PubMed] [Google Scholar]
- 17.Thomas T., Evans F.F., Schleheck D., Mai-Prochnow A., Burke C., Penesyan A., Dalisay D.S., Stelzer-Braid S., Saunders N., Johnson J., et al. Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS ONE. 2008;3:e3252. doi: 10.1371/journal.pone.0003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Ikawa A., Okaue S., Taniguchi S., Osaka I., Yoshimoto A., Kishida Y., Arakawa R., Enomoto K. Quorum sensing signaling molecules involved in the production of violacein by Pseudoalteromonas. Biosci. Biotechnol. Biochem. 2008;72:1958–1961. doi: 10.1271/bbb.80090. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Enomoto K. Characterization of a gene cluster and its putative promoter region for violacein biosynthesis in Pseudoalteromonas sp. 520P1. Appl. Microbiol. Biotechnol. 2011;90:1963–1971. doi: 10.1007/s00253-011-3203-9. [DOI] [PubMed] [Google Scholar]
- 20.Mi Z.H., Yu Z.C., Su H.N., Wang L., Chen X.L., Pang X., Qin Q.L., Xie B.B., Zhang X.Y., Zhou B.C., et al. Physiological and genetic analyses reveal a mechanistic insight into the multifaceted lifestyles of Pseudoalteromonas sp. SM9913 adapted to the deep-sea sediment. Environ. Microbiol. 2015;17:3795–3806. doi: 10.1111/1462-2920.12823. [DOI] [PubMed] [Google Scholar]
- 21.Aye A.M., Bonnin-Jusserand M., Brian-Jaisson F., Ortalo-Magne A., Culioli G., Nevry R.K., Rabah N., Blache Y., Molmeret M. Modulation of violacein production and phenotypes associated with biofilm by exogenous quorum sensing N-acylhomoserine lactones in the marine bacterium Pseudoalteromonas ulvae TC14. Microbiology. 2015;161:2039–2051. doi: 10.1099/mic.0.000147. [DOI] [PubMed] [Google Scholar]
- 22.Dang H.T., Komatsu S., Masuda H., Enomoto K. Characterization of LuxI and LuxR protein homologs of N-acylhomoserine lactone-dependent quorum sensing system in Pseudoalteromonas sp. 520P1. Mar. Biotechnol. 2017;19:1–10. doi: 10.1007/s10126-016-9726-4. [DOI] [PubMed] [Google Scholar]
- 23.Huang Y., Zeng Y., Yu Z., Zhang J., Feng H., Lin X. In silico and experimental methods revealed highly diverse bacteria with quorum sensing and aromatics biodegradation systems—A potential broad application on bioremediation. Bioresour. Technol. 2013;148:311–316. doi: 10.1016/j.biortech.2013.08.155. [DOI] [PubMed] [Google Scholar]
- 24.Yu Z., Ma Y., Zhong W., Qiu J., Li J. Comparative genomics of Methanopyrus sp. SNP6 and KOL6 revealing genomic regions of plasticity implicated in extremely thermophilic profiles. Front. Microbiol. 2017;8:1278. doi: 10.3389/fmicb.2017.01278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belarmino L.C., Silva R.L., Cavalcanti Nda M., Krezdorn N., Kido E.A., Horres R., Winter P., Kahl G., Benko-Iseppon A.M. SymGRASS: A database of sugarcane orthologous genes involved in arbuscular mycorrhiza and root nodule symbiosis. BMC Bioinform. 2013;14:S2. doi: 10.1186/1471-2105-14-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn R.D., Clements J., Arndt W., Miller B.L., Wheeler T.J., Schreiber F., Bateman A., Eddy S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang P., Zeng Z., Wang W., Wen Z., Li J., Wang X. Dissemination and loss of a biofilm-related genomic island in marine Pseudoalteromonas mediated by integrative and conjugative elements. Environ. Microbiol. 2017;19:4620–4637. doi: 10.1111/1462-2920.13925. [DOI] [PubMed] [Google Scholar]
- 28.Shao Y., Bassler B.L. Quorum regulatory small RNAs repress type VI secretion in Vibrio cholerae. Mol. Microbiol. 2014;92:921–930. doi: 10.1111/mmi.12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Q., Defoirdt T. Quorum sensing positively regulates flagellar motility in pathogenic Vibrio harveyi. Environ. Microbiol. 2015;17:960–968. doi: 10.1111/1462-2920.12420. [DOI] [PubMed] [Google Scholar]
- 30.Viducic D., Murakami K., Amoh T., Ono T., Miyake Y. RpoN modulates carbapenem tolerance in Pseudomonas aeruginosa through Pseudomonas quinolone signal and PqsE. Antimicrob. Agents Chemother. 2016;60:5752–5764. doi: 10.1128/AAC.00260-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai Z., Liu Y., Chen Y., Yam J.K., Chew S.C., Chua S.L., Wang K., Givskov M., Yang L. RpoN regulates virulence factors of Pseudomonas aeruginosa via modulating the PqsR quorum sensing regulator. Int. J. Mol. Sci. 2015;16:28311–28319. doi: 10.3390/ijms161226103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Totten P.A., Lara J.C., Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 1990;172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papenfort K., Bassler B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Case R.J., Labbate M., Kjelleberg S. AHL-driven quorum-sensing circuits: Their frequency and function among the Proteobacteria. ISME J. 2008;2:345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- 35.Miller M.B., Skorupski K., Lenz D.H., Taylor R.K., Bassler B.L. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell. 2002;110:303–314. doi: 10.1016/S0092-8674(02)00829-2. [DOI] [PubMed] [Google Scholar]
- 36.van Kessel J.C., Rutherford S.T., Shao Y., Utria A.F., Bassler B.L. Individual and combined roles of the master regulators AphA and LuxR in control of the Vibrio harveyi quorum-sensing regulon. J. Bacteriol. 2013;195:436–443. doi: 10.1128/JB.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Higgins D.A., Pomianek M.E., Kraml C.M., Taylor R.K., Semmelhack M.F., Bassler B.L. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450:883–886. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 38.Ng W.L., Perez L.J., Wei Y., Kraml C., Semmelhack M.F., Bassler B.L. Signal production and detection specificity in Vibrio CqsA/CqsS quorum-sensing systems. Mol. Microbiol. 2011;79:1407–1417. doi: 10.1111/j.1365-2958.2011.07548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papenfort K., Silpe J.E., Schramma K.R., Cong J.P., Seyedsayamdost M.R., Bassler B.L. A Vibrio cholerae autoinducer-receptor pair that controls biofilm formation. Nat. Chem. Biol. 2017;13:551–557. doi: 10.1038/nchembio.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee J., Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell. 2015;6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lintz M.J., Oinuma K., Wysoczynski C.L., Greenberg E.P., Churchill M.E. Crystal structure of QscR, a Pseudomonas aeruginosa quorum sensing signal receptor. Proc. Natl. Acad. Sci. USA. 2011;108:15763–15768. doi: 10.1073/pnas.1112398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bottomley M.J., Muraglia E., Bazzo R., Carfi A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 2007;282:13592–13600. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 43.Welsh M.A., Blackwell H.E. Chemical genetics reveals environment-specific roles for quorum sensing circuits in Pseudomonas aeruginosa. Cell Chem. Biol. 2016;23:361–369. doi: 10.1016/j.chembiol.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J., Tai C., Deng Z., Zhong W., He Y., Ou H.Y. VRprofile: Gene-cluster-detection-based profiling of virulence and antibiotic resistance traits encoded within genome sequences of pathogenic bacteria. Brief. Bioinform. 2017;19:566–574. doi: 10.1093/bib/bbw141. [DOI] [PubMed] [Google Scholar]
- 45.Liang H., Deng X., Li X., Ye Y., Wu M. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res. 2014;42:10307–10320. doi: 10.1093/nar/gku586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hmelo L.R. Quorum sensing in marine microbial environments. Annu. Rev. Mar. Sci. 2017;9:257–281. doi: 10.1146/annurev-marine-010816-060656. [DOI] [PubMed] [Google Scholar]
- 47.Schauder S., Bassler B.L. The languages of bacteria. Genes Dev. 2001;15:1468–1480. doi: 10.1101/gad.899601. [DOI] [PubMed] [Google Scholar]
- 48.Bowman J.P. Bioactive compound synthetic capacity and ecological significance of marine bacterial genus Pseudoalteromonas. Mar. Drugs. 2007;5:220–241. doi: 10.3390/md504220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim W.J., Kim Y.O., Kim J.H., Nam B.H., Kim D.G., An C.M., Lee J.S., Kim P.S., Lee H.M., Oh J.S., et al. Liquid chromatography-mass spectrometry-based rapid secondary-metabolite profiling of marine Pseudoalteromonas sp. M2. Mar. Drugs. 2016;14:24. doi: 10.3390/md14010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J., Qian S., Xiong L., Zhu C., Shu M., Wang J., Jiao Y., He H., Zhang F., Linhardt R.J., et al. Comparative genomics reveals specific genetic architectures in nicotine metabolism of Pseudomonas sp. JY-Q. Front. Microbiol. 2017;8:2085. doi: 10.3389/fmicb.2017.02085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., Yao Y., Xu H.H., Hao L., Deng Z., Rajakumar K., Ou H.Y. SecReT6: A web-based resource for type VI secretion systems found in bacteria. Environ. Microbiol. 2015;17:2196–2202. doi: 10.1111/1462-2920.12794. [DOI] [PubMed] [Google Scholar]
- 52.Aschtgen M.S., Gavioli M., Dessen A., Lloubes R., Cascales E. The SciZ protein anchors the enteroaggregative Escherichia coli Type VI secretion system to the cell wall. Mol. Microbiol. 2010;75:886–899. doi: 10.1111/j.1365-2958.2009.07028.x. [DOI] [PubMed] [Google Scholar]
- 53.Jia Q., Lee B.Y., Bowen R., Dillon B.J., Som S.M., Horwitz M.A. A Francisella tularensis live vaccine strain (LVS) mutant with a deletion in capB, encoding a putative capsular biosynthesis protein, is significantly more attenuated than LVS yet induces potent protective immunity in mice against F. tularensis challenge. Infect. Immun. 2010;78:4341–4355. doi: 10.1128/IAI.00192-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ravn L., Christensen A.B., Molin S., Givskov M., Gram L. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods. 2001;44:239–251. doi: 10.1016/S0167-7012(01)00217-2. [DOI] [PubMed] [Google Scholar]
- 55.Steindler L., Venturi V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol. Lett. 2007;266:1–9. doi: 10.1111/j.1574-6968.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 56.Zeng Y., Wang Y., Yu Z., Huang Y. Hypersensitive response of plasmid-encoded AHL synthase gene to lifestyle and nutrient by Ensifer adhaerens X097. Front. Microbiol. 2017;8:1160. doi: 10.3389/fmicb.2017.01160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y., Zeng Y., Yu Z., Zhang J. Distribution and diversity of acyl homoserine lactone producing bacteria from four different soils. Curr. Microbiol. 2013;66:10–15. doi: 10.1007/s00284-012-0234-0. [DOI] [PubMed] [Google Scholar]
- 58.Quail M.A., Smith M., Coupland P., Otto T.D., Harris S.R., Connor T.R., Bertoni A., Swerdlow H.P., Gu Y. A tale of three next generation sequencing platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genom. 2012;13:341. doi: 10.1186/1471-2164-13-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zerbino D.R., Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 61.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Junker V., Contrino S., Fleischmann W., Hermjakob H., Lang F., Magrane M., Martin M.J., Mitaritonna N., O’Donovan C., Apweiler R. The role SWISS-PROT and TrEMBL play in the genome research environment. J. Biotechnol. 2000;78:221–234. doi: 10.1016/S0168-1656(00)00198-X. [DOI] [PubMed] [Google Scholar]
- 63.Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chalmel F., Lardenois A., Thompson J.D., Muller J., Sahel J.A., Leveillard T., Poch O. GOAnno: GO annotation based on multiple alignment. Bioinformatics. 2005;21:2095–2096. doi: 10.1093/bioinformatics/bti252. [DOI] [PubMed] [Google Scholar]
- 65.Lagesen K., Hallin P., Rodland E.A., Staerfeldt H.H., Rognes T., Ussery D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lowe T.M., Chan P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu Z., Hao B. CVTree update: A newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009;37:W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Y.L., Li M., Yu Z., Qian P.Y. Correlation between pigmentation and larval settlement deterrence by Pseudoalteromonas sp. sf57. Biofouling. 2011;27:287–293. doi: 10.1080/08927014.2011.562978. [DOI] [PubMed] [Google Scholar]
- 69.Solano F., Lucas-Elio P., Fernandez E., Sanchez-Amat A. Marinomonas mediterranea MMB-1 transposon mutagenesis: Isolation of a multipotent polyphenol oxidase mutant. J. Bacteriol. 2000;182:3754–3760. doi: 10.1128/JB.182.13.3754-3760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu Z., Wang J., Lin J., Zhao M., Qiu J. Exploring regulation genes involved in the expression of L-amino acid oxidase in Pseudoalteromonas sp. Rf-1. PLoS ONE. 2015;10:e0122741. doi: 10.1371/journal.pone.0122741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu Z., Yang Y., Wang Y., Yin J., Qiu J. Reactive oxygen species-scavenging system is involved in L-amino acid oxidase accumulation in Pseudoalteromonas sp. B3. 3 Biotech. 2017;7:326. doi: 10.1007/s13205-017-0976-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The annotated draft genomes of Pseudoalteromonas isolates under this study have been submitted to WGS database under NCBI Accession Numbers listed in Table 2. The remaining data that support the findings of this study are available from the corresponding author upon request.