Abstract
Cytochrome P450 monooxygenases (CYPs/P450s) are among the most catalytically-diverse enzymes, capable of performing enzymatic reactions with chemo-, regio-, and stereo-selectivity. Our understanding of P450s’ role in secondary metabolite biosynthesis is becoming broader. Among bacteria, Bacillus species are known to produce secondary metabolites, and recent studies have revealed the presence of secondary metabolite biosynthetic gene clusters (BGCs) in these species. However, a comprehensive comparative analysis of P450s and P450s involved in the synthesis of secondary metabolites in Bacillus species has not been reported. This study intends to address these two research gaps. In silico analysis of P450s in 128 Bacillus species revealed the presence of 507 P450s that can be grouped into 13 P450 families and 28 subfamilies. No P450 family was found to be conserved in Bacillus species. Bacillus species were found to have lower numbers of P450s, P450 families and subfamilies, and a lower P450 diversity percentage compared to mycobacterial species. This study revealed that a large number of P450s (112 P450s) are part of different secondary metabolite BGCs, and also identified an association between a specific P450 family and secondary metabolite BGCs in Bacillus species. This study opened new vistas for further characterization of secondary metabolite BGCs, especially P450s in Bacillus species.
Keywords: Antibiotics, Bacillus, biosynthetic gene clusters, comparative analysis, cytochrome P450 monooxygenase, Mycobacterium, P450 diversity percentage, P450 profiling, secondary metabolites
1. Introduction
Cytochrome P450 monooxygenases, also known as CYPs/P450s, are undoubtedly among the most catalytically-diverse enzymes, performing enzymatic reactions with chemo-, regio- and stereo-selectivity [1,2,3,4,5,6]. The catalytic diversity combined with chemo-, regio- and stereo-specific oxidation of substrates exerted by P450s are used in diverse biotechnological applications ranging from drug discovery to bioethanol production and synthesis of different secondary metabolites [7,8,9,10,11,12]. P450s are heme-thiolate proteins ubiquitously found in species belonging to different biological kingdoms, including non-living entities such as viruses [13,14]. In bacteria, P450s have been found to play a key role in enzymatic reactions, leading to the biosynthesis of physiological compounds or the biodegradation of xenobiotics [9,11,15,16].
P450s’ role in the synthesis of a diverse array of secondary metabolites has been thoroughly reviewed [8,12]. Secondary metabolites are natural products that are widely-used in human and veterinary medicine, agriculture, and manufacturing, and are known to mediate a variety of microbe-host and microbe-microbe interactions [17]. P450s were found to play a key role in the synthesis of different secondary metabolites, including terpenes, alkaloids, shikimates, polyketides, and peptides [12]. The coding sequences (genes) of enzymes involved in the synthesis of different secondary metabolites, including P450s, were found to be part of gene clusters named biosynthetic gene clusters (BGCs) [17]. Bacterial species have been found to have more than 1000 different types of BGCs involved in the synthesis of known and unknown secondary metabolites [17].
Among bacteria, species belonging to the genus Bacillus are ubiquitously present in the biosphere, and are well known for their distinct features with one common characteristic, i.e., making dormant endospores during unfavorable growth conditions [18,19]. Applications of Bacillus species across different spectra have been well explored in the industrial, agricultural, and ecological fields, and by academics, against a backdrop of being a well-known human pathogen [18,19]. Comprehensive in silico studies detailing Bacillus species’ ability to produce different secondary metabolites and different types of BGCs have frequently been reported [20,21]. Analysis of 1566 Bacillus species’ genomes revealed the presence of 20,000 BGCs, most of which were found to produce known secondary metabolites that play a key role in the physiology and development of Bacillus species [21]. The study by Grubbs et al. [21] also reported that secondary metabolite alkylpyrones play a key role in inhibiting spore development in Bacillus species.
Despite comprehensive analysis of Bacillus species’ secondary metabolite BGCs, P450s that are part of different BGCs have not been reported. Analysis of P450s in Bacillus species date back to 2009, when the authors performed a comparative analysis of P450s in 29 Bacillus species, and identified a few P450s belonging to a limited number of P450 families such as CYP102, CYP106, CYP107, CYP109, CYP134, CYP152, and CYP197 [22]. Among the P450 families identified in Bacillus species, the CYP102 P450 family has a special place in P450 research, being one of the most extensively-studied bacterial P450s, for its structural, functional, and biotechnological potential. Even the Bacillus species B. megaterium has become very famous owing to the identification of CYP102 P450 from this organism [23,24]. In silico comparative analysis of P450s in bacterial species is gaining momentum. Recently, a comprehensive comparative analysis of P450s in 60 mycobacterial species has been reported; the authors identified that mycobacteria possess a large number of P450s, and that different mycobacterial categories have characteristic P450 families that can be used as biomarkers to identify different mycobacterial species [25].
The current trend of whole-genome sequencing of organisms resulted in the genome sequencing of a large number of Bacillus species genomes. Quite a large number of Bacillus species genome sequences are available for public use at Kyoto Encyclopedia of Genes and Genomes—GenomeNet (KEGG) [26]. This gives us the opportunity to perform comprehensive a comparative analysis of P450s in Bacillus species as per international P450 nomenclature committee rules [27,28,29], and to identify P450s involved in the synthesis of different secondary metabolites. Here, we report genome data mining, annotation, phylogenetic and comparative analysis of P450s in 128 Bacillus species, including identification of P450s involved in the synthesis of different secondary metabolites. This study also reports comparative analysis of P450s between the genera Bacillus and Mycobacterium. Last but not least, a previous study reporting BGCs in Bacillus species did not clearly indicate BGCs on genomic DNA (gDNA) and plasmid DNA [21]; thus, in this study, gDNA and plasmid DNAs were individually subjected to BGC analysis.
2. Results and Discussion
2.1. Bacillus Species Have the Lowest Number of P450s
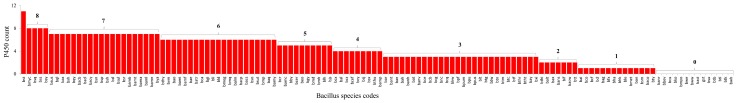
Genome data-mining and annotation of P450s in 128 Bacillus species revealed the presence of the lowest number of P450s in their genomes (Figure 1 and Figure 2). In total, 507 P450s were found in 114 Bacillus species, where 14 species did not have P450s in their genomes (Figure 1 and Figure 2). On average, four P450s were found in Bacillus species, with the highest number, 11, found in Bacillus subtilis subsp. spizizenii TU-B-10. The number of P450s found in Bacillus species is very low compared to mycobacterial species (60 species); the latter species have, on average, 35 P450s in their genomes [25]. The P450 count in Bacillus species, apart from B. subtilis subsp. spizizenii TU-B-10, which has 16, is as follows: 9 in 2 species, 8 in 4 species, 7 in 20 species, 6 in 21 species, 5 in 10 species, 4 in 9 species, 3 in 28 species, 2 in 7 species, and a single in 14 species (Figure 1 and Figure 2). This indicates that most Bacillus species (28 species) have three P450s in their genomes. P450s identified in each Bacillus species and respective P450 sequences were presented in Supplementary datasets 1 and 2.
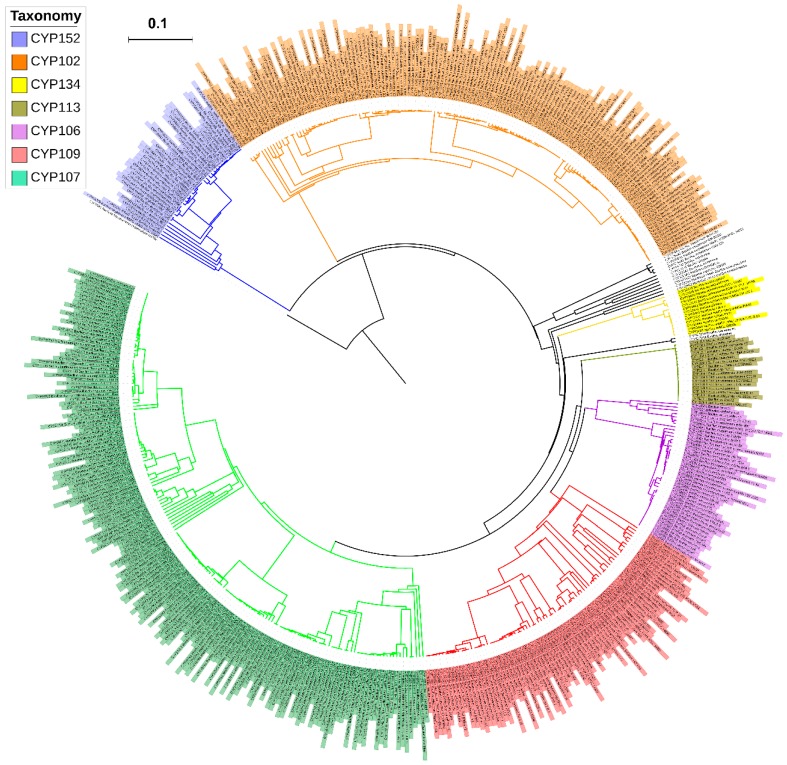
Figure 1.
Phylogenetic analysis of Bacillus species P450s. Dominant P450 families were indicated in different colors. CYP51B1 from Mycobacterium tuberculosis H37Rv is used as an outgroup. A high-resolution phylogenetic tree is provided in the supplementary Figure S1.
Figure 2.
Comparative analysis of P450s in Bacillus species. The numbers next to bars indicate the number of P450s in Bacillus species. Bacillus species’ names with respect to their codes can be found in Supplementary Dataset 1.
2.2. Bacillus Species Have the Lowest Number of P450 Families and Subfamilies’
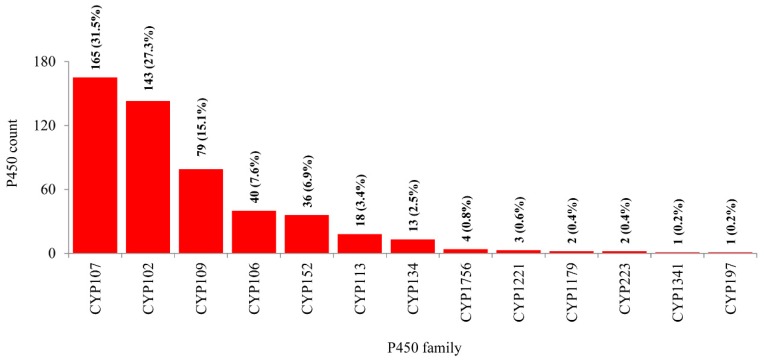
As per the International P450 Nomenclature Committee rules [27,28,29], all 507 P450s found in 114 Bacillus species can be grouped into 13 P450 families and 28 subfamilies (Figure 3 and Figure 4). Phylogenetic analysis of Bacillus P450s revealed P450s belonging to the same family grouped together, suggesting that the annotation of P450s in this study is accurate (Figure 1). The number of P450 families and subfamilies found in Bacillus species is lower compared to mycobacterial species (60 species), which have 77 P450 families and 132 subfamilies [25]. Because of the presence of the lowest number of P450 families, the P450 diversity percentage in Bacillus species was found to be lowest (3.9%) compared to mycobacterial species (72%) [25]. Among 13 P450 families, the CYP107 P450 family has the highest number of P450s (165 P450s) contributing 31.5% of 507 P450s (Figure 3), followed by CYP102 (143 P450s), CYP109 (79 P450s), CYP106 (40 P450s), CYP152 (36 P450s), CYP113 (18 P450s), CYP134 (13 P450s), CYP1756 (4 P450s), CYP1221 (3 P450s), CYP1179 and CYP223 (2 P450s), CYP1341 and CYP197 (single P450s) (Figure 3).
Figure 3.
Comparative analysis of P450 families in Bacillus species. The numbers next to the family bar indicate the total number of P450s and percentage contribution (parenthesis) by a respective family to the total number of P450s. The data on the number of P450s in each P450 family in Bacillus species is presented in Table S1.
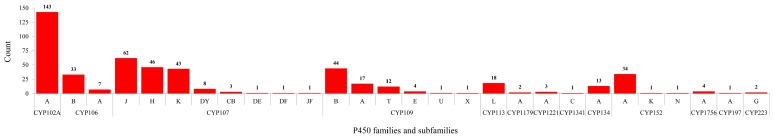
Figure 4.
Comparative analysis of P450 subfamilies in Bacillus species. The numbers next to bars indicate the total number of members in a particular subfamily. Data on the number of P450s in each P450 subfamily in Bacillus species is presented in Table S1.
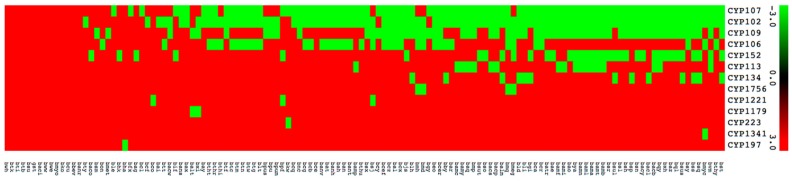
P450 subfamily analysis revealed that most P450 families have a single subfamily (Figure 4). Among P450 families, CYP107 has the highest number of P450 subfamilies (eight subfamilies) followed by CYP109 (six subfamilies), CYP152 (three subfamilies), and CYP106 (two subfamilies) (Figure 4). The remaining nine P450 families, CYP102, CYP113, CYP1179, CYP1221, CYP1341, CYP134, CYP1756, CYP197, and CYP223, all have a single subfamily (Figure 4). It is interesting to note that the CYP102 P450 family, despite having the second largest number of P450s, has a single subfamily “A”. Analysis of P450 subfamily profiles revealed that specific subfamilies are dominant in a particular family (Figure 4). Subfamily “J” is dominant in the CYP107 family, subfamily “B” is dominant in the CYP109 family and subfamily “A” is dominant in the CYP152 family (Figure 4). Analysis of P450 family profiles revealed that no P450 family is conserved across Bacillus species (Figure 5). Most Bacillus species have CYP102, CYP107, CYP109, CYP106, and CYP152 P450 families (Figure 5). CYP197, CYP223, and CYP1341 are present in a single Bacillus species (Supplementary Dataset 1).
Figure 5.
Heatmap of presence or absence of cytochrome P450 families in 128 species of Bacillus. The data have been represented as −3 for family presence (green) and 3 for family absence (red). A hundred and twenty-eight Bacillus species form the horizontal axis and CYP family numbers form the vertical axis. The respective data used in the generation of Figure 5 is presented in Supplementary Dataset 3.
2.3. Bacillus Species Have the Lowest Number of Secondary Metabolite BGCs
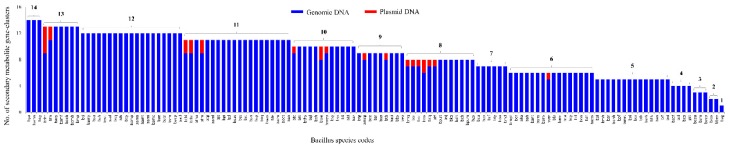
In order to identify P450s involved in the biosynthesis of secondary metabolites, the gDNA and plasmid DNA of each Bacillus species (Table S2) has been subjected to secondary metabolite BGCs analysis using anti-SMASH [30]. In total, 203 plasmids were identified in 60 Bacillus species (Table S2). Analysis of 128 Bacillus species genomes revealed the presence of 1098 and 26 secondary metabolite BGCs on gDNA and plasmid DNA, respectively (Figure 6 and Table S3).
Figure 6.
Comparative analysis of secondary metabolite BGCs in 128 Bacillus species (gDNA and plasmid DNA). Numbers next to bars indicate the number of secondary metabolite BGCs. Detailed analysis of secondary metabolite BGCs in each species is presented in Table S3.
The number of secondary metabolite BGCs varied from a maximum of 14 to one in Bacillus species gDNA. Interestingly, among 203 plasmid DNAs from 60 Bacillus species (Table S3), only 21 plasmid DNAs from 18 Bacillus species were found to have secondary metabolite BGCs (Figure 6 and Table S3). The number of secondary metabolites BGCs on plasmid DNAs varied from a maximum of four to one (Figure 6 and Table S3).
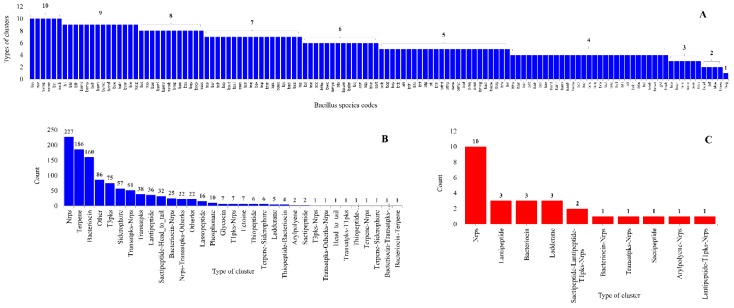
Analysis of types of secondary metabolite BGCs revealed the presence of 33 and 10 types of BGCs on gDNA and plasmid DNAs in Bacillus species (Figure 7 and Supplementary Dataset 4). The types of BGCs in individual Bacillus species varied from a maximum of 10 types to one (Figure 7). Among types of BGCs, Nonribosomal peptides secondary metabolite (Nrps) BGCs were dominant in Bacillus species, both on gDNA and plasmid DNAs (Figure 7).
Figure 7.
Comparative analysis of types of secondary metabolite BGCs in Bacillus species. (A) The number of types of secondary metabolite BGCs in Bacillus species. (B,C). Comparative analysis of types of secondary metabolite BGCs on gDNA and plasmid DNAs. Standard abbreviations representing secondary metabolite BGCs as indicated in anti-SMASH [30] were used in the figure.
Analysis of types of BGCs on gDNA and plasmid DNAs revealed the presence of seven types of common BGCs between gDNA and plasmid DNAs (Figure 7). This suggests that these plasmids might be involved in horizontal gene transfer of different BGCs among Bacillus species. It is important to note that horizontal gene transfer of BGCs is a common phenomenon among Bacillus species [21]. Interestingly, three distinct types of secondary metabolite gene clusters, namely Sactipeptide-Lantipeptide-T1pks-Nrps, Arylpolyene-Nrps, and Lantipeptide-T1pks-Nrps, were only identified on plasmid DNAs (Figure 7).
2.4. Large Number of P450s Found to Be Part of Secondary Metabolites BGCs in Bacillus Species
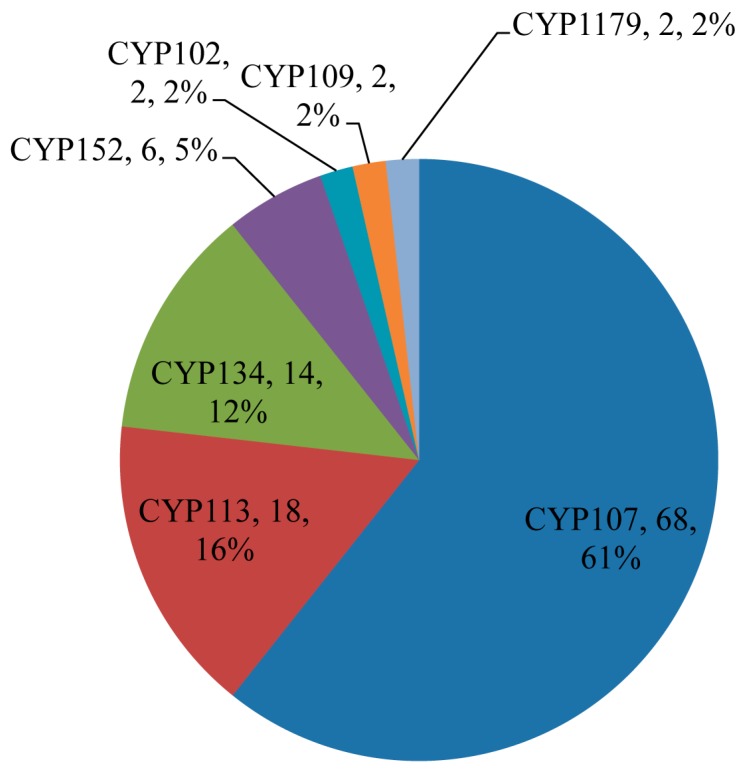
Among 507 P450s identified in 128 Bacillus species, 112 P450s (22%) from 50 Bacillus species were found to be part of secondary metabolite BGCs (Table 1). Among 13 P450 families, only seven families, namely CYP107, CYP113, CYP134, CYP152, CYP102, CYP109, and CYP1179, were found to be part of different secondary metabolite BGCs (Figure 8). P450 subfamily level analysis revealed that P450s belonging to the subfamilies H and K in the CYP107 family were part of secondary metabolite BGCs, despite subfamily J being dominant in that family (Figure 4). In the CYP152 family, P450s belonging to subfamily A were found to be part of the secondary metabolite BGCs. Analysis of P450s involving secondary metabolite biosynthesis revealed that P450s belonging to the CYP107 family are dominant by 61% (68 P450s) of all P450s (112 P450s) involved in secondary metabolite BGCs, followed by CYP113, CYP152, CYP102, CYP109, and CYP1179 (Figure 8). It is interesting to note that these P450 families are highly-populated in Bacillus species (Figure 3 and Figure 5). This further supports the previous hypothesis that species populate specific P450s if they are useful in their adaptation to certain ecological niches or useful in their physiology [31,32,33,34,35]. Considering the large number of P450s, their widespread nature, and part in secondary metabolite BGCs, it can be hypothesized that the P450s belonging to the CYP107, CYP102, CYP109, CYP152, and CYP113 families play a key role in Bacillus species’ physiology, including synthesis of different secondary metabolites. Despite secondary metabolite BGCs being found on plasmid DNA, no P450 was found to be part of these clusters. Analysis of association between P450 families and secondary metabolite BGCs revealed that CYP107 family P450s were mostly associated with BGCs Nrps-Transatpks-Otherks and Transatpks-Nrps; CYP113 family P450s are associated with Transatpks BGC, and CYP134 family P450s are associated with other, a putative gene cluster (Table 1).
Table 1.
Identification of P450s that are involved in secondary metabolite BGCs in Bacillus species. BGCs in each species and P450 identified as part of a particular cluster are presented in Supplementary Dataset 5.
| Species Name | Cluster Number | Type of BGCs | P450 Name |
|---|---|---|---|
| Bacillus subtilis subsp. subtilis 168 | 4 | Nrps-Transatpks-Otherks | CYP134A1 |
| 10 | Other | CYP134A1 | |
| Bacillus subtilis subsp. subtilis RO-NN-1 | 3 | Nrps-Transatpks-Otherks | CYP107K1 |
| Bacillus subtilis subsp. subtilis BSP1 | 8 | Transatpks-Otherks-Nrps | CYP107K1 |
| Bacillus subtilis subsp. subtilis 6051-HGW | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| 10 | Other | CYP107K1 | |
| Bacillus subtilis subsp. subtilis BAB-1 | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| Bacillus subtilis subsp. subtilis AG1839 | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| 10 | Other | CYP107K1 | |
| Bacillus subtilis subsp. subtilis JH642 | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| 10 | Other | CYP134A1 | |
| Bacillus subtilis subsp. subtilis OH 131.1 | 1 | Lantipeptide | CYP152A1 |
| 4 | Nrps-Transatpks-Otherks | CYP107K1 | |
| 9 | Other | CYP134A1 | |
| Bacillus subtilis subsp. spizizenii W23 | 1 | Phosphonate | CYP152A1 |
| 4 | Nrps-Transatpks-Otherks | CYP107K1 | |
| 10 | Other | CYP134A1 | |
| Bacillus subtilis subsp. spizizenii TU-B-10 | 3 | Nrps-Transatpks-Otherks | CYP107K1 |
| 9 | Other | CYP134A1 | |
| Bacillus subtilis BSn5 | 4 | Other | CYP102A48 |
| 8 | Lantipeptide | CYP152A1 | |
| 11 | Nrps-Transatpks-Otherks | CYP107K1 | |
| Bacillus subtilis QB928 | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| 10 | Other | CYP134A1 | |
| Bacillus subtilis XF-1 | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| Bacillus subtilis PY79 | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| 9 | Other | CYP134A1 | |
| Bacillus licheniformis ATCC 14580 | 7 | Other | CYP134A5 |
| Bacillus licheniformis DSM 13 = ATCC 14580 | 7 | Other | CYP134A5 |
| Bacillus paralicheniformis | 10 | Other | CYP134A5 |
| Bacillus velezensis FZB42 | 5 | Transatpks-Nrps | CYP107K3 |
| 6 | Transatpks-Nrps | CYP107H4 | |
| 9 | Transatpks | CYP113L1 | |
| Bacillus velezensis CAU B946 | 5 | Transatpks-Nrps | CYP107K3 |
| 6 | Transatpks-Nrps | CYP107H4 | |
| 9 | Transatpks | CYP113L1 | |
| Bacillus velezensis YAU B9601-Y2 | 5 | Transatpks | CYP107K3 |
| 6 | Transatpks-Nrps | CYP107K3 | |
| 7 | Transatpks-Nrps | CYP107H4 | |
| 10 | Transatpks | CYP113L1 | |
| Bacillus velezensis AS43.3 | 6 | Transatpks | CYP107K3 |
| 7 | Transatpks-Nrps | CYP113L1 | |
| 10 | Transatpks | CYP113L1 | |
| Bacillus velezensis UCMB5036 | 5 | Transatpks-Nrps | CYP107K3 |
| 6 | Transatpks-Nrps | CYP107H4 | |
| 9 | Bacteriocin-Nrps | CYP113L1 | |
| Bacillus velezensis UCMB5033 | 6 | Transatpks-Nrps | CYP107K3 |
| 7 | Transatpks-Nrps | CYP107H4 | |
| 10 | Transatpks | CYP113L1 | |
| Bacillus velezensis UCMB5113 | 7 | Transatpks-Nrps | CYP107K3 |
| 8 | Transatpks-Nrps | CYP107H4 | |
| 11 | Transatpks | CYP113L1 | |
| Bacillus velezensis NAU-B3 | 3 | Transatpks | CYP113L1 |
| 6 | Transatpks-Nrps | CYP107H4 | |
| 7 | Transatpks-Nrps | CYP107K3 | |
| Bacillus velezensis TrigoCor1448 | 5 | Transatpks-Nrps | CYP107K3 |
| 6 | Transatpks-Nrps | CYP107H4 | |
| Bacillus velezensis SQR9 | 6 | Transatpks-Nrps | CYP107K3 |
| 7 | Transatpks-Nrps | CYP107H4 | |
| 10 | Transatpks | CYP113L1 | |
| Bacillus velezensis | 6 | Transatpks-Nrps | CYP107K3 |
| 7 | Transatpks-Nrps | CYP107H4 | |
| 10 | Transatpks | CYP113L1 | |
| Bacillus amyloliquefaciens DSM 7 | 5 | Transatpks-Nrps | CYP107K3 |
| 6 | Transatpks-Nrps | CYP107H2 | |
| Bacillus amyloliquefaciens TA208 | 7 | Transatpks-Nrps | CYP107H2 |
| 8 | Transatpks-Nrps | CYP107K3 | |
| Bacillus amyloliquefaciens LL3 | 5 | Transatpks-Nrps | CYP107K3 |
| 6 | Transatpks-Nrps | CYP107H2 | |
| Bacillus amyloliquefaciens XH7 | 7 | Transatpks-Nrps | CYP107H2 |
| 8 | Transatpks-Nrps | CYP107K3 | |
| Bacillus amyloliquefaciens Y2 | 6 | Transatpks-Nrps | CYP107K3 |
| 7 | Transatpks-Nrps | CYP107H4 | |
| 10 | Transatpks | CYP113L1 | |
| Bacillus amyloliquefaciens IT-45 | 3 | Transatpks | CYP113L1 |
| 6 | Transatpks-Nrps | CYP107H4 | |
| 7 | Transatpks-Nrps | CYP107K3 | |
| Bacillus amyloliquefaciens CC178 | 6 | Transatpks-Nrps | CYP107H4 |
| 9 | Transatpks | CYP113L1 | |
| Bacillus amyloliquefaciens LFB112 | 7 | Transatpks-Nrps | CYP107K3 |
| 8 | Transatpks-Nrps | CYP107H4 | |
| 11 | Transatpks | CYP113L1 | |
| Bacillus atrophaeus 1942 | 3 | Nrps-Transatpks-Otherks | CYP107K2 |
| 10 | Nrps | CYP152A9 | |
| Bacillus atrophaeus NRS 1221A | 3 | Nrps-Transatpks-Otherks | CYP107K2 |
| 10 | Nrps | CYP152A9 | |
| Bacillus vallismortis | 6 | Transatpks-Nrps | CYP107K3 |
| 7 | Transatpks-Nrps | CYP107H4 | |
| 10 | Transatpks | CYP113L1 | |
| Bacillus pumilus SH-B9 | 8 | Nrps | CYP109B6 |
| Bacillus sp. JS | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| Bacillus sp. Pc3 | 1 | Bacteriocin-Transatpks-Nrps | CYP107H4 |
| 2 | Transatpks-Nrps | CYP107K3 | |
| 10 | Transatpks | CYP113L1 | |
| Bacillus sp. BH072 | 8 | Transatpks-Nrps | CYP107K3 |
| 9 | Transatpks-Nrps | CYP107H4 | |
| 12 | Transatpks | CYP107H4 | |
| Bacillus sp. YP1 | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| Bacillus sp. BS34A | 4 | Nrps-Transatpks-Otherks | CYP107K1 |
| 10 | Other | CYP134A1 | |
| Bacillus sp. LM 4-2 | 3 | Nrps-Transatpks-Otherks | CYP107K1 |
| 7 | Other | CYP102A48 | |
| 9 | Other | CYP134A1 | |
| Bacillus gibsonii | 1 | Nrps-Transatpks-Otherks | CYP107K1 |
| 6 | Other | CYP134A1 | |
| 9 | Lantipeptide | CYP152A1 | |
| Bacillus xiamenensis | 2 | Nrps | CYP1179A4 |
| Bacillus altitudinis | 2 | Nrps | CYP1179A4 |
| 8 | Nrps | CYP109B5 | |
| Bacillus sp. SDLI1 | 3 | Transatpks-Nrps | CYP107H4 |
| 4 | Transatpks-Nrps | CYP107K3 | |
| 11 | Transatpks | CYP113L1 |
Figure 8.
Comparative analysis of P450 families involved in secondary metabolite biosynthesis. The P450 family name, number of P450s and their percentage of the total number of 112 P450s are presented in the figure.
2.5. Bacillus P450s Indeed Involved in the Synthesis of Secondary Metabolites
Based on in silico analysis (in this study), seven P450 families, namely CYP107, CYP113, CYP134, CYP152, CYP102, CYP109, and CYP1179, were identified as part of secondary metabolite BGCs in Bacillus species (Figure 8). Functional data available for some P450s confirms that the predicted P450s, in this study, are indeed involved in biosynthesis of different secondary metabolites, and some of the P450 families, such as CYP105, CYP107, and CYP109, have been found to display highly-diverse functions [9,12,36]. CYP102A1 from B. megaterium [24,37,38] and CYP152A1 from B. subtilis [39,40] were found to be fatty acid hydroxylases. P450s belonging to the CYP106, CYP107, CYP109, and CYP134 families were found to hydroxylate different steroids, albeit with different substrate specificities [22]. CYP134A1 is involved in the synthesis of pulcherriminic acid, a natural product [41], and CYP107H1 (P450 biol) is involved in the synthesis of polyketides [42]. Based on functionally characterized homolog P450s from other organisms, CYP105, CYP107, and CYP109 family P450s have been found to be associated with the degradation and biotransformation of a diverse array of xenobiotics and secondary metabolites [36,43,44]. CYP113 P450s are involved in the biosynthesis of secondary metabolites such as erythromycin [45,46] and tylosin [47,48]. Despite CYP102 and CYP152 P450s being found in secondary metabolite BGCs (in this study), their role in secondary metabolites biosynthesis has not been yet elucidated.
3. Materials and Methods
3.1. Species and Database
In total, 128 Bacillus species genomes available for public use at KEGG (https://www.genome.jp/kegg-bin/show_organism?category=Bacillus) were used in this study (Table S4). Bacillus species used in this study, along with their names, species codes, and individual genome database links, were presented in Table S4.
3.2. Genome Data Mining and Annotation of P450s
P450 mining in Bacillus species was carried out following the methods described elsewhere [25]. Briefly, the whole proteome of Bacillus species was downloaded from the databases listed in Table S4, and subjected to the NCBI Batch Web CD-Search Tool (http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). Proteins that belong to a P450 superfamily were selected and based on the International P450 Nomenclature Committee rule; proteins with >40% identity and >55% identity were grouped under the same family and subfamily, respectively [27,28,29]. Proteins with less than 40% identity were assigned to a new P450 family.
3.3. Phylogenetic Analysis of P450s
The phylogenetic tree of Bacillus species P450s was built as described elsewhere [25], with slight modifications. Briefly, the Bacillus P450s protein sequences along with the outgroup M. tuberculosis CYP51B1 (Rv0764c) protein were aligned by MAFFT v6.864 [49], embedded on the Trex web server [50]. Then, the alignments were automatically subjected to tree inferring and optimization by the Trex web server. Finally, the best-inferred trees were visualized, colored, and generated by iTOL (http://itol.embl.de/about.cgi) [51].
3.4. P450 Diversity Percentage Analysis
P450 diversity percentage analysis was carried out as described elsewhere [25,34]. Briefly, the P450 diversity percentage in Bacillus species was measured as a percentage contribution of the number of P450 families in the total number of P450s.
3.5. Generation of P450 Profile Heat-Maps
The presence or absence of P450s in Bacillus species was shown with heat-maps generated using P450 family data. The data was represented as −3 for family presence (green) and 3 for family absence (red). A tab-delimited file was imported into Mev (Multi-experiment viewer) [52]. Hierarchical clustering using a Euclidean distance metric was used to cluster the data. A hundred and twenty-eight Bacillus species formed the horizontal axis (see Supplementary dataset 3 for codes) and CYP family numbers formed the vertical axis.
3.6. Secondary Metabolite BGCs Analysis
Individual Bacillus species genome ID and plasmids IDs from the various species databases (Table S2) were submitted to anti-SMASH [30] for identification of secondary metabolite BGCs. Results were downloaded both in the form of gene cluster sequences and Excel spreadsheets representing species-wise cluster information, and finally, P450s that are part of a specific gene cluster were identified. Standard gene cluster abbreviation terminology available at anti-SMASH database [30] was maintained in this study.
3.7. Comparative Analysis of P450s
Mycobacterial P450s were retrieved from a published article [25] and used for comparative analysis with Bacillus species P450s. P450 families and subfamilies and the P450 diversity percentage were compared between the genera Mycobacterium and Bacillus.
4. Conclusions
Comparative analysis of P450s in bacterial species is gaining momentum and the availability of a large number of bacterial genome sequences is fueling this process. This study is an attempt to perform a comprehensive comparative analysis of P450s and to identify the P450s involved in secondary metabolite synthesis in Bacillus species. Future work involves understanding the role of different Bacillus P450s, identified in this study, in the synthesis of various secondary metabolites.
Acknowledgments
The authors want to thank Barbara Bradley, Pretoria, South Africa for English language editing.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/11/3623/s1.
Author Contributions
K.S. conceived and designed the experiments; all the authors were involved in performing the experiments, analysis of the data and writing of the manuscript. All the authors reviewed and approved the manuscript.
Funding
Khajamohiddin Syed expresses sincere gratitude to the University of Zululand Research Committee for funding (Grant No. C686) and to the National Research Foundation (NRF), South Africa for a research grant (Grant No. 114159). Puleng Rosinah Syed thanks NRF, South Africa for DST-NRF Innovation Master’s Scholarship (Grant No. 114575). Abidemi Paul Kappo is grateful to South African Medical Research Council (SAMRC) for research grant (Grant No. PC57009).
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study, the collection, analysis, or interpretation of data, the writing of the manuscript or the decision to publish the results.
References
- 1.Sono M., Roach M.P., Coulter E.D., Dawson J.H. Heme-containing oxygenases. Chem. Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 2.Bernhardt R. Cytochromes P450 as versatile biocatalysts. J. Biotechnol. 2006;124:128–145. doi: 10.1016/j.jbiotec.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 3.Isin E.M., Guengerich F.P. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim. Biophys. Acta (BBA) Gen. Subj. 2007;1770:314–329. doi: 10.1016/j.bbagen.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Fasan R. Tuning P450 enzymes as oxidation catalysts. ACS Catal. 2012;2:647–666. doi: 10.1021/cs300001x. [DOI] [Google Scholar]
- 5.Syed K., Porollo A., Lam Y.W., Grimmett P.E., Yadav J.S. CYP63A2, a catalytically versatile fungal P450 monooxygenase capable of oxidizing higher-molecular-weight polycyclic aromatic hydrocarbons, alkylphenols, and alkanes. Appl. Environ. Microbiol. 2013;79:2692–2702. doi: 10.1128/AEM.03767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le-Huu P., Heidt T., Claasen B., Laschat S., Urlacher V.B. Chemo-, regio-, and stereoselective oxidation of the monocyclic diterpenoid β-cembrenediol by P450 BM3. ACS Catal. 2015;5:1772–1780. doi: 10.1021/cs5020404. [DOI] [Google Scholar]
- 7.Syed K., Yadav J.S. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit. Rev. Microbiol. 2012;38:339–363. doi: 10.3109/1040841X.2012.682050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Podust L.M., Sherman D.H. Diversity of P450 enzymes in the biosynthesis of natural products. Nat. Prod. Rep. 2012;29:1251–1266. doi: 10.1039/c2np20020a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLean K.J., Leys D., Munro A.W. Microbial cytochrome P450s. In: Montellano P.R.O., editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th ed. Springer; Basel, Switzerland: 2015. pp. 261–407. [Google Scholar]
- 10.Guengerich F.P. Human cytochrome P450 enzymes. In: Montellano P.R.O., editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th ed. Springer; Basel, Switzerland: 2015. pp. 523–785. [Google Scholar]
- 11.Girhard M., Bakkes P.J., Mahmoud O., Urlacher V.B. P450 Biotechnology. In: Montellano P.R.O., editor. Cytochrome P450: Structure, Mechanism, and Biochemistry. 4th ed. Springer; Basel, Switzerland: 2015. pp. 451–520. [Google Scholar]
- 12.Greule A., Stok J.E., De Voss J.J., Cryle M.J. Unrivalled diversity: The many roles and reactions of bacterial cytochromes P450 in secondary metabolism. Nat. Prod. Rep. 2018;35:757–791. doi: 10.1039/C7NP00063D. [DOI] [PubMed] [Google Scholar]
- 13.Lamb D.C., Lei L., Warrilow A.G., Lepesheva G.I., Mullins J.G., Waterman M.R., Kelly S.L. The first virally encoded cytochrome p450. J. Virol. 2009;83:8266–8269. doi: 10.1128/JVI.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson D.R. Cytochrome P450 diversity in the tree of life. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2018;1866:141–154. doi: 10.1016/j.bbapap.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urlacher V.B., Eiben S. Cytochrome P450 monooxygenases: Perspectives for synthetic application. Trends Biochem. Sci. 2006;24:324–330. doi: 10.1016/j.tibtech.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Urlacher V.B., Girhard M. Cytochrome P450 monooxygenases: An update on perspectives for synthetic application. Trends Biochem. Sci. 2012;30:26–36. doi: 10.1016/j.tibtech.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 17.Cimermancic P., Medema M.H., Claesen J., Kurita K., Brown L.C.W., Mavrommatis K., Pati A., Godfrey P.A., Koehrsen M., Clardy J., et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeigler D.R., Perkins J.B. The genus Bacillus. In: Goldman E., Green L.H., editors. Practical Handbook of Microbiology. 2nd ed. CRC Press, Taylor & Francis Group; Boca Raton, FL, USA: 2009. pp. 309–337. [DOI] [Google Scholar]
- 19.Graumann P. Bacillus: Cellular and Molecular Biology. 2nd ed. Caister Academic Press; Haverhill, UK: 2012. [Google Scholar]
- 20.Zhao X., Kuipers O.P. Identification and classification of known and putative antimicrobial compounds produced by a wide variety of Bacillales species. BMC Genom. 2016;17:882. doi: 10.1186/s12864-016-3224-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grubbs K.J., Bleich R.M., Santa Maria K.C., Allen S.E., Farag S., Team A., Shank E.A., Bowers A.A. Large-scale bioinformatics analysis of Bacillus genomes uncovers conserved roles of natural products in Bacterial physiology. MSystems. 2017;2:e00040-17. doi: 10.1128/mSystems.00040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuya T., Shibata D., Kino K. Phylogenetic analysis of Bacillus P450 monooxygenases and evaluation of their activity towards steroids. Steroid. 2009;74:906–912. doi: 10.1016/j.steroids.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Ruettinger R.T., Wen L.P., Fulco A.J. Coding nucleotide, 5′ regulatory, and deduced amino acid sequences of P-450BM-3, a single peptide cytochrome P-450: NADPH-P-450 reductase from Bacillus megaterium. J. Biol. Chem. 1989;264:10987–10995. [PubMed] [Google Scholar]
- 24.Munro A.W., Leys D.G., McLean K.J., Marshall K.R., Ost T.W., Daff S., Miles C.S., Chapman S.K., Lysek D.A., Moser C.C., et al. P450 BM3: The very model of a modern flavocytochrome. Trends Biochem. Sci. 2002;27:250–257. doi: 10.1016/S0968-0004(02)02086-8. [DOI] [PubMed] [Google Scholar]
- 25.Parvez M., Qhanya L.B., Mthakathi N.T., Kgosiemang I.K.R., Bamal H.D., Pagadala N.S., Xie T., Yang H., Chen H., Theron C.W., et al. Molecular evolutionary dynamics of cytochrome P450 monooxygenases across kingdoms: Special focus on mycobacterial P450s. Sci. Rep. 2016;6:33099. doi: 10.1038/srep33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson D.R., Kamataki T., Waxman D.J., Guengerich F.P., Estabrook R.W., Feyereisen R., Gonzalez F.J., Coon M.J., Gunsalus I.C., Gotoh O., et al. The P450 superfamily: Update on new sequences, gene mapping, accession numbers, early trivial names of enzymes, and nomenclature. DNA Cell Biol. 1993;12:1–51. doi: 10.1089/dna.1993.12.1. [DOI] [PubMed] [Google Scholar]
- 28.Nelson D.R. Cytochrome P450 nomenclature. Methods Mol. Biol. 1998;107:15–24. doi: 10.1385/0-89603-519-0:15. [DOI] [PubMed] [Google Scholar]
- 29.Nelson D.R. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006;320:1–10. doi: 10.1385/1-59259-998-2:1. [DOI] [PubMed] [Google Scholar]
- 30.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R., Lee S.Y., Fischbach M.A., Müller R., Wohlleben W., et al. AntiSMASH 3.0—A comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Syed K., Shale K., Pagadala N.S., Tuszynski J. Systematic identification and evolutionary analysis of catalytically versatile cytochrome P450 monooxygenase families enriched in model basidiomycete fungi. PLoS ONE. 2014;9:e86683. doi: 10.1371/journal.pone.0086683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kgosiemang I.K.R., Mashele S.S., Syed K. Comparative genomics and evolutionary analysis of cytochrome P450 monooxygenases in fungal subphylum Saccharomycotina. J. Pure Appl. Microbiol. 2014;8:291–302. [Google Scholar]
- 33.Qhanya L.B., Matowane G., Chen W., Sun Y., Letsimo E.M., Parvez M., Yu J.H., Mashele S.S., Syed K. Genome-wide annotation and comparative analysis of cytochrome P450 monooxygenases in Basidiomycete biotrophic plant pathogens. PLoS ONE. 2015;10:e0142100. doi: 10.1371/journal.pone.0142100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ngwenya M.L., Chen W., Basson A.K., Shandu J.S., Yu J.H., Nelson D.R., Syed K. Blooming of unusual cytochrome P450s by tandem duplication in the pathogenic fungus Conidiobolus coronatus. Int. J. Mol. Sci. 2018;19:1711. doi: 10.3390/ijms19061711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sello M.M., Jafta N., Nelson D.R., Chen W., Yu J.H., Parvez M., Kgosiemang I.K.R., Monyaki R., Raselemane S.C., Qhanya L.B., et al. Diversity and evolution of cytochrome P450 monooxygenases in Oomycetes. Sci. Rep. 2015;5:11572. doi: 10.1038/srep11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moody S.C., Loveridge E.J. CYP 105—Diverse structures, functions and roles in an intriguing family of enzymes in Streptomyces. J. Appl. Microbiol. 2014;117:1549–1563. doi: 10.1111/jam.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., Poulos T.L. The structure of the cytochrome p450BM-3 haem domain complexed with the fatty acid substrate, palmitoleic acid. Nat. Struct. Biol. 1997;4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 38.Noble M.A., Miles C.S., Reid G.A., Chapman S.K., Munro A.W. Catalytic properties of key active site mutants of flavocytochrome P-450 BM3. Biochem. Soc. Trans. 1999;27:A44. doi: 10.1042/bst027a044b. [DOI] [PubMed] [Google Scholar]
- 39.Lee D.S., Yamada A., Matsunaga I., Ichihara K., Adachi S.I., Park S.Y., Shiro Y. Crystallization and preliminary X-ray diffraction analysis of fatty-acid hydroxylase cytochrome P450BSβ from Bacillus subtilis. Acta Crystallogr. D Struct. Biol. 2002;58:687–689. doi: 10.1107/S0907444902001762. [DOI] [PubMed] [Google Scholar]
- 40.Lee D.S., Yamada A., Sugimoto H., Matsunaga I., Ogura H., Ichihara K., Adachi S.I., Park S.Y., Shiro Y. Substrate recognition and molecular mechanism of fatty acid hydroxylation by cytochrome P450 from Bacillus subtilis: Crystallographic, spectroscopic and mutational studies. J. Biol. Chem. 2003;278:9761–9767. doi: 10.1074/jbc.M211575200. [DOI] [PubMed] [Google Scholar]
- 41.Cryle M.J., Bell S.G., Schlichting I. Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: A cyclo-l-leucyl-l-leucyl dipeptide oxidase. Biochemistry. 2010;49:7282–7296. doi: 10.1021/bi100910y. [DOI] [PubMed] [Google Scholar]
- 42.Cryle M.J., Schlichting I. Structural insights from a P450 carrier protein complex reveal how specificity is achieved in the P450BioI ACP complex. Proc. Natl. Acad. Sci. USA. 2008;105:15696–15701. doi: 10.1073/pnas.0805983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z.Z., Li X.F., Yang W., Dong X., Yu J., Zhu S.L., Li M., Xie L., Tong W.Y. Identification and functional analysis of cytochrome P450 complement in Streptomyces virginiae IBL14. BMC Genom. 2013;14:130. doi: 10.1186/1471-2164-14-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang A., Zhang T., Hall E.A., Hutchinson S., Cryle M.J., Wong L.L., Zhou W., Bell S.G. The crystal structure of the versatile cytochrome P450 enzyme CYP109B1 from Bacillus subtilis. Mol. Biosyst. 2015;11:869–881. doi: 10.1039/C4MB00665H. [DOI] [PubMed] [Google Scholar]
- 45.Savino C., Montemiglio L.C., Sciara G., Miele A.E., Kendrew S.G., Jemth P., Gianni S., Vallone B. Investigating the structural plasticity of a cytochrome P450: Three dimensional structures of P450 EryK and binding to its physiological substrate. J. Biol. Chem. 2009 doi: 10.1074/jbc.M109.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montemiglio L.C., Gianni S., Vallone B., Savino C. Azole drugs trap cytochrome P450 EryK in alternative conformational states. Biochemistry. 2010;49:9199–9206. doi: 10.1021/bi101062v. [DOI] [PubMed] [Google Scholar]
- 47.Merson-Davies L.A., Cundiiffe E. Analysis of five tyiosin biosynthetic genes from the tyllBA region of the Streptomyces fradiae genome. Mol. Microbiol. 1994;13:349–355. doi: 10.1111/j.1365-2958.1994.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 48.Fouces R., Mellado E., Díez B., Barredo J.L. The tylosin biosynthetic cluster from Streptomyces fradiae: Genetic organization of the left region. Microbiology. 1999;145:855–868. doi: 10.1099/13500872-145-4-855. [DOI] [PubMed] [Google Scholar]
- 49.Katoh K., Kuma K., Toh H., Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boc A., Diallo A.B., Makarenkov V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012;40:W573–W579. doi: 10.1093/nar/gks485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Letunic I., Bork P. Interactive Tree of Life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.