Abstract
Modifications in the anticodon loop of transfer RNAs (tRNAs) have been shown to ensure optimal codon translation rates and prevent protein homeostasis defects that arise in response to translational pausing. Consequently, several yeast mutants lacking important anticodon loop modifications were shown to accumulate protein aggregates. Here we analyze whether this includes the activation of the unfolded protein response (UPR), which is commonly triggered by protein aggregation within the endoplasmic reticulum (ER). We demonstrate that two different aggregation prone tRNA modification mutants (elp6 ncs2; elp3 deg1) lacking combinations of 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U: elp3; elp6; ncs2) and pseudouridine (Ψ: deg1) reduce, rather than increase, splicing of HAC1 mRNA, an event normally occurring as a precondition of UPR induction. In addition, tunicamycin (TM) induced HAC1 splicing is strongly impaired in the elp3 deg1 mutant. Strikingly, this mutant displays UPR independent resistance against TM, a phenotype we found to be rescued by overexpression of tRNAGln(UUG), the tRNA species usually carrying the mcm5s2U34 and Ψ38 modifications. Our data indicate that proper tRNA anticodon loop modifications promote rather than impair UPR activation and reveal that protein synthesis and homeostasis defects in their absence do not routinely result in UPR induction but may relieve endogenous ER stress.
Keywords: tRNA anticodon modifications, unfolded protein response, tunicamycin, yeast, elongator complex, Deg1
1. Introduction
Transfer RNA (tRNA) is extensively modified to fine-tune the efficiency of translation. Various tRNA modifications are known to contribute to the maintenance of optimal codon translation rates in yeast by preventing ribosomal pausing during the decoding process [1,2]. For example, 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) is present at the wobble position in tRNAGln(UUG) and tRNALys(UUU) and is required for efficient translation of the cognate A-ending codons by both tRNAs [2,3,4]. Formation of the mcm5s2U modification requires the Elongator complex and regulatory proteins, as well as Trm9/Trm112 methyl-transferase [5,6,7,8,9]. The thiomodification is separately installed via a sulfur transfer pathway involving Nfs1, Tum1, Uba4, Urm1 and thiolase Ncs2-Ncs6 [10,11,12]. In budding yeast, wobble uridine thiolation and mcm5U modification occur partially independent of each other. Loss of each part (mcm5U or s2U) alone induces shared pleiotropic phenotypes which are aggravated upon combined loss of both [2,13,14,15,16]. Similarly, ribosomal pausing is increased in double mutants lacking mcm5U in tandem with s2U (ncs2 elp6), which goes along with a severe protein homeostasis defect including the accumulation of protein aggregates [2].
In mice, loss of the mcm5 moiety alone was shown to induce ribosomal pausing and trigger the unfolded protein response (UPR) within the endoplasmic reticulum (ER), which is assumed to contribute to associated defects in neurodevelopment causing microcephaly [17]. In humans, various mutations in Elongator subunit genes are correlated with different pathologies involving neurodegeneration [18,19], pointing to a positive role of the Elongator in neurodevelopment and in protection against neurodegeneration. Since protein misfolding plays a key role in various neurodegenerative diseases and proteopathies [20], maintenance of translational efficiency and protein homeostasis might represent a functional link between tRNA modification and neurogenesis.
Another tRNA modification recently linked to human neuropathies is Deg1/Pus3 dependent formation of pseudouridine (Ψ38/39), since homozygous DEG1/PUS3 mutations are linked to a severe form of intellectual disability [21], a syndrome which can also be caused by human Elongator defects [22]. In yeast, a combination of Elongator and deg1 mutations was shown to cause severe growth impairment and was associated with the formation of protein aggregates [16]. Furthermore, defects in Deg1 or Elongator dependent modification and tRNA thiolation copy pleiotropic phenotypes in yeast, including temperature and rapamycin sensitivity [3,13,23,24], which is consistent with shared or related cellular roles for these tRNA modifications. For the elp3 deg1 mutant, severe translational inefficiency and growth phenotypes were shown to be suppressible by elevated copy numbers of tRNAGln(UUG), suggesting this tRNA critically relies on the joint presence of mcm5s2U and Ψ38 [16,25]. Hence, both anticodon loop modifications appear to collaborate in tRNAGln(UUG) functioning and translational efficiency [16,19].
Since protein aggregation may represent an important pathomechanism of tRNA modification defects in humans, a further characterization of tRNA linked protein homeostasis defects in the yeast model system appears justified. In this study, we aimed to clarify whether combined tRNA modification defects resulting in protein aggregation in yeast occur concomitantly with an induction of the UPR system as observed in the mice model system. Onset of UPR involves detection of ER-based protein homeostasis defects via the endonuclease Ire1, which mediates unconventional splicing of the HAC1 mRNA [26,27,28,29]. Only after HAC1 splicing is the functional Hac1 transcription factor produced, which is subsequently involved in the induction of ER localized chaperones to restore ER protein homeostasis [29].
Here, we demonstrate that two different combined tRNA modification defects known to result in protein aggregation in yeast, do so without concomitant induction of UPR. Moreover, basal UPR induction levels are decreased rather than increased, and genetic evidence points to the relief of basal ER stress levels due to severe translational defects as observed in the composite elp3 deg1 tRNA modification mutant. Hence, yeast models might differ considerably from metazoan neuronal cells in their cellular responses upon interference with tRNA anticodon modifications and inappropriate decoding.
2. Materials and Methods
2.1. Strains, Plasmids and Cultivation
The strains used and generated in this study are listed in Table 1. Genomic deletions were conducted by PCR mediated protocols based on the pUG plasmid set [30] with oligonucleotides listed in Table 2. Gene replacements were verified with forward/reverse primers positioned outside of the target loci (Table 2). Cultivation of the different strains with yeast nitrogen base (YNB)/yeast peptone dextrose (YPD) was performed using standard methods [31]. Overexpression of the tRNAGln(UUG) utilized pRK55, a YEplac195-based construct [16] and an empty vector served as a control. The designated strains were transformed as previously published [32].
Table 1.
Saccharomycescerevisiae strains used in this study.
| Strain | Genotype | References/Sources |
|---|---|---|
| BY4741 | MATa, his3Δ, leu2Δ, met15Δ, ura3Δ | Euroscarf, Frankfurt |
| AB43 | BY4741 ire1Δ::SpHIS5 | This study |
| RK520 | BY4741 ncs2Δ::SpHIS5 elp6Δ::KanMX4 | This study |
| RK206 | BY4741 urm1Δ::KanMX4 deg1Δ::SpHIS5 | [16] |
| RK220 | BY4741 elp3Δ::KanMX4 deg1Δ::SpHIS5 | [16] |
| AB97 | BY4741 elp3Δ::KanMX4 deg1Δ::SpHIS5 ire1Δ::KlURA3 | This study |
Table 2.
Oligonucleotides used in this study.
| Oligonucleotide | Sequence (5′–3′) | Target | References/Sources |
|---|---|---|---|
| Ire1_KO_Fwd | CATTAAAAAAACAGCATATCTGAGGAATTAATATTTTAGCACTTTGAAAACAGCTGAAGCTTCGTACGC | pUG27, pUG72 /IRE1 |
This study |
| Ire1_KO_Rev | TAACATTAATGCAATAATCAACCAAGAAGAAGCAGAGGGGCATGAACATGGCATAGGCCACTAGTGGATCTG | pUG27, pUG72 /IRE1 |
This study |
| Ire1_KO+_Fwd | CTTCGGGCAATACCTTCGACT | IRE1 | This study |
| Ire1_KO+_Rev | CAACCAAGAAGAAGCAGAGGG | IRE1 | This study |
| KO_NCS2_FW | TGCTATTGTCCATCCCTATCCTAGTTTTAAAAATATAATTCTATCAAGTTCAGCTGAAGCTTCGTACGC | pUG27 /NCS2 |
This study |
| KO_NCS2_RV | TAAATAAATAAATACATAACCATTGGAATAGCGAAGCCTTTGACATTTCAGCATAGGCCACTAGTGGATCTG | pUG27 /NCS2 |
This study |
| N_NCS2_FW | ACCGATGAGATGAGTGAGAC | NCS2 | This study |
| pUG27/SpHIS rev | GTCCAAAGCGATGGCAACGC | SpHIS5 | This study |
| HAC1_qPCR_Fwd | GACGACGCTACCTGCCG | HAC1i | This study |
| HAC1_qPCR_Rev | ACTGCGCTTCTGGATTACG | HAC1i | This study |
| qPCR_ACT1_FW | TTCCAGCCTTCTACGTTTCC | ACT1 | [16] |
| qPCR_ACT1_RV | AATCTCTACCGGCCAAATCG | ACT1 | [16] |
| HAC1F | CTGGCTGACCACGAAGACGC | HAC1u/i | [34] |
| HAC1R | TTGTCTTCATGAAGTGATGA | HAC1u/i | [34] |
2.2. Liquid Growth Inhibition Assays
The mutants as well as the transformants used in this study were adjusted to OD600 = 0.025 and grown in a 96-well plate for 24 h in YNB or YPD media with serially increasing concentrations of tunicamycin (TM) used in every well. For each strain, three independent cultures were incubated at the above-mentioned conditions. After the incubation, optical density (OD600) was measured using an Epoch Micro–Volume Spectrophotometer System (BioTek, Winooski, VT, USA). Relative growth of the different yeast strains was calculated by normalizing the optical densities with TM against those without.
2.3. Isolation of Total RNA
Yeast cultures were incubated until OD600 = 1.0 was reached, and total RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) following the instructions of the manufacturer. The isolated RNA was then used for qRT-PCR or RT-PCR experiments.
2.4. Quantification of mRNA Levels by qRT-PCR
As a control of UPR induction, splicing of the HAC1 mRNA in the indicated strains was induced in cultures after OD600 = 1.0 was reached, by the addition of 0.5 µg/mL TM for 3 h. To quantify HAC1i mRNA levels from three biological replicates, the SensiFAST SYBRR® No-ROX Kit (Bioline, Luckenwalde, Germany) and Mastercycler ep realplex (Eppendorf, Hamburg, Germany) were used, applying technical triplicates for each sample, following the manufacturers’ protocols and primers listed in Table 2. Calculation of the relative mRNA level of HAC1i in the different strains and under various conditions was achieved by normalizing against the quantified amount of ACT1 mRNA according to [33] whereas a two-tailed t-test was used for statistical analyses.
2.5. RT-PCR of HAC1 mRNA
For the cDNA synthesis, the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, Waltham, MA, USA) was used applying 1 µg of total RNA. The synthesized cDNA was conducted for PCR following established procedures [34] and using the primer HAC1F/R or qPCR_ACT1_FW/RV (PCR-protocol add-on: annealing temperature 60 °C for the control (Table 2), respectively. To separate the PCR products a 2% agarose gel was used. The expected RT-PCR product sizes of unspliced and spliced HAC1 mRNA are 720 bp and 470 bp, or 168 bp for the ACT1 mRNA, respectively [34,35].
3. Results and Discussion
3.1. Dysfunction of the UPR System in tRNA Modification Mutants
Loss of different critical tRNA modifications in yeast induces accumulation of protein aggregates [2,16,36]. However, it has remained unknown whether protein aggregation induced by tRNA defects occurs solely in the cytoplasm, or also within the ER. Importantly, protein aggregation in the ER is triggered in mice and flies in response to tRNA defects, and activates a transcriptomic change termed the unfolded protein response (UPR) [17]. UPR is mediated by the transcription factor Hac1 and its activation requires splicing of the HAC1 mRNA [26,28,29,37]. To test whether the UPR system is induced in yeast tRNA modification mutants lacking mcm5U or s2U in combination or together with Ψ38/39, we used RT-PCR to detect HAC1 mRNA splicing. This Ire1 dependent processing step is a prerequisite for UPR induction in yeast [26,28,29].
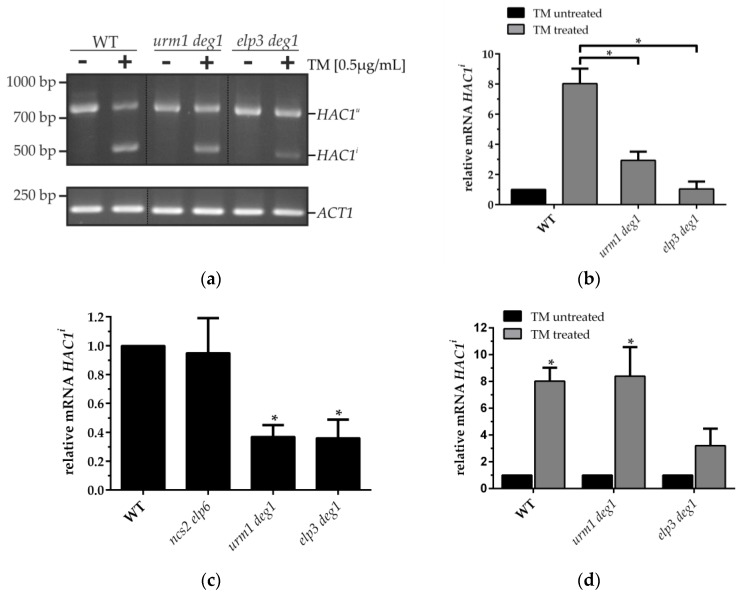
As seen in Figure 1a, the wild type (WT) and double tRNA modification mutants contained only unspliced HAC1u mRNA under optimal growth conditions basedon detection by the RT-PCR assay [34]. However, when WT cells were treated with TM, a well-known inhibitor of N-linked glycosylation [38] and therefore an UPR inducer in yeast, RT-PCR-based detection of spliced HAC1 mRNA was facilitated. Thus, absence of the tested tRNA modifications does not appear to induce splicing of HAC1 mRNA in a fashion, and to the extent, comparable to TM. To check for minor induction of HAC1 splicing in the tRNA modification mutants which may have escaped detection by the semi-quantitative RT-PCR assay, we devised a highly sensitive qRT-PCR based strategy to quantify spliced HAC1 mRNA (HAC1i). This strategy involves a set of oligonucleotides (Table 2), which generate a product only from spliced HAC1 mRNA. Indeed, TM exposure of WT cells resulted in an ~8-fold induction of spliced HAC1 levels, demonstrating the suitability of this assay to monitor UPR induction in yeast (Figure 1c).
Figure 1.
Analysis of basal and tunicamycin (TM) induced HAC1 splicing in various transfer RNA (tRNA) modification mutants. (a) To measure the splicing of HAC1 mRNA in wildtype (WT), elp3 deg1 and urm1 deg1 cells RT-PCR was conducted as described [34]. In each sample, ACT1 mRNA was detected as a control. Yeast strains were cultivated in yeast peptone dextrose (YPD) until OD600 = 1.0 and harvested for RNA extraction. As a control, WT was additionally treated with TM (0.5 µg/mL) for 3 h (+). HAC1u represents the unspliced HAC1 mRNA and HAC1i the mature spliced HAC1 mRNA. (b–d) Quantification of the HAC1i mRNA level without (b), and with TM treatment (c,d) via qRT-PCR of the indicated strains. Induction of HAC1 splicing with TM was carried out as described in (a). mRNA levels were normalized to ACT1 using the ΔΔCt method [33]. The results obtained with TM treated yeast strains were standardized against the HAC1i mRNA level of the untreated wild-type (c) or the corresponding untreated strains (d), respectively. Quantitative PCR was performed with at least three biological triplicates per strain and condition and statistical significance was determined using a two-tailed t-test and indicated in the bar charts (* p < 0.05, b–d).
When applying the quantitative assay to RNA from TM untreated tRNA modification mutants urm1 deg1 (s2U and Ψ38/39 deficient), elp3 deg1 (mcm5U and Ψ38/39 deficient) or ncs2 elp6 (mcm5s2U deficient) no elevated HAC1 splicing levels compared to the WT were observed (Figure 1b). In contrast, urm1 deg1 and elp3 deg1 mutants displayed significantly reduced HAC1 splicing levels, while the ncs2 elp6 mutant and the WT contained similar amounts of HAC1i (Figure 1b). Together these results indicate that observed protein homeostasis defects in ncs2 elp6 and elp3 deg1 [2,16] appear not to be associated with the induction of the UPR system. Hence, protein aggregation either does not include ER proteins, or is not sufficient to activate HAC1 splicing and subsequent transcriptional changes mediated by Hac1. In support of this notion, previous RNAseq data did not indicate an upregulation of UPR genes in the ncs2 elp6 mutant, and among the identified proteins in aggregates, cytosolic ones were strongly enriched whereas ER proteins were under-represented (Table S1) [2].
Since absence of UPR induction in aggregation prone tRNA modification mutants might be explainable either with absence of ER resident protein aggregates or with a UPR functional defect, we tested whether the UPR system is still functional in urm1 deg1 and elp3 deg1 mutants. We used TM to induce the HAC1 mRNA splicing [38], which we monitored using the RT-PCR approach. The WT showed after treatment with TM both spliced and unspliced HAC1 variants, respectively (Figure 1a, lane 2). The double mutants also displayed both HAC1u and HAC1i mRNAs following TM exposure (Figure 1a, lanes 3-6), indicating the functionality of the UPR system in these mutants. Interestingly, by comparing the intensity of the slower and faster migrating bands in urm1 deg1 mutant cells with the ones of the WT, it seemed that the HAC1i mRNA level was reduced in that mutant (Figure 1a, lanes 2 and 4). This effect was stronger for elp3 deg1 (Figure 1a, lanes 2 and 6).
To compare splicing efficiency in WT and tRNA modification mutant cells after TM exposure, we used qRT-PCR-based quantification of spliced HAC1 mRNA. Indeed, the double mutants showed a strong reduction in the amount of the TM-induced HAC1 splice product (Figure 1c, lanes 3 and 4). While TM treatment of WT cells induced HAC1i formation ~8-fold, this was reduced in urm1 deg1 (~3-fold) and elp3 deg1 (no induction), when standardized to the HAC1i level of the untreated WT, respectively. To further test if the double mutants additionally showed a reduced splicing rate of HAC1 mRNA, we set the HAC1i level of untreated strains as a standard and compared each to the amount after treatment, respectively (Figure 1d). While WT and urm1 deg1 displayed similar relative changes (~8-fold), the elp3 deg1 mutant showed decreased HAC1i inducibility (~3-fold) pointing to a reduced capacity to activate UPR.
Taken together, the tRNA modification double mutants are still capable of induced HAC1 mRNA splicing, but absolute HAC1i levels are reduced compared to the WT. Based on earlier studies, which revealed an accumulation of misfolded proteins in ncs2 elp6 and elp3 deg1 double mutants [2,16], it seems counterintuitive that these mutants exhibit reduced basal UPR activation and reduced capacity to initiate UPR upon exogenously induced ER stress. Possibly, protein aggregation in combined tRNA modification mutants is limited to the cytoplasm and reduced amounts of misfolded proteins are present in the ER. In support of this notion, it has been shown for the mcm5s2U deficient mutant ncs2 elp6 that only a small amount of the protein aggregates formed are ER-related [2].
3.2. Rescue of UPR Suppression by tRNA Overexpression
The above results revealed reduced HAC1i levels in combined tRNA modification mutants both before and after TM exposure, consistent with a general UPR defect. In several cases, downstream cellular effects resulting from translational defects and loss of critical tRNA anticodon loop modifications have been shown to be suppressed by overexpression of the tRNA substrates usually carrying the appropriate modifications [3,13,16,25,39,40]. Importantly, translational defects and phenotypes of the elp3 deg1 mutant were already shown to be suppressible by overexpression of tRNAGln(UUG), the single tRNA in yeast normally carrying the mcm5s2U and Ψ38 modifications in the anticodon stem loop [16]. Here we determined whether UPR defects of this mutant are similarly suppressible by tRNA overexpression. If so, UPR defects would likely occur as an indirect consequence of translational inefficiency, similar to the majority of pleiotropic phenotypes observed in Elongator and Deg1 deficient (elp3 deg1) cells.
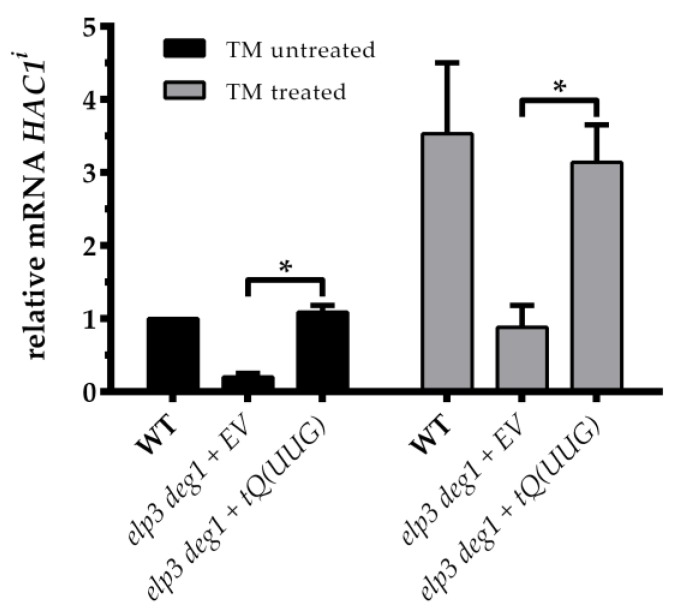
We thus first measured basal HAC1 splicing in elp3 deg1 mutant cells carrying either the empty vector or the tRNAGln(UUG) overexpression vector that had previously been shown to rescue multiple phenotypes of the double mutant. We found that the presence of the tRNAGln(UUG) expression vector resulted in a robust ~3-fold increase in basal HAC1 splicing in relation to the empty vector control (Figure 2). Basal HAC1 splicing levels were in fact restored to WT levels (Figure 2). Next, we analyzed whether tRNAGln(UUG) overexpression also suppresses the UPR induction defect of the double mutant after exposure to TM. Again, HAC1 splicing after TM treatment was significantly higher in response to the tRNAGln(UUG) overexpression construct compared to the empty vector control. Moreover, TM induced HAC1i levels of the elp3 deg1 mutant were restored nearly to WT levels in the presence of the tRNAGln(UUG) overexpression vector (Figure 2). Together, our results suggest that the observed UPR defect in the combined tRNA modification mutants highly likely lies with an indirect consequence of translational inefficiency, since it is suppressible by overexpression of the hypomodified tRNAGln(UUG), which causes the translational defect in the first place.
Figure 2.
Overexpression of tRNAGln(UUG) reverts the HAC1 mRNA splicing phenotype of the modification mutants. The indicated strains carrying either the empty vector (EV) or tRNAGln(UUG) (tQUUG) expressing vector were grown in YNB until OD600 = 1.0 before RNA extraction. HAC1i mRNA levels (before/after TM treatment) of the indicated transformants were quantified. Statistical significance was determined using a two-tailed t-test (* p < 0.05).
3.3. TM Phenotypes of tRNA Modification Mutants and Their Response to tRNAGln(UUG) Overexpression
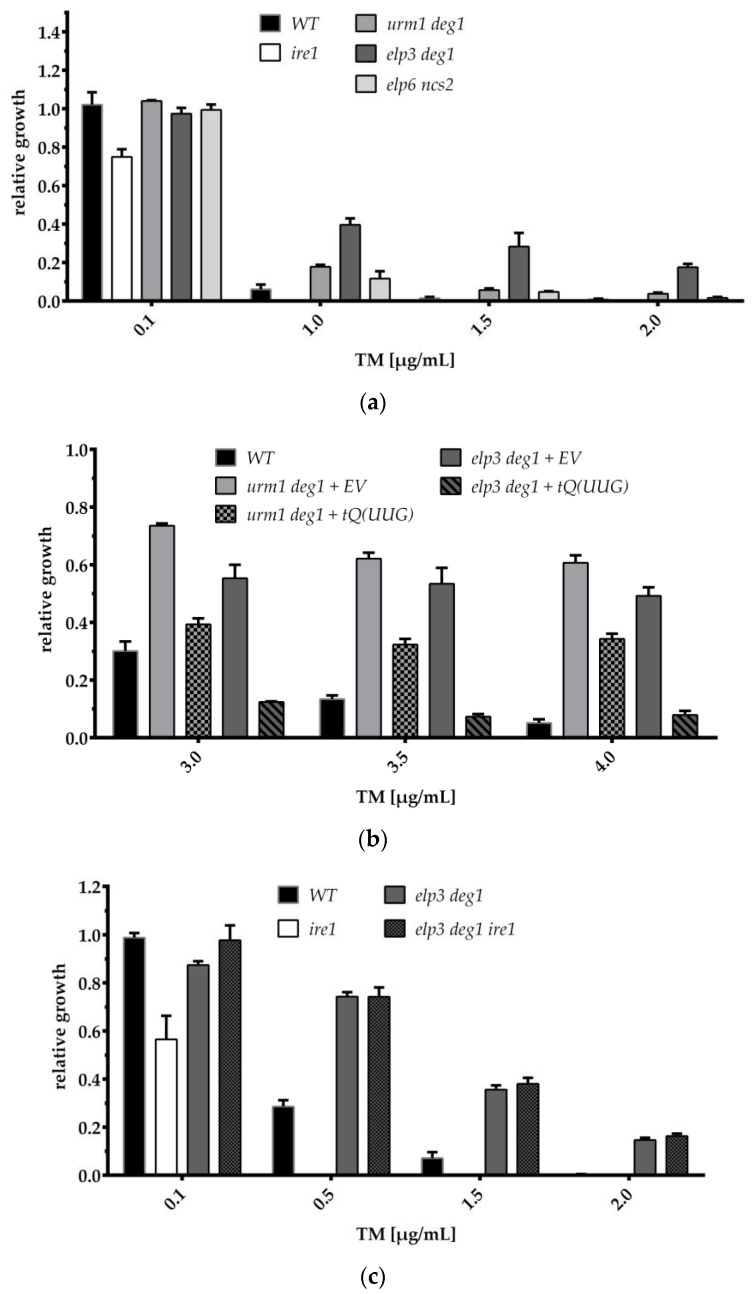
It has been demonstrated in different studies that the inability to induce the UPR system or downstream proteins results in a strong sensitivity against ER stress-inducing agents such as TM [41,42,43]. To test whether the tRNA modification mutants display altered TM sensitivity due to the reduced UPR induction we conducted liquid growth inhibition assays. At a TM concentration of 0.1 µg/mL, both the WT and the tRNA modification mutants (urm1 deg1; elp3 deg1; ncs2 elp6) showed a relative growth rate of about 100% (Figure 3a). At higher TM doses, the WT undergoes stepwise growth reduction, while urm1 deg1, elp3 deg1 and ncs2 elp6 mutants displayed partial TM resistance towards the ER stress-inducing agent. These phenotypes were unexpected concerning the strongly reduced HAC1i mRNA levels in the first two modification mutants (Figure 1c). Interestingly however, absence of the s2U part (of the mcm5s2U modification) in several Urm1 pathway mutants was already shown to confer resistance to ER stress induced by TM as well [44]. Moreover, it was demonstrated that overexpression of three tRNAs naturally carrying the s2U modification was sufficient to restore normal sensitivity to TM in a s2U deficient mutant [44]. Our observation that combined loss of tRNA modifications (including and excluding s2U) causes a HAC1 maturation defect and TM resistance, suggested a more general functional link between translational defects induced by inappropriate tRNA modifications and resistance to ER stress. However, it remained to be determined whether both, UPR induction and resistance to TM induced ER stress are jointly suppressible by tRNA overexpression. To test this, we compared TM resistance of urm1 deg1 and elp3 deg1 double mutants in the presence of either the empty vector or the tRNAGln(UUG) overexpression construct.
Figure 3.
TM phenotypes of tRNA modification mutants and their response to tRNAGln(UUG) overexpression. The indicated yeast strains were cultivated in YPD (a,c) or in YNB (b) 24 h with indicated concentrations of TM. Each experiment involved three biological replicates, and the resulting standard deviations are indicated on the bars. EV: empty vector; tQUUG: tRNAGln(UUG) expressing vector.
As shown in Figure 3b, at TM concentrations between 3 and 4 µg/mL, the presence of the tRNAGln(UUG) vector correlated with significantly reduced growth as compared to the empty vector control, for both urm1 deg1 and elp3 deg1 double mutants. Hence, tRNAGln(UUG) overexpression does not only restore HAC1 splicing in the absence and presence of TM (Figure 2), but also restores TM sensitivity of mutant strains. We further observed that the TM response differed depending on whether yeast cells were cultivated in rich or minimal medium with higher degrees of resistance observable in the latter condition (Figure 3a,b).
Interestingly, deletion of different ribosomal protein (RP) genes was shown to induce a similar TM resistance in yeast, and the strength of the TM effect correlated with the strength of growth inhibition due to loss of the individual RP gene [45]. Thus, reduced translational capacity and slower growth rates that result from either inappropriate tRNA modification or loss of RP function seem to generally improve the resistance against TM-induced ER stress. This possibly indicates that basal ER stress is already reduced due to the translational inefficiency and a concomitant reduction of protein traffic within the ER. In support of this assumption, it has already been demonstrated that translational inhibition by cycloheximide can induce partial TM resistance [45] and that different tRNA modification mutants indeed exhibit protein secretion defects [46].
Our results additionally argue against an important role of the canonical UPR pathway in the observed resistance against ER stress of tRNA modification mutants, since the latter goes along with a clear UPR defect rather than induction of the pathway. Most interestingly, TM resistance observed in several RP deficient mutant strains was at least partially independent of the presence of HAC1 and therefore occurs without participation of UPR induction [45]. Since our results revealed TM resistance in tRNA modification mutants with chronically reduced mature HAC1, we aimed to clarify whether both events are functionally linked or might occur independently of each other.
To this end we introduced an IRE1 deletion into the elp3 deg1 double mutant displaying the strongest TM resistance and subsequently scored changes in TM sensitivity by comparing WT, ire1, elp3 deg1 and elp3 deg1 ire1 mutants. Ire1 represents the endonuclease essential for HAC1 splicing [26,27,28,29], and therefore, its removal from elp3 deg1 and WT backgrounds allows us to clarify the role of HAC1 splicing in resistance against TM-induced ER stress in these mutants. As expected, the absence of Ire1 alone increased TM sensitivity in WT cells but did not alter the TM resistance of the elp3 deg1 strain, indicating that resistance against ER stress in tRNA modification mutants occurs largely independent of UPR (Figure 3c). We assume that interference with codon translation rates in yeast induces protein homeostasis defects mainly involving cytosolic proteins (Table S1), but at the same time decreases ER stress, resulting in a higher tolerance against ER stress-causing agents and decreased activation of the UPR. This may represent a distinct response as compared to the described effect of reduced codon translation rates in neuronal progenitor cells in mice where increased ER stress and UPR induction were observed [17]. Hence, such differences must be considered when yeast is utilized as a model system for the study of cellular effects of tRNA modification defects.
Acknowledgments
We thank Akif Ciftci (University of Freiburg, Freiburg, Germany) for generating the ncs2 elp6 double mutant.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/11/516/s1, Table S1: Gene ontology analysis of protein aggregates identified by mass spectrometry from tRNA modification mutant ncs2 elp6 §. § Cell compartment assignment of proteins identified in aggregates from modification mutant ncs2 elp6 by quantitative mass spectrometry [2] was analyzed with the gene ontology (GO) term function [47] of Saccharomyces genome database (SGD) [48].
Author Contributions
Conceptualization, A.B., R.K. and R.S.; investigation, A.B.; writing—original draft preparation, A.B., R.K. and R.S.; writing—review and editing, A.B., R.K. and R.S.; funding acquisition, R.K. and R.S.
Funding
We gratefully acknowledge support from the Deutsche Forschungsgemeinschaft (DFG) to R.S. (SCHA750/15, SCHA750/18) and their Priority Program SPP1784 Chemical Biology of Native Nucleic Acid Modifications to R.S. (SCHA750/20) and R.K. (KL2937/1). In addition, part of this work was carried out in the framework of the European Union Cost Action EPITRAN CA16120.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chou H.-J., Donnard E., Gustafsson H.T., Garber M., Rando O.J. Transcriptome-wide analysis of roles for tRNA modifications in translational regulation. Mol. Cell. 2017;68:978–992. doi: 10.1016/j.molcel.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nedialkova D.D., Leidel S.A. Optimization of Codon Translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esberg A., Huang B., Johansson M.J.O., Byström A.S. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 4.Schaffrath R., Leidel S.A. Wobble uridine modifications—A reason to live, a reason to die? RNA Biol. 2017;14:1209–1222. doi: 10.1080/15476286.2017.1295204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Studte P., Zink S., Jablonowski D., Bär C., von der Haar T., Tuite M.F., Schaffrath R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 2008;69:1266–1277. doi: 10.1111/j.1365-2958.2008.06358.x. [DOI] [PubMed] [Google Scholar]
- 6.Kalhor H.R., Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 2003;23:9283–9292. doi: 10.1128/MCB.23.24.9283-9292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B., Lu J., Byström A.S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA. 2008;14:2183–2194. doi: 10.1261/rna.1184108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang B., Johansson M.J.O., Byström A.S. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C., Huang B., Anderson J.T., Byström A.S. Unexpected accumulation of ncm5U and ncm5S2U in a trm9 mutant suggests an additional step in the synthesis of mcm5U and mcm5S2U. PLoS ONE. 2011;6:e20783. doi: 10.1371/journal.pone.0020783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leidel S., Pedrioli P.G.A., Bucher T., Brost R., Costanzo M., Schmidt A., Aebersold R., Boone C., Hofmann K., Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 11.Noma A., Sakaguchi Y., Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakai Y., Nakai M., Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J. Biol. Chem. 2008;283:27469–27476. doi: 10.1074/jbc.M804043200. [DOI] [PubMed] [Google Scholar]
- 13.Klassen R., Grunewald P., Thüring K.L., Eichler C., Helm M., Schaffrath R. Loss of anticodon wobble uridine modifications affects tRNALys function and protein levels in Saccharomyces cerevisiae. PLoS ONE. 2015;10:e0119261. doi: 10.1371/journal.pone.0119261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klassen R., Bruch A., Schaffrath R. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol. 2017;14:1252–1259. doi: 10.1080/15476286.2016.1267098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Björk G.R., Huang B., Persson O.P., Byström A.S. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA. 2007;13:1245–1255. doi: 10.1261/rna.558707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klassen R., Ciftci A., Funk J., Bruch A., Butter F., Schaffrath R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res. 2016;44:10946–10959. doi: 10.1093/nar/gkw705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laguesse S., Creppe C., Nedialkova D.D., Prévot P.-P., Borgs L., Huysseune S., Franco B., Duysens G., Krusy N., Lee G., et al. A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell. 2015;35:553–567. doi: 10.1016/j.devcel.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Kojic M., Wainwright B. The many faces of elongator in neurodevelopment and disease. Front. Mol. Neurosci. 2016;9:1–10. doi: 10.3389/fnmol.2016.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokołowski M., Klassen R., Bruch A., Schaffrath R., Glatt S. Cooperativity between different tRNA modifications and their modification pathways. Biochim. Biophys. Acta Gene Regul. Mech. 2017 doi: 10.1016/j.bbagrm.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Warren J.D., Rohrer J.D., Schott J.M., Fox N.C., Hardy J., Rossor M.N. Molecular nexopathies: A new paradigm of neurodegenerative disease. Trends Neurosci. 2013;36:561–569. doi: 10.1016/j.tins.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaheen R., Han L., Faqeih E., Ewida N., Alobeid E., Phizicky E.M., Alkuraya F.S. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum. Genet. 2016;135:707–713. doi: 10.1007/s00439-016-1665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen J.S., Srivastava S., Farwell K.D., Lu H.M., Zeng W., Lu H., Chao E.C., Fatemi A. ELP2 is a novel gene implicated in neurodevelopmental disabilities. Am. J. Med. Genet. Part A. 2015;167:1391–1395. doi: 10.1002/ajmg.a.36935. [DOI] [PubMed] [Google Scholar]
- 23.Klassen R., Schaffrath R. Role of pseudouridine formation by Deg1 for functionality of two glutamine isoacceptor tRNAs. Biomolecules. 2017;7:8. doi: 10.3390/biom7010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scheidt V., Jüdes A., Bär C., Klassen R., Schaffrath R. Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling. Microb. Cell. 2014;1:416–424. doi: 10.15698/mic2014.12.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han L., Kon Y., Phizicky E.M. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA. 2015;21:188–201. doi: 10.1261/rna.048173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/S0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 27.Mori K., Ma W., Gething M.J., Sambrook J. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-Q. [DOI] [PubMed] [Google Scholar]
- 28.Cox J.S., Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/S0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 29.Mori K., Kawahara T., Yoshida H., Yanagi H., Yura T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells. 1996;1:803–817. doi: 10.1046/j.1365-2443.1996.d01-274.x. [DOI] [PubMed] [Google Scholar]
- 30.Gueldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- 32.Chen D.C., Yang B.C., Kuo T.T. One-step transformation of yeast in stationary phase. Curr. Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 33.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De-Souza E.A., Pimentel F.S.A., Machado C.M., Martins L.S., da-Silva W.S., Montero-Lomeli M., Masuda C.A. The unfolded protein response has a protective role in yeast models of classic galactosemia. Dis. Model. Mech. 2014;7:55–61. doi: 10.1242/dmm.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori T., Ogasawara C., Inada T., Englert M., Beier H., Takezawa M., Endo T., Yoshihisa T. Dual functions of yeast tRNA ligase in the unfolded protein response: Unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol. Biol. Cell. 2010;21:3722–3734. doi: 10.1091/mbc.e10-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiaville P., Legendre R., Rojas-Benitez D., Baudin-Baillieu A., Hatin I., Chalancon G., Glavic A., Namy O., de Crecy-Lagard V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microb. Cell. 2016;3:29–45. doi: 10.15698/mic2016.01.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nojima H., Leem S.H., Araki H., Sakai A., Nakashima N., Kanaoka Y., Ono Y. Hac1: A novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic Acids Res. 1994;22:5279–5288. doi: 10.1093/nar/22.24.5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elbein A.D. Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu. Rev. Biochem. 1987;56:497–534. doi: 10.1146/annurev.bi.56.070187.002433. [DOI] [PubMed] [Google Scholar]
- 39.Alexandrov A., Chernyakov I., Gu W., Hiley S.L., Hughes T.R., Grayhack E.J., Phizicky E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. doi: 10.1016/j.molcel.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 40.Han L., Guy M.P., Kon Y., Phizicky E.M. Lack of 2’-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet. 2018;14:1–25. doi: 10.1371/journal.pgen.1007288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halbleib K., Pesek K., Covino R., Hofbauer H.F., Wunnicke D., Hänelt I., Hummer G., Ernst R. Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell. 2017;67:673–684. doi: 10.1016/j.molcel.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Sarkar D., Paira S., Das B. Nuclear mRNA degradation tunes the gain of the unfolded protein response in Saccharomyces cerevisiae. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizuno T., Masuda Y., Irie K. The Saccharomyces cerevisiae AMPK, Snf1, negatively regulates the Hog1 MAPK pathway in ER stress response. PLoS Genet. 2015;11:1–25. doi: 10.1371/journal.pgen.1005491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Damon J.R., Pincus D., Ploegh H.L. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol. Biol. Cell. 2014;26:270–282. doi: 10.1091/mbc.e14-06-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steffen K.K., McCormick M.A., Pham K.M., Mackay V.L., Delaney J.R., Murakami C.J., Kaeberlein M., Kennedy B.K. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012;191:107–118. doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abdullah U., Cullen P.J. The tRNA modification complex elongator regulates the Cdc42-dependent mitogen-activated protein kinase pathway that controls filamentous growth in yeast. Eukaryot. Cell. 2009;8:1362–1372. doi: 10.1128/EC.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris M.A., Deegan J.I., Ireland A., Lomax J., Ashburner M., Tweedie S., Carbon S., Lewis S., Mungall C., Day-Richter J., et al. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:440–444. doi: 10.1093/nar/gkm883. [DOI] [Google Scholar]
- 48.Skrzypek M.S., Hirschman J. Using the Saccharomyces Genome Database (SGD) for analysis of genomic information. Curr. Protoc. Bioinform. Chapter. 2011:1. doi: 10.1002/0471250953.bi0120s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.