Abstract
Background:
Fgf10 is expressed in various tissues and organs, such as the limb bud, heart, inner ear and head mesenchyme. Previous studies identified Fgf10 enhancers for the inner ear and heart. However, Fgf10 enhancers for other tissues have not been identified.
Results:
By using primary culture chick embryo lateral plate mesoderm cells, we compared activities of deletion constructs of the Fgf10 promoter region, cloned into a promoter-less luciferase reporter vector. We identified a 0.34kb proximal promoter that can activate luciferase expression. Then, we cloned 11 evolutionarily conserved sequences located within or outside of the Fgf10 gene into the 0.34kb-promoter-luciferase vector, and tested their activities in vitro using primary cultured cells. Two sequences showed the highest activities. By using the Tol2 system and electroporation into chick embryos, activities of the 0.34 kb promoter with and without the two sequences were tested in vivo. No activities were detected in limb buds. However, the 0.34kb-promoter exhibited activities in the dorsal midline of the brain, while Fgf10 was detected in broader region in the brain. The two non-coding sequences negatively acted on the 0.34kb-promoter in the brain.
Conclusions:
The proximal 0.34kb promoter has activities to drive expression in restricted areas of the brain.
Keywords: Fgf10, enhancer, primary culture, chick embryos, in vivo electroporation
INTRODUCTION
Fibroblast growth factor 10 (Fgf10), which encodes a secreted factor, is one of the earliest genes to be expressed during limb development (Ohuchi et al., 1997), and has multiple important roles. During initiation of limb development, both cellular and molecular events are coordinated to form nascent limb buds. More specifically, localized epithelial-to-mesenchymal transition of the coelomic epithelium within the presumptive limb field generates mesenchymal limb progenitors (Gros and Tabin, 2014). Subsequently, localized regulation of proliferation at the limb-forming region (Searls and Janners, 1971), as well as oriented cell motility and division (Wyngaarden et al., 2010; Gros and Tabin, 2014), lead to initial outgrowth of limb buds. During these processes, Fgf10 is induced in the limb-forming region at Hamburger-Hamilton (HH) stage 13/14 in chick embryos (embryonic day 2) (Hamburger and Hamilton, 1951; Ohuchi et al., 1997), and acts on the somatopleure epithelium to contribute to the epithelial-to-mesenchymal transition process (Gros and Tabin, 2014).
Fgf10 also initiates the molecular events for limb outgrowth. At HH stage 16 (embryonic day 2.25) in chick embryos, mesenchymal FGF10 induces Fgf8 expression in the overlying ectoderm that develops into the apical ectodermal ridge, a specialized ectodermal tissue (Crossley et al., 1996; Vogel et al., 1996; Ohuchi et al., 1997). This process involves WNT/ß-catenin signaling (Kengaku et al., 1998; Kawakami et al., 2001; Barrow et al., 2003; Soshnikova et al., 2003) and the Sp transcription factor family members, SP6 and SP8 (Haro et al., 2014). FGF8, together with other FGFs produced by the apical ectodermal ridge, signals to the mesenchyme to promote mesenchymal survival and proliferation and also maintains Fgf10 expression (Ohuchi et al., 1997; Sun et al., 2002; Boulet et al., 2004; Mariani et al., 2008). The FGF10-FGF8 feedback loop interacts with other genes, such as Shh, Gremlin1 and Bmp4 (Delgado and Torres, 2017), to maintain limb outgrowth until tissue growth disconnects the feedback loop (Scherz et al., 2004; Verheyden and Sun, 2008).
During the stages of Fgf10 induction in limb progenitors, Fgf10 is also expressed in other tissues. For instance, Fgf10 is expressed in the secondary heart field (Watanabe et al., 2012) and the lung bud (Teshima et al., 2016) in mouse embryos, and is required for the development of the heart and lung (Sekine et al., 1999; Watanabe et al., 2010). In chick embryos, Fgf10 expression is detected in the proepicardium (Kruithof et al., 2006) and paraxial mesoderm (Ohuchi et al., 1997; Karabagli et al., 2002). Fgf10 is also expressed in various tissues in craniofacial structures, such as the inner ear in chick and mouse embryos (Alvarez et al., 2003; Kruithof et al., 2006; Sanchez-Calderon et al., 2007; Bothe et al., 2011) and in pharyngeal arches (Havens et al., 2006; Bothe et al., 2011). Genetic experiments in mice revealed that Fgf10 is required in the cranial neural crest cells that contribute to the development of the palate and salivary gland (Teshima et al., 2016; Chatzeli et al., 2017). In addition, Fgf10 expression has been reported in chick craniofacial development (Bothe et al., 2011).
Cis-regulatory elements are DNA sequence elements where transcription factors and chromatin modifying enzymes act to control gene expression (Rister and Desplan, 2010; Suryamohan and Halfon, 2015; Peter and Davidson, 2016). For instance, promoter sequences are cis-regulatory elements that are bound by basal transcriptional machinery, including RNA polymerase II, and determine transcriptional initiation sites of genes. Distal cis-regulatory modules include insulators and enhancers, where multiple transcription factors bind and cooperate. While insulators function for isolating regulatory domains of the genome, enhancers function to determine tissue or cell type-specific gene expression in a spatial-temporal manner during embryonic development (Spielmann and Mundlos, 2016; Petit et al., 2017). Tissue-specific enhancers, bound by transcription factors, contact corresponding gene promoters to stabilize RNA polymerase II and the general transcription factors, leading to regulation of tissue-specific expression for proper tissue and organ development (Amano et al., 2009; Birnbaum et al., 2012; Williamson et al., 2016; Hu and Tee, 2017). Identifying and characterizing such enhancers is an important approach in understanding the mechanisms of tissue-specific gene expression.
Our knowledge about the location and function of tissue-specific enhancers is limited, compared to genes and proteins they encode in animal genome (Visel et al., 2007). Evolutionary conservation of non-coding sequences and analysis of chromatin modifications offer prediction of enhancer sequences, yet experimental testing in vivo is needed to determine enhancers (Visel et al., 2007; Cunningham et al., 2018; Osterwalder et al., 2018). VISTA Enhancer Browser (https://enhancer.lbl.gov/) has been established to generate functional in vivo datasets with standardized methodology and consistent functional annotation (Visel et al., 2007). This web site provides a centralized resource for in vivo enhancer data of human and mouse sequences, assayed in E11.5 mouse embryos by transgenesis; however, researchers still need to assay activities of putative enhancers for tissue- and stage-specific developmental expression.
In the case of Fgf10, an in vitro study characterized the Fgf10 promoter region. A sequence of 6.5 kb upstream of the Fgf10 gene contains three T-box protein binding elements and a LEF/TCF binding motif, which are conserved between human and mouse. The 6.5 kb sequence could be transcriptionally activated in luciferase reporter assays by co-transfection of a Tbx5-expressing construct or by activation of ß-catenin signaling (Agarwal et al., 2003). An in vivo LacZ-reporter transgenesis experiments in mouse embryos also characterized Fgf10 promoter region (Sasaki et al., 2002). A 2.0 kb fragment, consisting of 1.3 kb proximal promoter sequence and 0.7 kb 5’ untranslated region (UTR), did not have activities to regulate Fgf10 expression in the limb buds at E11.5. In contrast, the same study determined that a 0.7 kb sequence upstream of initiation ATG codon (i.e. 5’ UTR sequence) contained activities to drive reporter expression at the distal tip of condensing digit primordia at E12.5. In addition to these promoter characterization studies, several studies have identified and characterized tissue-specific enhancers for Fgf10 expression in vivo. These include an inner ear enhancer (Ohuchi et al., 2005; Economou et al., 2013) and a cardiac enhancer (Watanabe et al., 2012). In the limb, a recent report identified that evolutionarily conserved AGAAAR clusters in the Fgf10 promoter region act to regulate limb bud mesenchyme-specific Fgf10 expression in the FGF10-FGF8 feedback loop maintenance during limb outgrowth (Yamamoto-Shiraishi et al., 2014). However, cis-regulatory modules for Fgf10 expression during limb initiation and expression in other tissues remain unknown.
In an attempt to better understand the transcriptional regulation of Fgf10, we compared genomic sequences surrounding the Fgf10 gene in vertebrates and selected conserved non-coding sequences as candidates of cis-regulatory modules of Fgf10. Through luciferase-reporter assays using a chick lateral plate mesoderm (LPM)-derived primary culture system, we identified and characterized the activities of non-coding sequences in stimulating reporter expression. We tested the activities of the proximal promoter and two cis elements in developing embryos by electroporating reporter constructs in chick embryos. Our in vivo data suggest that, rather than acting in limb progenitors, the proximal promoter and the two cis elements might function in regulation of Fgf10 expression in the craniofacial region.
RESULTS
Activities of Fgf10 promoter region
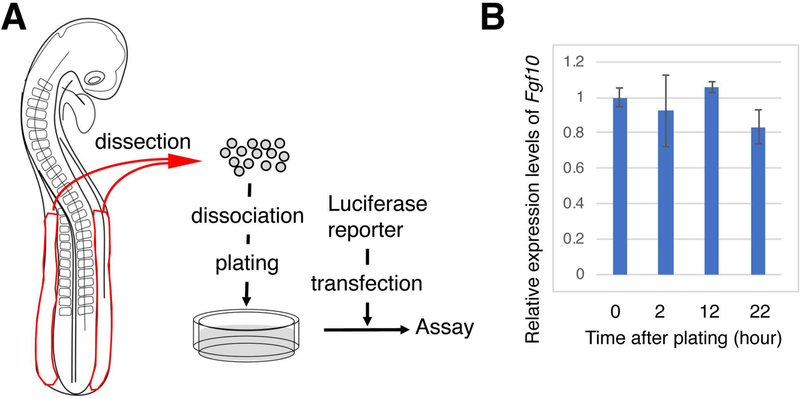
We sought to characterize candidate sequences in the regulation of Fgf10 expression in limb progenitor cells by reporter assays using primary cultured LPM cells from chick embryos. In the chick embryo, forelimb buds and hindlimb buds develop at 15–20th and 20–25th somite levels, respectively (Burke et al., 1995). Previous experiments have demonstrated that flank tissues can respond to stimuli, such as ectopic FGF8, which mimic the formation of the FGF10-FGF8 feedback loop, and initiate ectopic Fgf10 expression and ectopic limb formation (Cohn et al., 1995; Gibson-Brown et al., 1998; Isaac et al., 1998; Logan et al., 1998; Ohuchi et al., 1998; Isaac et al., 2000). In order to test activities of cis-elements to drive Fgf10 expression, we used LPM cells from HH stage 14–16 chick embryos that possess endogenous factors to drive expression of Fgf10. We dissected LPM posterior to the 14th to 15th somite levels, and set up primary cultures (Figure 1). In order to test whether the LPM cells in culture maintain endogenous Fgf10 expression, we performed a qunatitative reverse transcription-polymerase chain reaction (qRT-PCR) assay (Figure 1B). The LPM cells in culture maintained Fgf10 expression for up to 22 hours. This result suggest that cultured primary LPM cells could be used to test activities of cis-elements for Fgf10 expression.
Figure 1. Schematic of luciferase reporter assay using primary cultured cells from LPM of chick embryos.
(A) Chick embryo LPM was dissected, and cell were plated after dissociation. Luciferase reporter constructs were transfected and the luciferase activities in the LPM cells were measured.
(B) Quantitative RT-PCR for Fgf10 at indicated time points of LPM cell culture. Gapdh was used as a reference. The y axis represents relative levels of Fgf10 expression compared to time point 0 (cells just before plating). Average ± standard deviation with n=3 is shown at each time point. No statistical difference was observed at each time point, compared to time point 0 by t-test.
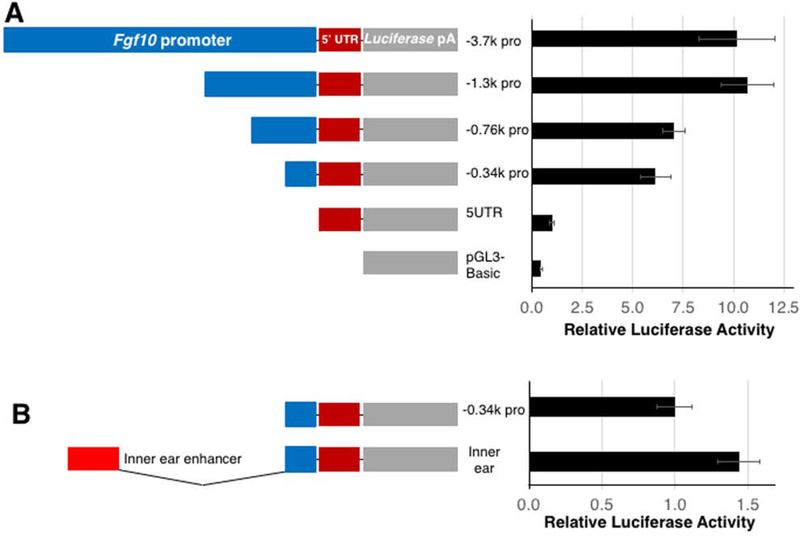
A previous study has shown that a 3.7 kb sequence upstream of the Fgf10 gene is conserved among humans, mice and chickens (Ohuchi et al., 2005). In previous studies, tissue-specific regulatory elements are identified by their conservation between humans and non-mammalian vertebrates across long evolutionary distances (Pennacchio et al., 2006; Visel et al., 2007). Therefore, we tested mouse DNA sequences in the chick system in this study. We cloned the mouse sequence of −3.7kb to +384 bp position, relative to the transcription start site (TSS), into a promoter-less luciferase vector. To characterize the activities of the sequence upstream of the Fgf10 gene, we generated deletion mutants from the −3.7 kb-luciferase construct, consisting of −1.3 kb, −757 bp and −341 bp to +384 bp sequences (Figure 2). We also generated a luciferase construct without a promoter but with a 5’ untranslated region (UTR) sequence.
Figure 2. Activities of Fgf10 promoter regions and inner ear enhancer in LPM cells in vitro.
(A) Luciferase reporter activities of indicated promoter constructs. Blue bars indicate various deletion versions of the proximal promoter of the Fgf10 gene. The red and gray bars represent 5’ UTR of Fgf10 and luciferase-polyA cassette, respectively.
(B) Comparison of activities of −0.34k promoter construct with and without previously-characterized inner ear enhancer.
Luciferase activities are shown as mean ± S.D.
By transfecting primary culture LPM cells with the promoter-luciferrase constructs, we compared the activities of sequences upstream of the Fgf10 gene. Transfecting a reporter that lacked the promoter sequence but possessed a 384 bp 5’ UTR sequence resulted in only an approximately 2 fold activation of luciferase activities, compared to the promoter-less luciferase vector. Therefore, we compared activities of promoter-containing constructs to the promoter-less 5’ UTR-luciferase construct. We detected approximately 10 and 10.5 fold luciferase activity with the −3.7 kb and −1.3 kb promoter reporters, respectively. Deletions to −757 bp and to −341 bp promoters decreased the activities to 6.8 and 5.9 fold, respectively. These results suggest that the 341 bp sequence upstream of the Fgf10 TSS is able to to drive luciferase reporter expression, and extending the sequence to −1.3 kb position further enhances this activity.
Activities of a previously characterized cis element within the 3.7 kb promoter region
Next, we sought to characterize potential Fgf10 cis-elements using the LPM primary culture-luciferase assay system. Because enhancers interact with promoters for activation of gene expression (Amano et al., 2009; Shlyueva et al., 2014; Williamson et al., 2016), we cloned candidate cis-elements into the luciferase vector with the Fgf10 promoter sequence. In this setting, putative enhancers are cloned in close proximity to the Fgf10 promoter, which do not test potential contribution of distance between the promoter and an enhancer to transcriptional regulation. We chose the 341 bp promoter sequence plus the 384 bp 5’ UTR sequence, which we referred to as −0.34k pro (Figure 2B) for the following reasons. The VISTA Enhancer Browser (https://enhancer.lbl.gov/) contains enhancer activities examined by LacZ transgenesis in mouse embryos (Visel et al., 2007). One of sequences, termed hs516, covered the −562 to +696 bp position of the human FGF10 gene, which exhibited LacZ reporter staining in a part of hindlimb buds, but not forelimb buds, at E11.5. Although this data does not provide information about hs516 activities at the time of limb bud initiation, the result implies that the proximal promoter region of the FGF10 gene contributes to FGF10 expression. Moreover, the proximal promoter covering the −341 to +384 bp position showed significant activity in the LPM cells in culture (Figure 2A). Therefore, we chose to use this sequence as a proximal promoter in combination with cis-elements.
We first tested activities of the inner ear-specific cis-element. Previous reports demonstrated that the sequence located approximately 3 kb upstream from the mouse Fgf10 TSS is able to drive reporter expression in a inner ear-specific manner (Ohuchi et al., 2005; Economou et al., 2013). In four independent assays (n=3 each), the inner ear element exhibited 1.6 ± 0.4 fold activation. As an example, Figure 2B shows a result with 1.4 fold activation of the luciferase activities in the LPM cells. This weak activation may be due to the weak activity of the inner ear enhancer to drive a LacZ reporter expression in the LPM in mouse embryos, as observed in a previous study (Ohuchi et al., 2005). Therefore, we consider that activation below 2 fold is weak and unlikely contributes to Fgf10 expression during limb initiation.
Activities of candidate cis elements located in the intron sequence
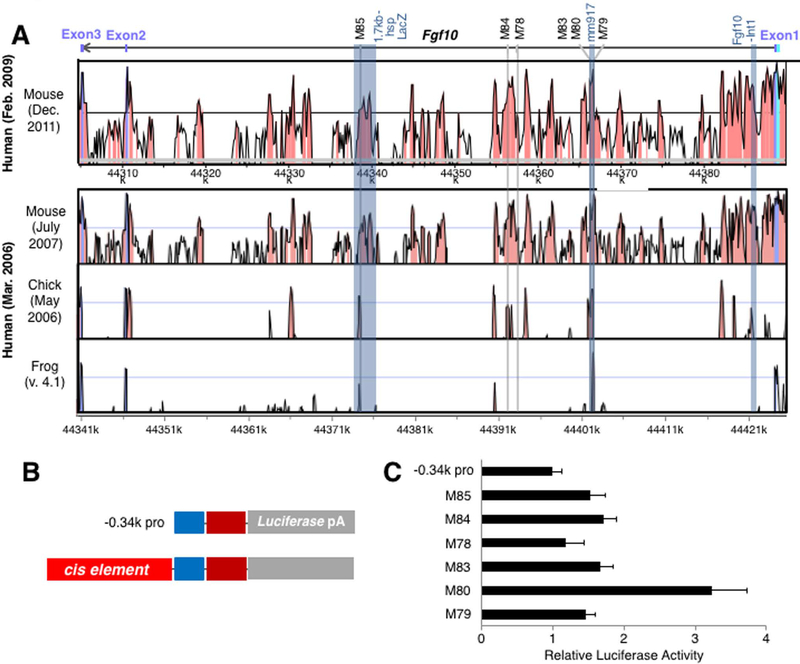
Next, we compared genome sequences in order to identify evolutionarily conserved sequences which may function in regulating Fgf10 expression. By using the VISTA Brower, we compared the human sequence with sequences of other limb-developing animals. Due to the high degree of conservation of non-coding sequence among mammals, which makes it difficult to select evolutionarily-conserved sequences, we limited our comparison of the human genome to mice, chickens, and frogs. First, we analyzed intronic sequences using genomic information from these animals. The FGF10 gene consists of three exons, as well as two introns: the first intronic sequence is approximately 83 kb in length, while the second intronic sequence is approximately 5.5 kb in length (Figure 3). We chose six intronic sequences from the mouse genome based on conservation of the sequences between human and mouse genomes. Figure 3 and Table 1 show the positions of these sequences, and nucleotide sequence alighments are shown in Supplemental information. The VISTA Enhancer Browser shows the activities of one cis-element, mm917, located in intron 1 of the mouse Fgf10 gene. The mm917 sequence showed LacZ reporter expression in the dorsal root ganglion and the limb at E11.5 by LacZ transgenesis in mouse embryos. The mm917 sequence covers three of the cis-elements we analyzed; M79, M80 and M83. Among these three sequences, M80 exhibited approximately 3.2 fold activation, comparred to the −0.34kb promoter construct (Figure 3C), while the M79 and M83 elements showed 1.4 and 1.7 fold activation, respectively.
Figure 3. Activities of conserved non-coding sequences in the introns of the Fgf10 gene in vitro.
(A) VISTA comparison of the Fgf10 locus in human vs mouse (upper) and human vs mouse vs chick vs frog (lower). Peaks with orange color indicate conserved non-coding sequences. The Fgf10 gene and exons are indicated with a bar and boxes at the top. Previously characterized sequences are shown with blue columns with their names at the top. The six sequences in intron 1 used in this study are indicated by light gray columns with their names on the top.
(B) Schematic presentation of −0.34k promoter-luciferase vector (upper) and a construct with a tested cis-element.
(C) Comparison of activities of −0.34k promoter construct and the six constructs with indicated cis-elements. Luciferase activities are shown as mean ± S.D.
Table 1.
Locations of enhancer elements examined in this study
| Analyzed enhancers located 5’ to the Fgf10 gene | ||
|---|---|---|
| Sequence ID | Distance from Fgf10 TSS | Chromosome position |
| M81 | 123 kb | 118,592,098–118,592,348 |
| M82 | 71.9 kb | 118,643,131–118,643,472 |
| Analyzed enhancers located 3’ to the Fgf10 gene | ||
|---|---|---|
| Sequence ID | Distance from 3’ end of the Fgf10 gene | Chromosome position |
| M86 | 233.3 kb | 119,025,892–119,026,225 |
| M87 | 2.6 kb | 118,789,402–118,790,008 |
| Analyzed enhancers located in the first intron of the Fgf10 gene | ||
|---|---|---|
| Sequence ID | Distance from TSS | Chromosome position |
| M85 | 44.5 kb | 118,760,110–118,759,911 |
| M84 | 29.8 kb | 118,745,259–118,745,148 |
| M78 | 28.8 kb | 118,744,148–118,744,391 |
| M83 | 20.8 kb | 118, 736,178–118,736,267 |
| M80 | 20.7 kb | 118,736,470–118,736,111 |
| M79 | 19.7 kb | 118,735,505–118,735,984 |
Fgf10 TSS position is 118,715,384
3’ end of the Fgf10 is 118,792,573
Fgf10 TSS position is 118,715,384 on chromosome 13 in the UCSC Genome Browser on Mouse Dec. 2011 (GRCm38/mm10) Assembly
The M78 and M84 sequences, also located in intron 1, exhibited subtle (1.2 fold) and weak activation (1.7 fold), respectively. We also tested the activities of the M85 sequence, located 44.5 kb downstream of the Fgf10 TSS. This sequence is a part of the 1.7 kb element that has been shown to act as cardiac enhancer (Watanabe et al., 2012). The M85 sequence also exhibited only weak (1.5 fold) activity in LPM cells. These results show only weak activation of the luciferase reporter in LPM primary cell culture, and suggest that these sequences may not function in the activation of Fgf10 during limb development.
Activities of candidate cis elements located in the 5’ or 3’ of the Fgf10 gene
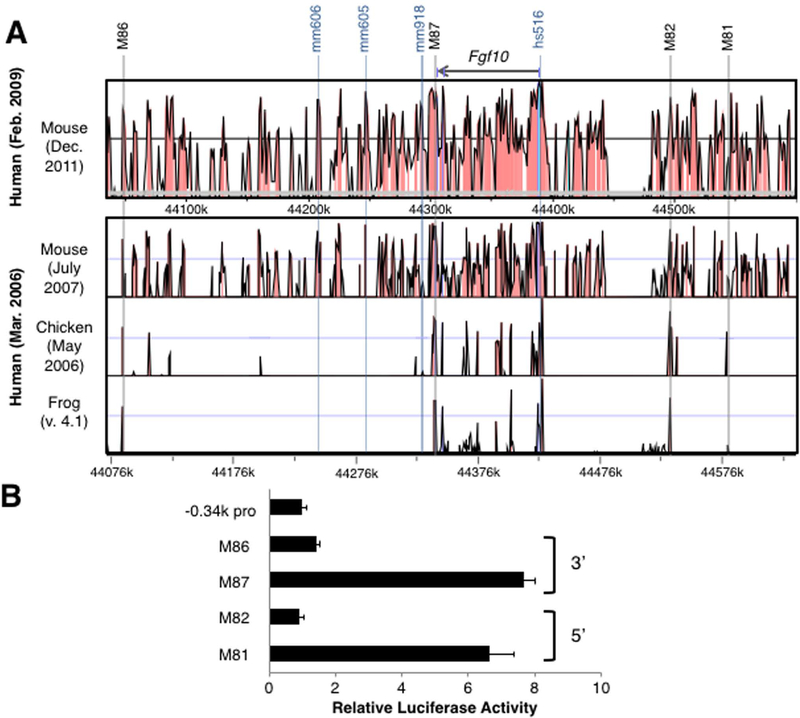
Next, we searched for conserved sequences in the upstream and downstream regions of the FGF10 gene. In the 5’ upstream region, we chose two sequences, M81 and M82 (Figure 4A, Table 1, Supplemental information). The human M81 sequence is conserved among mice and chickens, but not in frogs. In contrast, the M82 sequence showed conservation among all four species. Luciferase assays in primary chick LPM cells showed 6.6 fold activation by the M81 sequence, while M82 sequence did not show any activation (Figure 4B).
Figure 4. Activities of conserved non-coding sequences in the 5’ and 3’ of the Fgf10 gene in vitro.
(A) VISTA comparison of the Fgf10 locus in human vs mouse (upper) and human vs mouse vs chick vs frog (lower). The Fgf10 gene is indicated as a bar at the top. Previously characterized sequences are shown with blue columns with their names at the top. Two sequences in the 5’ and two sequences in the 3’ to the Fgf10 gene, tested in this study are indicated by light gray columns with their names on the top.
(B) Comparison of activities of −0.34k promoter construct and the four constructs with indicated cis-elements. Luciferase activities are shown as mean ± S.D.
We also searched sequences in the 3’ region of the FGF10 gene. In this region, three sequences were analyzed by the VISTA Enhancer Browser (Figure 4A). Among the three sequences, the mm606 and mm918 sequences did not show any activity at E11.5 by the LacZ reporter transgenesis assay in the mouse embryos. The mm605 sequence showed activity in the facial mesenchyme and cranial ganglion at E11.5. Furthermore, we analyzed two sequences, M87 and M86, which are disinct from the sequences shown in the VISTA Enhancer Browser (Table 1, Supplemental information). Both M87 and M86 show conservation among mice, chickens and frogs. Although the M86 sequence showed only weak activity (1.4 fold), the M87 sequence exhibited a 7.7 fold activation of the luciferase reporter.
These results suggest that the M81 sequence, located 5’ of the Fgf10 gene, and the M87 sequence, located 3’, showed the highest activation of luciferase activity among the tested sequences in primary cultured LPM cells.
Activities of cis elements in limb buds in vivo
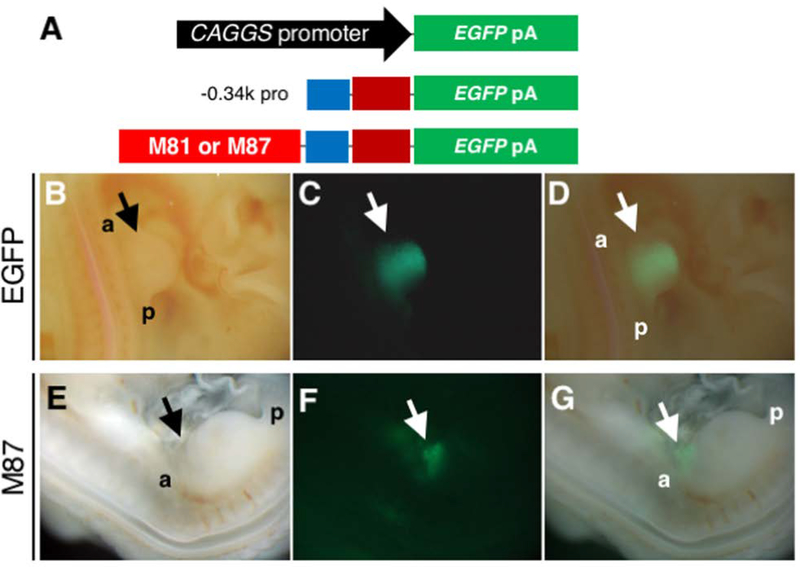
We next tested the activities of the M81 and M87 sequences in vivo, which showed the highest level of activity in primary culture LPM cells. We chose to use the Tol2 transposone system in combination with electroporation in chick embryos, which has been shown to be efficient to stably transduce constructs in developing embryos (Sato et al., 2007). The constructs were electroporated into the LPM including presumptive limb field of HH stage 11–13 embryos. When the pCAGGS-EGFP, a constitutive EGFP-expression construct (Figure 5A), was electroporated into the LPM of hindlimb-forming region, the EGFP signal was observed within 24 hours. The signal was maintained in the broad region of the hindlimb bud at 48 hours post electroporation (Figure 5B-D, n=11/15). By electroporating a construct that contained the −0.34k promoter and EGFP (Figure 5A), we did not detect the EGFP signal in the LPM and limb bud (n=10), suggesting that the −0.34 kb proximal promoter sequence does not have sufficient activity to induce downstream reporter gene expression in the LPM and limb bud.
Figure 5. Activities of proximal promoter with and without two cis-elements in the developing chick limb bud.
(A) Schematics of constructs used in the Tol2 vectors. Top: the pCAGGS-EGFP. Middle: −0.34k promoter-EGFP. Bottom: −0.34k promoter-EGFP with either M81 sequence or M87 sequence.
(B-D) Images of the hindlimb bud of an embryo in which the Tol2-pCAGGS-EGFP and pCAGGS-transposase constructs were electroporated into the LPM of hindlimb-forming region. Bright field (B), EGFP (C) and a merged image (D) of the hindlimb bud. EGFP signal was observed throughout the limb mesenchyme at 48 hours after electroporation (n=11/15).
(E-G) Images of an embryo in which the Tol2–0.34k promoter-EGFP with the M87 sequence and pCAGGS-transposase constructs were electroporated into the LPM. Bright field (E), EGFP (F) and a merged image (G) of the hindlimb bud. Weak EGFP signal in the anterior edge of the hindlimb bud and the flank region at 48hr after electroporation.
We next tested whether the 5’ element (M81) or the 3’ element (M87) had any activity when combined with the proximal promoter. We performed electroporation of constructs which contained the −0.34k promoter, an EGFP reporter, and either the 5’ (M81) sequence (n=34) or the 3’ (M87) sequence (n=64) (Figure 5A). We also tested co-electroporation of these constructs together (n=24). In two out of the 64 embryos with the 3’ sequence (M87), we observed weak EGFP signals in the anterior edge of the hindlimb bud and the flank region (Figure 5E-G). However, we did not detect EGFP signals in all other embryos. We also did not detect the EGFP signal by plasmid introduction and electroporation at an earlier stage (HH stage 9–10, n=20).
Activities of cis elements in the craniofacial region in vivo
Fgf10 is expressed in tissues outside of limb buds, and we reasoned that the cis elements identified in our in vitro assay may have activities outside of the limb bud. Therefore, we tested whether the 5’ element (M81) or the 3’ element (M87) had activity in other tissues. The developing face and brain are other regions where Fgf10 is expressed around the time of its expression in limb progenitors (Havens et al., 2006; Teshima et al., 2016; Chatzeli et al., 2017).
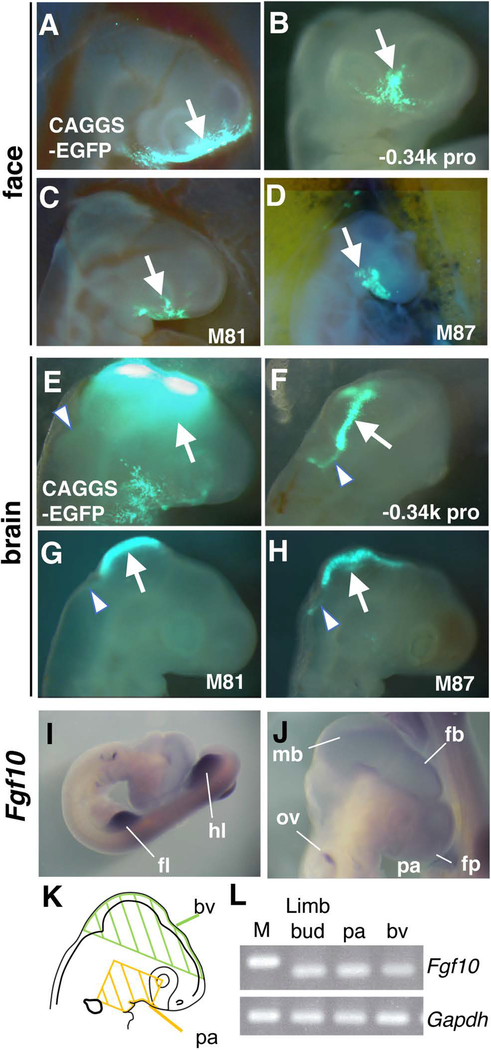
By electroporation of the pCAGGS-EGFP construct into the frontonasal region of HH stage 14–15 embryos, a strong EGFP signal was detected in the frontonasal prominence, maxirally prominence and ventral region of the optic vesicle within 24 hours (n=5/5, Figure 6A), showing successful introduction of a reporter construct in the frontonasal region. We next performed electroporation using the construct with the −0.34k promoter and EGFP reporter. We detected the EGFP signal in the frontonasal and maxillary prominence (n=3/4, Figure 6B, arrow), showing the activity of the −0.34k proximal promoter to drive a reporter expression in the frontonsal and maxillary region. The EGFP signal was restricted to a narrow region of the face compared to the broad signal by the constitutive pCAGGS-EGFP construct, suggesting that the activity of the proximal promoter shows spatial specificity in the facial region. Moreover, the reproducible reporter expression also suggests that the reporter signals are unlikely caused by integration-position effects. Next, we electroporated −0.34k promoter + EGFP with either the 5’ (M81) or the 3’ (M87) sequences into the facial prominence. We found that each construct induced EGFP reporter expression (n=9/12, in each experiment) (Figure 6C, D, arrows), which was comparable to that induced by the −0.34k promoter construct. These results suggest that the 5’ element (M81) and the 3’ element (M87) do not have detectable activity in the facial region.
Figure 6. Activities of proximal promoter with and without two cis-elements in the developing craniofacial region.
(A) An EGFP image of the face 24 hours after electroporation of the pCAGGS-EGFP and pCAGGS-transposase into the frontonasal region (n=5/5). Arrow points to broad signals in the frontonasal and maxillary region.
(B-D) EGFP images of the face 24 hours after electroporation of indicated constructs using the Tol2 system. Arrows point to the EGFP signals in the frontonasal and maxillary region.
(E) An EGFP image of the dorsal head region 24 hours after electroporation of the pCAGGS-EGFP and pCAGGS-transposase into the brain vesicle (n=5/5). Arrow points to strong signals in the dorsal side of the posterior forebrain to the midbrain, and arrowhead points to weak signals in the hindbrain.
(F-H) EGFP images of the dorsal regions of the brain 24 hours after electroporation of indicated constructs using the Tol2 system. Arrows point to the strong EGFP signals in the dorsal midline of the posterior forebrain – midbrain. Arrowheads point to weak EGFP signals in the dorsal midline of the anterior hindbrain.
(I, J) in situ hybridization of Fgf10 in HH stage 19 embryo. J shows close up of the craniofacial region. fb, forebrain; fl, forelimb bud; fp, frontonasal prominence; hl, hindlimb bud; mb, midbrain; ov, otic vesicle; pa, pharyngeal arch.
(K) A schematic of the regions dissected from chick embryos for RT-PCR assays.
(L) RT-PCR analysis of expression of Fgf10 and Gapdh. The expression in the limb bud, pharyngeal arch (pa) and brain vesicle (bv) at HH stage 16–17 is shown. M, DNA marker.
We also tested activity in the brain vesicle by injecting constructs into the anterior region of the neural tube. By introducing the pCAGGS-EGFP construct into the anterior region of the neural tube, we detected broad and strong EGFP signals in the dorsal region of the posterior forebrain and midbrain (n=5/5, Figure 6E, arrow). In addition, weak EGFP signals were also detected in the hindbrain in this experimental setting (Figure 6E, arrowhead). Introduction of the −0.34k promoter + EGFP reporter led to strong EGFP expression in the dorsal midline of the midbrain (Figure 6F, arrow) and weak expression in the hindbrain (Figure 6F, arrowhead) in a restricted manner (n=4/8). Introduction of the −0.34k promoter + EGFP with either the 5’ (M81) or the 3’ (M87) sequences resulted in the expression of the EGFP reporter in the same domain as −0.34k promoter + EGFP construct (Figure 6G, H), but the frequency of EGFP-positive embryos dropped significantly (n=1/10 for both the 5’ and 3’ sequences). The lower frequencies of EGFP-positive embryos with reporters containing the 5’ or 3’ sequernces suggest that these sequences negatively affect the activities of the −0.34k proximal promoter.
Prior genetic experiments demonstrated that Fgf10 is expressed in the cranial neural crest (Teshima et al., 2016). Gene expression experiments in chick embryos also showed Fgf10 expression in head development (Bothe et al., 2011). Accordingly, we detected weak expression of Fgf10 signals by whole mount in situ hybridization in the forebrain (fb) and midbrain (mb), the frontonasal prominence (fp), the 1st pharyngenal arch (pa), the otic vesicle (ov), in addition to strong expression in the limb bud at HH stage 19–20 (Figure 6I, J). As the brain vesicle tends to get non-specific signals by in situ hybridization analysis, we dissected the brain vesicle, pharyngeal arches and limb bud (Figure 6K), and examined Fgf10 expression by RT-PCR. This analysis showed evident amplicons of Fgf10 transcripts from the brain vesicle, similar to limb buds and pharyngeal arches (Figure 6L). This data further provides evidence that Fgf10 is expressed in the brain vesicle. Although our initial analysis was intended to characterize Fgf10 enhancer in limb progenitors, our in vivo analysis suggests that the −0.34k promoter region has an activity to drive expression in the dorsal midline of the midbrain and anterior hindbrain. Furthermore, the significantly reduced frequency of EGFP reporter expression with the 5’ (M81) and 3’ (M87) sequences suggests that these sequences may negatively regulate the activity of the −0.34k promoter sequence in the brain.
DISCUSSION
In the past decade, several studies attempted to characterize cis-regulatory elements controlling Fgf10 expression. In particular, several previous in vitro experiments also characterized activities of the Fgf10 promoter region. For instance, a luciferase construct containing −6.5 kb to TSS of the Fgf10 gene can be activated by Tbx5 when transfected into COS7 cells, a fibroblast-like cell line derived from African green monkey kidney (Agarwal et al., 2003). Another study showed that a 500 bp sequence, located approximately 2kb upstream of the TSS, can be activated by Hoxd9 when transfected into P19, a pluripotent mouse embryonic carcinoma cell line (Sheth et al., 2013). Such an approach, co-transfection of transcription factors of interest and luciferase reporter constructs with putative regulatory elements, is widely used to study transcriptional regulation. Results from such experiments can provide insights into how specific transcription factors of interest act on the regulatory elements, if the transcription factors of interest are not endogenously expressed in the cell lines. Compared to such an approach, our experiment was designed to test enhancer activities by endogenous factors in LPM cells without transfecting exogenous transcription factors. In this experimental setting, cells may change gene expression pattern due to alterations of the native molecular environment. Because the levels of Fgf10 expression did not show significant changes during culture of LPM cells, these cells would maintain LPM/limb progenitor status during the in vitro assay. Although a limb bud cell line that represent E11.5 limb bud mesenchyme status has been recently reported (Peluso et al., 2017), so far, there is no cell line that represents LPM and/or early limb progenitor status for in vitro assays. Therefore, the use of primary cultured cells from developing embryos could offer an assay of activities of putative regulatory elements in limb progenitor cells in vitro.
Our characterization of the Fgf10 promoter region show higher activities when longer sequences are used in the assay. This observation suggests that, while the proximal 0.34 kb sequence has promoter activities, further upstream sequences in the promoter region may receive more input for transcriptional activation. A recent study in mouse embryos demonstrated that a proximal promoter of the mouse Fgf10 gene regulates Fgf10 expression in developing forelimb buds and hindlimb buds at E10.75 through the FGF10-FGF8 feedback loop (Yamamoto-Shiraishi et al., 2014). This element is located between −768 to + 14 bp relative to the TSS of the Fgf10 gene, and exhibited transcriptional activities in the distal limb mesenchyme. In our in vitro experiment, the −0.78k construct (−781 to +384) and −0.34k construct (−341 to +384) exhibited similar levels of activity. However, the −0.34k construct did not show activity in our in vivo electroporation experiment. The difference between our data and the data by the recent publication suggest that sequences between −781 and −341 may be needed during limb outgrowth in vivo.
In the last two decades, technologies to introduce plasmid DNA into developing chick embryos have been developed. These methods include in ovo electroporation and sonoporation (Itasaki et al., 1999; Nakamura et al., 2000; Ohta et al., 2008). By in ovo transfection methods, enhancer-reporter constructs were introduced into tissues in developing chick embryos to test enhancer activities in vivo (Timmer et al., 2001). Such applications lead to identification of enhancers of the chick Sox2 gene, which regulate distinct spatial-temporal specificities of Sox2 expression (Uchikawa et al., 2003). More recently, activities of enhancer sequences obtained from mouse and catshark genomic sequences were tested in the developing chick limb bud system (Onimaru et al., 2015; Haro et al., 2017). These studies demonstrated the usefulness of developing chick embryos as a platform to test putative enhancer activities of other species. Although previous reports demonstrated robust reporter expression in the in ovo enhancer assays, introduced plasmid DNA can be diluted by active cell proliferation. Therefore, we combined the Tol2-based integration of plasmids (Kawakami, 2007; Sato et al., 2007), so the reporter constructs could be stably inherited to daughter cells. While our study focused on enhancer activities only after short period of time from introduction of plasmid DNA, such stable-integration approach may be beneficial for longer period of analysis.
We characterized activities of a total of 11 cis-elements by in vitro assays, two of which exhibited higher activities than others. One element is located far upstream of the TSS of the Fgf10 gene and the other is located immediately 3’ to the Fgf10 gene. In vivo characterization of these cis-elements using an EGFP reporter and the Tol2 system in the LPM did not show evident activity. One possible reason for the lack of activity is that the activity by these elements alone are weak, and may be dependent on the activity of other sequences to elicit higher activities. Interestingly, when the −0.34k promoter-reporter construct was introduced into the craniofacial region, EGFP reporter expression was successfully driven. Fgf10 is expressed in the craniofacial region (Havens et al., 2006; Chatzeli et al., 2017). Moreover, genetic inactivation of Fgf10 in cranial neural crest cells results in defects in the palate, salivary gland, ocular gland and circumvallate papilla of the tongue (Teshima et al., 2016), demonstrating the requirement of Fgf10 in neural crest cells in the development of these tissues. Accordingly, we also detected expression of Fgf10 in the brain vesicle by RT-PCR analysis. The in vivo analysis by electroporation in chick embryos suggests that the −0.34k promoter sequence possesses an activity in the dorsal midline of the brain. Although we initially characterized the 5’ (M81) and the 3’ (M87) sequences by using LPM cells, these elements might negatively function on the −0.34k promoter sequences in the brain.
Given the importance of Fgf10 functions in vivo, several studies reported cis-elements that regulate the spatial-temporal expression of Fgf10 in sites other than the developing limb bud. Those studies identified that the sequence located approximately 3 kb upstream of the Fgf10 transcription start site regulate inner ear expression (Ohuchi et al., 2005). In addition, a cis-element in intron 1 of the Fgf10 gene regulates Fgf10 expression in the heart (Golzio et al., 2012; Watanabe et al., 2012). Further studies in the future to elucidate the detailed mechanisms regulating Fgf10 expression will enhance the understanding of the upstream regulation of Fgf10 during limb initiation and outgrowth, as well as during the development of other tissues.
EXPERIMENTAL PROCEDURES
Comparative genome analysis
A comparative genome analysis of the Fgf10 genomic sequence, including regions covering 420kb upstream of the TSS and 600kb downstream of the transcriptional stop site, was performed using the VISTA Genome Pipeline (http://pipeline.lbl.gov). The browser allowed comparison between Feb 2009/GFCh37/hg19 version human sequences only with mammalian sequences. Therefore, we also used the Mar. 2006/NCBI36/hg18 version human sequence, which allowed for comparison with mouse (July 2007/NCBI37/mm9), chick (May 2006/WCUGSC 2.1/galGal3), and frog (Xenopus tropicalis, ver. JGI 4.1.XenTro2) sequences in order to identify evolutionarily-conserved regions.
Fgf10 promoter reporter constructs
A 3.7 kb mouse Fgf10 promoter region, including 384 bp of the 5’ UTR, were cloned into pGL3-Basic vector. Serial deletion constructs containing 1.3 kb, 781 bp and 341 bp proximal promoters were generated. A deletion construct without the promoter but the 5’ UTR clone was also generated.
Fgf10 enhancer reporter constructs
Consensus sequences conserved among three (human, mouse, chick) or four (human, mouse, chick, frog) species were amplified from mouse genome by polymerase chain reaction using the Expand High Fidelity PCR System (Roche) (Table 1). We primarily chose sequences with more than 70% conservation between human, mouse, and chick and/or frog sequences. We also selected sequences that are less conserved, when compared to chick and/or frog sequences, in the case that human and mouse sequences show high conservation.
Amplified DNA fragments were subcloned into the pGL3-Basic luciferase vector containing the 341 bp Fgf10 proximal promoter and 384 bp 5’ UTR sequences, with a short spacer sequence, CTCTTACGCGTGCTAGCCCG. The constructs were verified by Sanger sequencing.
Luciferase reporter assays
Chick embryos at HH stage 15–16 (staging according to (Hamburger and Hamilton, 1951)) were isolated into plastic dishes containing Hank’s Balanced Salt Solution with antibiotics. The lateral plate mesoderm (LPM), posterior to 14th - 15th somite levels, was dissected by cutting the tissue from dorsal side between the paraxial mesoderm and LPM using fine tungsten needles (Figure 1). The dissected tissues were kept in a 15-ml tube with Hank’s Balanced Salt Solution with antibiotics on ice until desired amount of the tissue was collected. The collected tissues were then dissociated by TripLE digestion (Invitrogen) and pipetting, and the reaction was stopped by adding D-MEM plus 10% fetal bovine serum. Cells were filtered through 100 μm Cell Strainer (BD Falcon), pelleted by centrifugation, washed with PBS twice, and plated at 1.2×105 cells/well in a 96 well plates (BD Falcon). Cells were cultured in Opti-MEM supplemented with 1% fetal bovine serum and antibiotics. Immediately after plating, cells were transfected with 100 ng of the luciferase reporter construct and 10 ng of pRL-TK using FuGENE 6 (Promega). Cells were cultured 24 hours after transfection, and luciferase activity was measured using the Dual-Luciferase reporter assay system according to the manufacturer’s instructions (Promega). Luciferase activity was normalized to the Renilla luciferase internal control. Experiments were repeated in triplicate in three independent assays. We repeated experiments at least three times to ensure the results.
Quantitative RT-PCR analysis of primary culture LPM cells
Chick LPM cells, immediately before plating, were harvested with Trizol (Invitrogen) as the time point 0 hour. LPM cells in culture were harvested at time points, 2, 12 and 22 hours after plating using Trizol. cDNA was prepared using one microgram of total RNA and oligo dT primers using Super Script III (Invitrogen). Quantitative PCR was done by using cDNA that corresponds to 20 ng RNA per reaction by using GoTaq qPCR master mix (Promega). The primers for Fgf10 are GTTGTGGCAGTTAAGTCCATC (nucleotide 610 – 630 of NM_204696, in the exon 2) and TCCCCTTCCATTCAGAGCAAC (nucleotide 784 – 804 of NM_204696, in the exon 3). Gapdh expression was used as a reference with the primers, GTCATCCATGACAACTTTGGC (nucleotide 538 – 558 of NM_204305, exon 6) and CAGGTCAACAACAGAGACATTG (nucleotide 765 – 786, NM_204305, exon 8).
In vivo analysis of enhancer activities in developing chick embryos
To test the activity of cis-elements and the promoter regions to drive expression in developing chick embryos, we used the Tol2 system (Kawakami, 2007). The 341 bp proximal promoter and 381 bp 5’ UTR were cloned into the Tol2 transposable vector that contains EGFP and SV40 polyA signal sequences. An enhancer element was cloned in front of the Fgf10 promoter-5’UTR sequence.
Plasmid solutions were adjusted to a final concentration of 2.5–4.0 μg/μl with distilled water colored with Fast Green. The amnion membrane on the posterior half of the embryo was removed from HH stage 11–13 chick embryos, then, 50–100 nl of plasmid solution (Tol2 reporter plasmid and pCAGGS-Transposase, 1:1 in volume) was injected into the LPM of the presumptive hindlimb field. Electroporation into the LPM was performed as previously described (Suzuki and Ogura, 2008). In short, the cathode electrode was placed under the LPM, and the anode electrode was positioned on top of the LMP, and electric pulse was charged. The pulse was combined with one short pulse (25v, 0.05 msec pulse-on, 0.1 msec pulse-off), and five long pulses (5v, 10.0 msec pulse-on, 1.0 msec pulse-off). Then, eggs were re-incubated, and the GFP signals were detected 24–48hr after electroporation.
Electroporation into the facial region was performed as previously described (Nakamura et al., 2000) with a slight modification. The plasmid solution was injected beneath the ectoderm of the frontonasal prominence of HH stage 14–15 embryos. The cathode electrode was placed along the frontonasal prominence, and the anode electrode was set along the dorsal side of the brain. Then an electric pulse (25v, 50 msec pulse-on, 950 msec pulse-off, five repetitions) was charged. Electroporation into the brain vesicle was also performed in a similar manner with the plasmid solution injection into the anterior lumen of the neural tube. The pulse for the brain vesicle was the same as for the facial region.
RT-PCR for Fgf10, expressed in tissue primodia
Chick brain vesicle, pharyngeal arch and limb buds were dissected from HH stage 16–17 embryos (Figure 6K). Total RNA was prepared using the NucleoSpin Gel and PCR clean-up kit (Takara/Macherey-Nagel), and cDNA was prepared by Superscript III with oligo dT primer. Fgf10 expression was confirmed by RT-PCR with the same primers used in the qRT-PCR experiment for LPM cell culture. The PCR products were further confirmed to be a part of Fgf10 and Gapdh by Sanger sequencing.
Supplementary Material
Key findings:
Primary culture of lateral plate mesoderm from chick embryos is used to test activities of cis-elements of Fgf10 by luciferase reporter assays.
Among 11 sequences tested, two sequences in combination with a proximal promoter showed higher luciferase reporter activities than others.
The sequences with high activity in vitro did not show significant activities in developing limb buds in chick embryos.
The proximal promoter showed activity in the dorsal midline of the brain in developing chick embryos
ACKNOWLEDGEMENTS
We are grateful to Katherine Chen for editorial assistance, to Drs. Naoko Koyano-Nakagawa and Michael O'Connor for sharing their equipment, and to Dr. Yoshiko Takahashi for the pCAGGS-Transposase construct. This study was supported by the National Institute of Arthritis, Musculoskeletal and Skin Diseases to YK (R01AR064195) and the Japan Society for the Promotion of Science KAKENHI to NW (17K08498). AS was supported by Carleton College's Kolenkow-Reitz Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The authors have no conflicts to disclose.
Grant sponsors and numbers:
• National Institute of Arthritis, Musculoskeletal and Skin Diseases of the National Institutes of Health, USA (Grant number: R01AR064195)
• Japan Society for the Promotion of Science (KAKENHI, 17K08498)
Abbreviations:
- Fgf
Fibroblast growth factor
- HH
Hamburger-Hamilton
- LPM
lateral plate mesoderm
- TSS
transcription start site
- UTR
untranslated regio
REFERENCES
- Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG. 2003. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130:623–633. [DOI] [PubMed] [Google Scholar]
- Alvarez Y, Alonso MT, Vendrell V, Zelarayan LC, Chamero P, Theil T, Bosl MR, Kato S, Maconochie M, Riethmacher D, Schimmang T. 2003. Requirements for FGF3 and FGF10 during inner ear formation. Development 130:6329–6338. [DOI] [PubMed] [Google Scholar]
- Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. 2009. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev Cell 16:47–57. [DOI] [PubMed] [Google Scholar]
- Barrow JR, Thomas KR, Boussadia-Zahui O, Moore R, Kemler R, Capecchi MR, McMahon AP. 2003. Ectodermal Wnt3/beta-catenin signaling is required for the establishment and maintenance of the apical ectodermal ridge. Genes Dev 17:394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum RY, Everman DB, Murphy KK, Gurrieri F, Schwartz CE, Ahituv N. 2012. Functional characterization of tissue-specific enhancers in the DLX5/6 locus. Hum Mol Genet 21:4930–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe I, Tenin G, Oseni A, Dietrich S. 2011. Dynamic control of head mesoderm patterning. Development 138:2807–2821. [DOI] [PubMed] [Google Scholar]
- Boulet AM, Moon AM, Arenkiel BR, Capecchi MR. 2004. The roles of Fgf4 and Fgf8 in limb bud initiation and outgrowth. Dev Biol 273:361–372. [DOI] [PubMed] [Google Scholar]
- Burke AC, Nelson CE, Morgan BA, Tabin C. 1995. Hox genes and the evolution of vertebrate axial morphology. Development 121:333–346. [DOI] [PubMed] [Google Scholar]
- Chatzeli L, Gaete M, Tucker AS. 2017. Fgf10 and Sox9 are essential for the establishment of distal progenitor cells during mouse salivary gland development. Development 144:2294–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MJ, Izpisua-Belmonte JC, Abud H, Heath JK, Tickle C. 1995. Fibroblast growth factors induce additional limb development from the flank of chick embryos. Cell 80:739–746. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Minowada G, MacArthur CA, Martin GR. 1996. Roles for FGF8 in the induction, initiation, and maintenance of chick limb development. Cell 84:127–136. [DOI] [PubMed] [Google Scholar]
- Cunningham TJ, Lancman JJ, Berenguer M, Dong PDS, Duester G. 2018. Genomic Knockout of Two Presumed Forelimb Tbx5 Enhancers Reveals They Are Nonessential for Limb Development. Cell Rep 23:3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado I, Torres M. 2017. Coordination of limb development by crosstalk among axial patterning pathways. Dev Biol 429:382–386. [DOI] [PubMed] [Google Scholar]
- Economou A, Datta P, Georgiadis V, Cadot S, Frenz D, Maconochie M. 2013. Gata3 directly regulates early inner ear expression of Fgf10. Dev Biol 374:210–222. [DOI] [PubMed] [Google Scholar]
- Gibson-Brown JJ, Agulnik SI, Silver LM, Niswander L, Papaioannou VE. 1998. Involvement of T-box genes Tbx2-Tbx5 in vertebrate limb specification and development. Development 125:2499–2509. [DOI] [PubMed] [Google Scholar]
- Golzio C, Havis E, Daubas P, Nuel G, Babarit C, Munnich A, Vekemans M, Zaffran S, Lyonnet S, Etchevers HC. 2012. ISL1 directly regulates FGF10 transcription during human cardiac outflow formation. PLoS One 7:e30677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Tabin CJ. 2014. Vertebrate limb bud formation is initiated by localized epithelial-to-mesenchymal transition. Science 343:1253–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. 1951. A series of normal stages in the development of the chick embryo. J. Morph. 88:49–92. [PubMed] [Google Scholar]
- Haro E, Delgado I, Junco M, Yamada Y, Mansouri A, Oberg KC, Ros MA. 2014. Sp6 and Sp8 transcription factors control AER formation and dorsal-ventral patterning in limb development. PLoS Genet 10:e1004468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haro E, Watson BA, Feenstra JM, Tegeler L, Pira CU, Mohan S, Oberg KC. 2017. Lmx1b-targeted cis-regulatory modules involved in limb dorsalization. Development 144:2009–2020. [DOI] [PubMed] [Google Scholar]
- Havens BA, Rodgers B, Mina M. 2006. Tissue-specific expression of Fgfr2b and Fgfr2c isoforms, Fgf10 and Fgf9 in the developing chick mandible. Arch Oral Biol 51:134–145. [DOI] [PubMed] [Google Scholar]
- Hu Z, Tee WW. 2017. Enhancers and chromatin structures: regulatory hubs in gene expression and diseases. Biosci Rep 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac A, Cohn MJ, Ashby P, Ataliotis P, Spicer DB, Cooke J, Tickle C. 2000. FGF and genes encoding transcription factors in early limb specification. Mech Dev 93:41–48. [DOI] [PubMed] [Google Scholar]
- Isaac A, Rodriguez-Esteban C, Ryan A, Altabef M, Tsukui T, Patel K, Tickle C, Izpisua-Belmonte JC. 1998. Tbx genes and limb identity in chick embryo development. Development 125:1867–1875. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Bel-Vialar S, Krumlauf R. 1999. ‘Shocking’ developments in chick embryology: electroporation and in ovo gene expression. Nat Cell Biol 1:E203–207. [DOI] [PubMed] [Google Scholar]
- Karabagli H, Karabagli P, Ladher RK, Schoenwolf GC. 2002. Comparison of the expression patterns of several fibroblast growth factors during chick gastrulation and neurulation. Anat Embryol (Berl) 205:365–370. [DOI] [PubMed] [Google Scholar]
- Kawakami K 2007. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol 8 Suppl 1:S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Capdevila J, Buscher D, Itoh T, Rodriguez Esteban C, Izpisua Belmonte JC. 2001. WNT signals control FGF-dependent limb initiation and AER induction in the chick embryo. Cell 104:891–900. [DOI] [PubMed] [Google Scholar]
- Kengaku M, Capdevila J, Rodriguez-Esteban C, De La Pena J, Johnson RL, Belmonte JC, Tabin CJ. 1998. Distinct WNT pathways regulating AER formation and dorsoventral polarity in the chick limb bud. Science 280:1274–1277. [DOI] [PubMed] [Google Scholar]
- Kruithof BP, van Wijk B, Somi S, Kruithof-de Julio M, Perez Pomares JM, Weesie F, Wessels A, Moorman AF, van den Hoff MJ. 2006. BMP and FGF regulate the differentiation of multipotential pericardial mesoderm into the myocardial or epicardial lineage. Dev Biol 295:507–522. [DOI] [PubMed] [Google Scholar]
- Logan M, Simon HG, Tabin C. 1998. Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development 125:2825–2835. [DOI] [PubMed] [Google Scholar]
- Mariani FV, Ahn CP, Martin GR. 2008. Genetic evidence that FGFs have an instructive role in limb proximal-distal patterning. Nature 453:401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H, Watanabe Y, Funahashi J. 2000. Misexpression of genes in brain vesicles by in ovo electroporation. Dev Growth Differ 42:199–201. [DOI] [PubMed] [Google Scholar]
- Ohta S, Suzuki K, Ogino Y, Miyagawa S, Murashima A, Matsumaru D, Yamada G. 2008. Gene transduction by sonoporation. Dev Growth Differ 50:517–520. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Nakagawa T, Yamamoto A, Araga A, Ohata T, Ishimaru Y, Yoshioka H, Kuwana T, Nohno T, Yamasaki M, Itoh N, Noji S. 1997. The mesenchymal factor, FGF10, initiates and maintains the outgrowth of the chick limb bud through interaction with FGF8, an apical ectodermal factor. Development 124:2235–2244. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Takeuchi J, Yoshioka H, Ishimaru Y, Ogura K, Takahashi N, Ogura T, Noji S. 1998. Correlation of wing-leg identity in ectopic FGF-induced chimeric limbs with the differential expression of chick Tbx5 and Tbx4. Development 125:51–60. [DOI] [PubMed] [Google Scholar]
- Ohuchi H, Yasue A, Ono K, Sasaoka S, Tomonari S, Takagi A, Itakura M, Moriyama K, Noji S, Nohno T. 2005. Identification of cis-element regulating expression of the mouse Fgf10 gene during inner ear development. Dev Dyn 233:177–187. [DOI] [PubMed] [Google Scholar]
- Onimaru K, Kuraku S, Takagi W, Hyodo S, Sharpe J, Tanaka M. 2015. A shift in anterior-posterior positional information underlies the fin-to-limb evolution. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissieres V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, Kato M, Garvin TH, Pham QT, Harrington AN, Akiyama JA, Afzal V, Lopez-Rios J, Dickel DE, Visel A, Pennacchio LA. 2018. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso S, Douglas A, Hill A, De Angelis C, Moore BL, Grimes G, Petrovich G, Essafi A, Hill RE. 2017. Fibroblast growth factors (FGFs) prime the limb specific Shh enhancer for chromatin changes that balance histone acetylation mediated by E26 transformation-specific (ETS) factors. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. 2006. In vivo enhancer analysis of human conserved non-coding sequences. Nature 444:499–502. [DOI] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. 2016. Implications of Developmental Gene Regulatory Networks Inside and Outside Developmental Biology. Curr Top Dev Biol 117:237–251. [DOI] [PubMed] [Google Scholar]
- Petit F, Sears KE, Ahituv N. 2017. Limb development: a paradigm of gene regulation. Nat Rev Genet 18:245–258. [DOI] [PubMed] [Google Scholar]
- Rister J, Desplan C. 2010. Deciphering the genome’s regulatory code: the many languages of DNA. Bioessays 32:381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Calderon H, Francisco-Morcillo J, Martin-Partido G, Hidalgo-Sanchez M. 2007. Fgf19 expression patterns in the developing chick inner ear. Gene Expr Patterns 7:30–38. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Yamaoka T, Ohuchi H, Yasue A, Nohno T, Kawano H, Kato S, Itakura M, Nagayama M, Noji S. 2002. Identification of cis-elements regulating expression of Fgf10 during limb development. Int J Dev Biol 46:963–967. [PubMed] [Google Scholar]
- Sato Y, Kasai T, Nakagawa S, Tanabe K, Watanabe T, Kawakami K, Takahashi Y. 2007. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev Biol 305:616–624. [DOI] [PubMed] [Google Scholar]
- Scherz PJ, Harfe BD, McMahon AP, Tabin CJ. 2004. The limb bud Shh-Fgf feedback loop is terminated by expansion of former ZPA cells. Science 305:396–399. [DOI] [PubMed] [Google Scholar]
- Searls RL, Janners MY. 1971. The initiation of limb bud outgrowth in the embryonic chick. Dev Biol 24:198–213. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. 1999. Fgf10 is essential for limb and lung formation. Nat Genet 21:138–141. [DOI] [PubMed] [Google Scholar]
- Sheth R, Gregoire D, Dumouchel A, Scotti M, Pham JM, Nemec S, Bastida MF, Ros MA, Kmita M. 2013. Decoupling the function of Hox and Shh in developing limb reveals multiple inputs of Hox genes on limb growth. Development 140:2130–2138. [DOI] [PubMed] [Google Scholar]
- Shlyueva D, Stampfel G, Stark A. 2014. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet 15:272–286. [DOI] [PubMed] [Google Scholar]
- Soshnikova N, Zechner D, Huelsken J, Mishina Y, Behringer RR, Taketo MM, Crenshaw EB, 3rd, Birchmeier W 2003. Genetic interaction between Wnt/beta-catenin and BMP receptor signaling during formation of the AER and the dorsal-ventral axis in the limb. Genes Dev 17:1963–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann M, Mundlos S. 2016. Looking beyond the genes: the role of non-coding variants in human disease. Hum Mol Genet 25:R157–R165. [DOI] [PubMed] [Google Scholar]
- Sun X, Mariani FV, Martin GR. 2002. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature 418:501–508. [DOI] [PubMed] [Google Scholar]
- Suryamohan K, Halfon MS. 2015. Identifying transcriptional cis-regulatory modules in animal genomes. Wiley Interdiscip Rev Dev Biol 4:59–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Ogura T. 2008. Congenic method in the chick limb buds by electroporation. Dev Growth Differ 50:459–465. [DOI] [PubMed] [Google Scholar]
- Teshima TH, Lourenco SV, Tucker AS. 2016. Multiple Cranial Organ Defects after Conditionally Knocking Out Fgf10 in the Neural Crest. Front Physiol 7:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmer J, Johnson J, Niswander L. 2001. The use of in ovo electroporation for the rapid analysis of neural-specific murine enhancers. Genesis 29:123–132. [DOI] [PubMed] [Google Scholar]
- Uchikawa M, Ishida Y, Takemoto T, Kamachi Y, Kondoh H. 2003. Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev Cell 4:509–519. [DOI] [PubMed] [Google Scholar]
- Verheyden JM, Sun X. 2008. An Fgf/Gremlin inhibitory feedback loop triggers termination of limb bud outgrowth. Nature 454:638–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res 35:D88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A, Rodriguez C, Izpisua-Belmonte JC. 1996. Involvement of FGF-8 in initiation, outgrowth and patterning of the vertebrate limb. Development 122:1737–1750. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Miyagawa-Tomita S, Vincent SD, Kelly RG, Moon AM, Buckingham ME. 2010. Role of mesodermal FGF8 and FGF10 overlaps in the development of the arterial pole of the heart and pharyngeal arch arteries. Circ Res 106:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Zaffran S, Kuroiwa A, Higuchi H, Ogura T, Harvey RP, Kelly RG, Buckingham M. 2012. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2–5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc Natl Acad Sci U S A 109:18273–18280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Lettice LA, Hill RE, Bickmore WA. 2016. Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development 143:2994–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyngaarden LA, Vogeli KM, Ciruna BG, Wells M, Hadjantonakis AK, Hopyan S. 2010. Oriented cell motility and division underlie early limb bud morphogenesis. Development 137:2551–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Shiraishi Y, Higuchi H, Yamamoto S, Hirano M, Kuroiwa A. 2014. Etv1 and Ewsr1 cooperatively regulate limb mesenchymal Fgf10 expression in response to apical ectodermal ridge-derived fibroblast growth factor signal. Dev Biol 394:181–190. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.