Abstract
To optimally perform the diversity of metabolic functions that occur within peroxisomes, cells must dynamically regulate peroxisome size, number and content in response to cell state and the environment. Except for transcriptional regulation little is known about the mechanisms used to perform this complicated fete. Here we used complementary high content screens to follow changes in most of the yeast, Saccharomyces cerevisiae, proteins during growth in oleate. We found extensive changes in cellular architecture and identified several proteins co-localizing with peroxisomes that have not previously been considered peroxisomal proteins. One of the newly identified peroxisomal proteins, Ymr018w, is a protein with an unknown function that is similar to the yeast and human peroxisomal targeting receptor Pex5. We demonstrate that Ymr018w is a novel peroxisomal targeting receptor that targets a subset of matrix proteins to peroxisomes. We have therefore renamed Ymr018w, Pex9, and suggest that Pex9 is a condition-specific targeting receptor that enables the dynamic rewiring of peroxisomes in response to metabolic needs. Moreover, we suggest that Pex5-like receptors may also exist in vertebrates.
Keywords: Peroxisome, Protein targeting, Glucose, Oleate, Saccharomyces cerevisiae, High content screen
Introduction
Peroxisomes are found in almost all eukaryotes and participate in central catabolic pathways such as β-oxidation of fatty acids, breakdown of amino acids and nucleotides and detoxification of reactive oxygen species (Islinger et al., 2010; Smith and Aitchison, 2013). It is therefore obvious why any change in peroxisome functions lead to severe diseases in humans (Waterham et al., 2016). The genetically powerful yeast S. cerevisiae has served as an attractive model to study peroxisomes due to the conservation of pathways required for biogenesis of the organelle, targeting of proteins and metabolic functions.
In S. cerevisiae (from here on termed yeast) peroxisomes are the sole organelles that perform β-oxidation of fatty acids making them essential for growth when oleic acid is the sole carbon source. Since yeasts usually grow while consuming sugar as a main carbon source, they must dramatically alter cellular organization to rely on lipids (Jung et al., 2013; Smith et al., 2002). Amongst others this includes optimizing peroxisomal functions as well as the functions of other organelles, such as mitochondria, where the metabolic products of the β-oxidation process are consumed (Trotter, 2001). Indeed it has been shown that during a transition to growth in oleate yeast actively alter their transcriptional response (Karpichev and Small, 1998; Smith et al., 2002; Smith et al., 2007) as well as the localization of several proteins (Jung et al., 2013). However, little is known about the extent of organellar and proteomic changes that occurs during the transition to oleate dependent growth.
To further uncover the extent of organelle and protein dynamics during growth in oleate we followed the localization of nearly all yeast proteins – tagged at either their N or C terminus (N’ or C’ respectively) with a Green Fluorescent Protein (GFP) (Huh et al., 2003; Yofe et al., 2016). We identified dramatic cellular changes such as alterations in organelle shapes and sizes as well as numerous changes in protein localizations. Furthermore, we identified several new potential peroxisomal proteins and demonstrated that Ymr018w, that we now name Pex9, is a peroxisomal targeting receptor that targets a subset of proteins that contain a Peroxisomal Targeting Signal 1 (PTS1) under specific conditions. We suggest that such a dedicated targeting receptor exists also in vertebrates. Our findings highlight the complexity of the targeting machinery to peroxisomes and provide an important step towards a deep understanding of peroxisomes, their roles and regulation under various metabolic conditions.
Results
A high content screen reveals localization dynamics of yeast proteins during growth in oleate
To characterize the proteome dynamics of yeast grown in oleate as a sole carbon source we performed a high content microscopic screen of collections of strains containing GFP-tagged proteins (Breker et al., 2014). An imaging based screen can be easily used to detect the organellar localization of GFP tagged proteins while avoiding subcellular fractionations of each organelle followed by mass spectrometry analysis (Breker and Schuldiner, 2014) which is laborious and might suffer from organelle contaminations. To ensure that we get an optimal coverage of all yeast proteins we used two complementary yeast libraries; the C’ tagged GFP library (Huh et al., 2003) and the N’ tagged SWAT-GFP library (Yofe et al., 2016). The C’ GFP library encompasses all yeast proteins expressed under their native promoter enabling us to follow changes both in transcriptional and post transcriptional levels. The N’ SWAT-GFP library contains ~1,800 strains focusing on endomembrane and peroxisomal proteins – all of which are expressed under a constitutive SpNOP1 promoter, which enables us to detect the localization of low abundance or condition-specific proteins. Importantly, tagging proteins at either terminus can mask targeting sequences as well as regulatory sequences that may affect their localization. Therefore, it is essential to utilize both libraries for increased accuracy. One relevant example is the peroxisomal matrix proteins that contain a PTS1 at their C’ thus only being correctly localized in the SWAT library (Yofe et al., 2016).
To identify the proteome changes that occur in oleate compared to glucose we imaged the strains after 20 hours of growth in media supplemented with oleate as the sole carbon source (Fig.1A). The localizations obtained were then compared to the previously described localizations in glucose (Breker et al., 2014; Huh et al., 2003; Yofe et al., 2016). Most of the peroxisomal proteins were observed in the expected punctate pattern only when the SWAT library (Yofe et al., 2016) was used. Hence, in order to differentiate between a peroxisomal localization and other punctate localizations (lipid droplets, Golgi, organelle contact sites etc.) we integrated a peroxisomal marker, Pex3-mCherry, into the SWAT strains (Fig.1A).
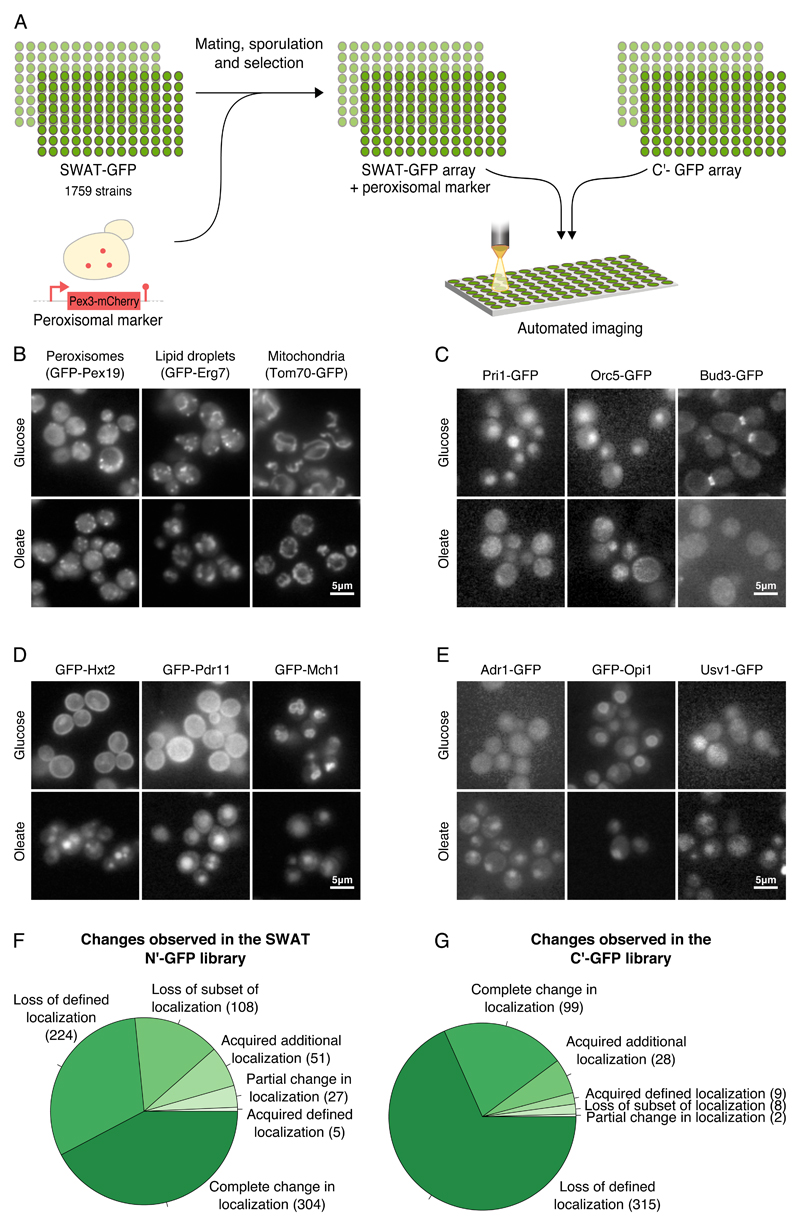
Figure 1. Unraveling the protein dynamics of yeast grown in oleate.
(A) A schematic diagram of the high content screen. Using an automated mating procedure we genomically integrated a peroxisomal marker, Pex3-mCherry to the SWAT N’ GFP library. Using a high-throughput fluorescence microscope, we screened this newly created library as well as the C’ GFP yeast library under growth in oleate-containing media. All images were taken at a single focal plane. We manually characterized the localization of each protein and compared it to its localization during growth on glucose-containing media. (B) Under growth in oleate more peroxisomes were observed, lipid droplets were larger and mitochondria were altered compared to growth in glucose. (C) Many changes that imply a general stress were observed in cells grown in oleate. Examples include loss of defined localization of Pri1, a DNA primase that is required for DNA synthesis and double-strand break repair, Orc5, a subunit of the origin recognition complex (ORC) which directs DNA replication and Bud3 that is involved in budding. (D) Many proteins were detected in the vacuole in oleate suggesting down regulation of various processes. Examples are Hxt2, a high affinity glucose transporter, and Pdr11, an ATP-binding cassette (ABC) transporter that moved from the plasma membrane to the vacuole. Mch1, a protein with similarity to mammalian monocarboxylate permeases that are involved in transport of monocarboxylic acids across the plasma membrane, moved from the vacuolar membrane to the vacuole. (E) Adr1, a transcription factor that regulates fatty acids utilization-related processes, Opi1, a negative transcriptional regulator of phospholipid biosynthetic genes, and Usv1 that affects transcriptional regulation of genes involved in growth on non-fermentable carbon sources, were localized to the nucleus under growth in oleate but not in glucose. (F) A pie chart summarizing the changes observed in the N’ GFP SWAT library. (G) A pie chart summarizing the changes observed in the C’ GFP library.
Following our analysis we observed that growth in oleate led to tremendous changes in the cell including changes in the abundance, size and shape of different organelles. The number of peroxisomes was increased as was previously reported (Veenhuis et al., 1987) (Fig.1B left), the size of lipid droplets was increased (Fig.1B middle and Fig.S1 A-C for a quantitative analysis) and mitochondrial morphology was dramatically altered (Fig.1B right and Fig.S1 D,E for a quantitative analysis). Interestingly, we found that Fzo1, that is involved in mitochondrial fusion, lost its defined localization in oleate (Fig.S1 F). Additionally, we could detect changes in localization that imply down regulation of DNA replication and budding, in line with the general stress that cells undergo when grown in oleate (Fig.1C).
One striking alteration in localization was the accumulation of proteins in the vacuole when cells were grown in oleate but not in glucose (Fig.1D) suggesting that many proteins, such as sugar transporters, that have no role during oleate growth, are degraded under this condition.
It is known that dramatic transcriptional changes occur during the transition to growth on oleate. In concordance with this we found transcription factors that we could detect in the nucleus only in oleate (Fig.1E). One such factor is Adr1 (Fig.1E, left) that enhances the expression of peroxisomal genes (Gurvitz et al., 2001; Karpichev et al., 2008). Another factor that changed localization is Opi1 (Fig.1E, middle) whose activity is known to be regulated by its localization. Interestingly, Opi1 was shown to move from the nuclear endoplasmic reticulum (ER) to the nucleus, as we saw, under conditions in which there is an excess of fatty acids in the yeast cell (Velazquez et al., 2016). However, we could not detect changes in peroxisomal size or abundance upon OPI1 deletion (Data not shown) thus it most probably regulates refined changes in cellular metabolism. Another protein, Usv1, that affects the transcriptional regulation of genes that are involved in growth on non-fermentable carbon sources, was also observed in the nucleus in oleate (Fig.1E, right).
In general - 719 proteins changed localization in the N’ library (supplementary Table 1 and Fig.1F) and 461 in the C’ library (supplementary Table 2 and Fig.1G) when cells were grown in oleate compared to glucose. Only 25 cellular changes overlapped between both GFP libraries emphasizing the power of using these complementary collections. Interestingly, the most frequent changes in localizations were the loss of a defined localization or movement to the vacuole (Fig.S2). These cellular changes suggest that many proteins are either downregulated or degraded when cells are grown in oleate. The large number of changes in protein localization highlights the dramatic cellular reorganization required to enable yeast to rely on lipids as a sole carbon source.
Identifying potential new peroxisomal proteins
We next focused on changes in peroxisomes, the sole organelle in cerevisiae that performs β oxidation (Trotter, 2001). It was previously suggested that the protein content of peroxisomes is highly dynamic (Jung et al., 2013; Smith and Aitchison, 2013) hence we first examined which known peroxisomal proteins changed their localization in oleate (supplementary Table 3). One nice example of the power of using two complementary libraries comes from Pxa2, a subunit of the peroxisomal fatty acid transporter. While Pxa2-GFP that is expressed under its native promoter was only observed in oleate (Fig.2A, left), the constitutively expressed GFP-Pxa2 could be visualized in peroxisomes in both conditions (Fig.2A, right) supporting the fact that Pxa2 levels are transcriptionally regulated (Karpichev and Small, 1998).
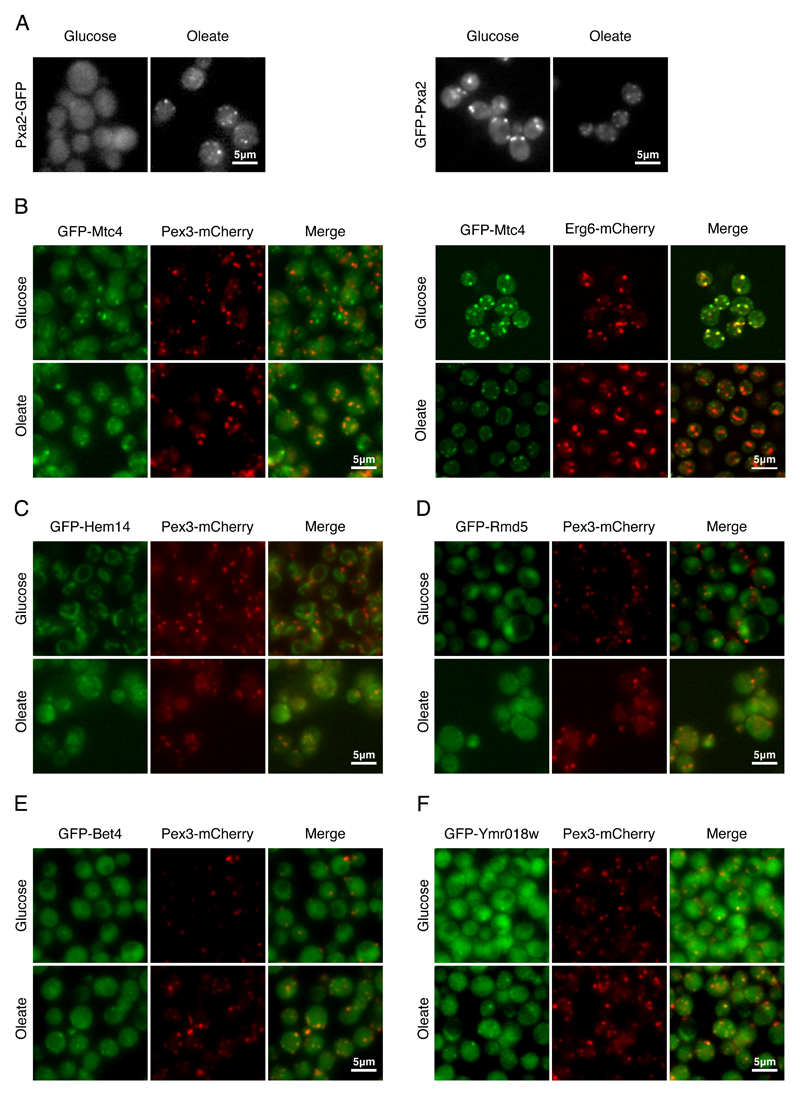
Figure 2. Identifying potential new peroxisomal proteins.
(A) When Pxa2-GFP was expressed under its native promoter it was observed in puncta only in oleate. However, when GFP-Pxa2 was expressed under a constitutive promoter, the protein was observed in puncta both in glucose and in oleate, implying that regulation is transcription-mediated. (B) Mtc4, a protein of unknown function, co-localized with a peroxisomal marker in oleate. In glucose Mtc4 co-localized with a lipid droplet marker, implying that Mtc4 localization is dependent on the carbon source of the cells. (C) Hem14, a mitochondrial enzyme that catalyzes the seventh step in the heme biosynthetic pathway, was the only mitochondrial protein that was partially co-localized with Pex3 in oleate. (D) Rmd5, a ubiquitin ligase, changed its localization from cytosol and nucleus in glucose to cytosol and puncta that co-localized with Pex3 in oleate. (E) Bet4, the α subunit of type II geranylgeranyltransferase, was co-localized with Pex3 during growth in glucose and oleate (F) Ymr018w, a protein whose function is unknown that shares similarity to the yeast Pex5, was co-localized with Pex3-mCherry both in glucose and in oleate. All images show a single focal plane.
Having integrated a Pex3-mCherry peroxisomal marker to the N’ GFP library (Fig.1A) we were able to identify proteins that were co-localized with peroxisomes in oleate, but were not reported before as peroxisomal proteins. We could detect that Mtc4, a protein with an unknown function, was mainly co-localized with Pex3 in oleate but not in glucose (Fig.2B, left). Intriguingly we found that Mtc4 co-localized with lipid droplets in glucose (Fig.2B, right). Since we did not see a general overlap between peroxisomes and lipid droplets in oleate (Fig.S3A) the change observed in Mtc4 implies that at least a fraction of Mtc4 is targeted to peroxisomes in oleate, plausibly to improve lipid consumption, as was previously suggested (Binns et al., 2006).
Hem14, a mitochondrial enzyme that catalyzes the seventh step in the heme biosynthetic pathway, was localized to mitochondria in glucose and was observed as puncta in oleate (Fig.2C). Since mitochondria were altered in oleate and often appeared more punctate, some of these puncta might simply be mitochondria. However, some of the GFP-Hem14 puncta co-localized with Pex3. The unique localization of Hem14 suggests that either Hem14 is dually localized to mitochondria and peroxisomes in oleate, or that subset of mitochondria, that are enriched with Hem14, are localized in the vicinity of peroxisomes (Cohen et al., 2014; Shai et al., 2016). Currently we cannot differentiate between these possibilities due to the limitations of fluorescence microscopy.
Finally, Rmd5, a component of the Glucose Induced degradation Deficient (GID) complex that degrades gluconeogenic enzymes (Braun et al., 2011), co-localized with Pex3 in oleate (Fig.2D). This suggests that gluconeogenic enzymes might be quality controlled proximally to peroxisomes.
We found several proteins, which were previously not suggested as peroxisomal proteins, co-localized to Pex3 in both glucose and oleate. One such protein, Bet4, is the α subunit of the Type II geranylgeranyltransferase (Fig.2E). While we did not detect Bet2, the β subunit, in peroxisomes (Data not shown), we further found that Bet4 localization to peroxisomes was dependent on the PTS1 targeting receptor, Pex5 (Fig.S3B). This was not expected since Bet4 does not contain a canonical PTS1 sequence. Hence, it would be intriguing to further examine if Bet4 is indeed localized to the matrix of peroxisomes and if so, if it directly binds Pex5 in a PTS1 independent manner or if it is inserted to the matrix by piggybacking on a PTS1 protein. Moreover, it now remains to further study whether Bet4 has a functional role in peroxisomes or whether one subunit is simply sequestered there under certain conditions.
Another newly identified peroxisomal protein that came up from our screen is Ymr018w, which co-localized with Pex3 in glucose and in oleate (Fig.2F). This protein was suggested to be a unique paralogue of Pex5 in S. cerevisiae (Kiel et al., 2006) as it displays 27% identity to the yeast Pex5 and 25% identity to the human Pex5 (Amery et al., 2001). Additionally, transcriptome profiling of yeast pointed out Ymr018w as having a role in peroxisome biogenesis or function (Smith et al., 2002). Interestingly, we could still detect GFP-Ymr018w co-localizing with peroxisomes when both targeting receptors, PEX5 and PEX7 (the targeting receptor for PTS2 containing proteins), were deleted (Fig.S3C). This suggests that the targeting and therefore maybe also the function of Ymr018w is independent of Pex5 and Pex7. Intrigued by its potential role in targeting proteins to peroxisomes we decided to further investigate Ymr018w.
Ymr018w affects targeting of peroxisomal proteins
To examine if Ymr018w is targeting proteins to the matrix of peroxisomes we first deleted YMR018W, PEX5 or PEX7 and examined if any of the N’ GFP-tagged peroxisomal proteins (N’ GFP peroxisomal library) lost their punctate localization in glucose or in oleate (Fig.3A). Although all known targets of Pex5 were affected by the absence of Pex5, and all Pex7 targets lost their peroxisomal localization upon loss of Pex7, we could not detect any obvious change in localization for any protein upon deletion of Ymr018w (Data not shown).
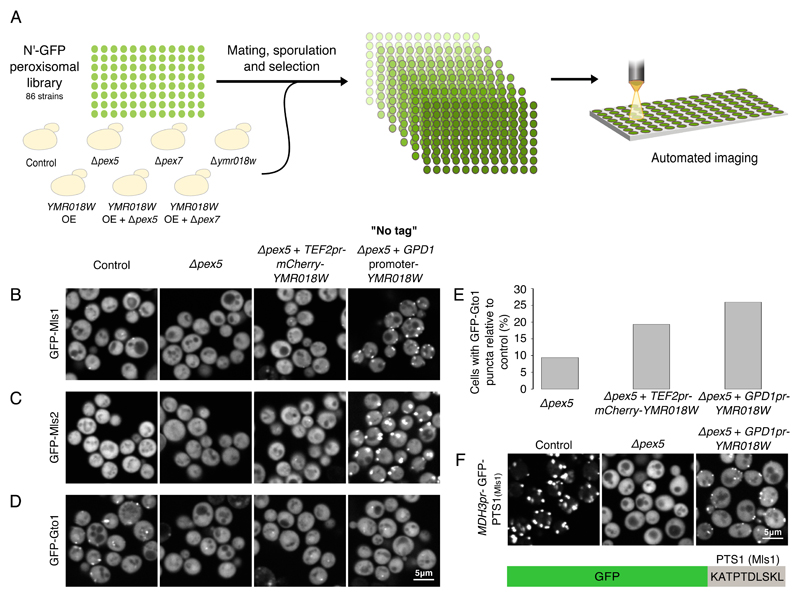
Figure 3. Ymr018w mediates targeting of a subset of PTS1 proteins to peroxisomes.
(A) Using an automated mating procedure we created an N’-tagged GFP-peroxisomal library with the deletion of YMR018W, PEX5 or PEX7 genes. Using a high content microscopy screen we examined the effect of the deletion on the localization of each of the peroxisomal proteins under growth on both glucose and oleate-containing media and could not detect an obvious effect of YMR018W deletion. Using the same automated mating procedure we created additional N’-GFP peroxisomal libraries with one of the following genomic modifications: mCherry-YMR018W overexpression (OE), Δpex5 + mCherry-YMR018W OE and Δpex7 + mCherry-YMR018W OE. A strain carrying a Neurseothricin selection was used as control for no modification in peroxisomal genes. We examined the effect of the different manipulations on the localization of each of the peroxisomal proteins and discovered three PTS1 proteins that were localized to peroxisomes when Ymr018w was over expressed either with a mCherry tag or when it was over expressed under a GPD promoter without a tag (no tag): (B) Mls1, (C) Mls2 and (D) Gto1. (E) A quantitative analysis of the percentage of cells (200 cells per strain) with GFP-Gto1 puncta in Δpex5, Δpex5+TEF2-mCherry-Ymr018w and Δpex5+GPDpr-Ymr018w strains, relative to a control strain with an intact PEX5 gene. Over expression of YMR018W increases the percentage of cells containing punctate Gto1 implying that Gto1 is a target of Ymr018w. (F) A strain expressing GFP with the last 10 amino acids of the Ymr018w-target Mls1 (MDH3pr-GFP-PTS1(Mls1)) was visualized. While GFP-PTS1(Mls1) appeared in puncta in the control strain and in the cytosol in the absence of PEX5, it resumed peroxisomal localization when Ymr018w was overexpressed. This indicates that the last 10 amino acids of Mls1 are sufficient to mediate targeting by Ymr018w. All images show a single focal plane.
Since both Pex5 and Pex7 were fully functional in the Δymr018w strain we assumed that their presence could compensate for any loss of Ymr018w activity. By similarity to Pex5 alone it was impossible to predict the function of Ymr018w since it could either target PTS1 proteins similar to Pex5, or might work as Pex5 Long (Pex5L), the mammalian Pex7 co-receptor (Schliebs and Kunau, 2006), and hence might target PTS2 proteins. Therefore, we created strains in which PEX5 or PEX7 were deleted and mCherry-Ymr018w was over expressed under a strong TEF2 promoter. After verifying that mCherry-Ymr018w could be localized to peroxisomes (Fig. S4A) we examined the localization of any of the GFP-tagged peroxisomal proteins in control strains (no mutation, Δpex5, Δpex7, TEF2-mCherry-Ymr018w) as well as in strains in which Ymr018w was overexpressed in the background of the absence of Pex5 or Pex7 (Fig. 3A). Indeed, under these conditions the substrate range of Ymr018w was exposed.
We could detect that Mls1, a malate synthase that contains a PTS1 signal, completely lost its peroxisomal localization upon deletion of PEX5 but regained peroxisomal targeting upon overexpression of Ymr018w (Fig. 3B) suggesting that Mls1 can use both Pex5 and Ymr018w for its targeting into peroxisomes. Importantly, the effect of Ymr018w was dependent on its levels and independent of the presence of the mCherry tag as it targeted substrates even more robustly when expressed under the GPD1 promoter (that drives approximately two-three fold stronger transcription than the TEF2pr (Janke et al., 2004)) and with no tag (Fig.3B “No tag”, Fig.S4B).
Mls2, the second PTS1-containing malate synthase, was targeted to peroxisomes only upon overexpression of Ymr018w (regardless of Pex5 presence). This suggests that Mls2, unlike Mls1, is completely dependent on Ymr018w for its targeting (Fig.3C).
Another Ymr018w target is Gto1, a glutathione transferase. Unlike the Mls proteins, Gto1 was the only PTS1 protein that we still observed in puncta when PEX5 was deleted and its punctate localization increased upon Ymr018w overexpression (Fig.3D and Fig.3E). Interestingly, we did not detect an obvious effect on GFP-Gto1 in the Δymr018w strain (Data not shown), most probably due to a backup function of Pex5.
Importantly, even when Ymr018w was over expressed under the GPD1 promoter it was not able to target any PTS1 protein to peroxisomes (Fig.S4C). Intrigued by this specificity we further studied the targeting specificity of Ymr018w and examined if a GFP protein to which we add the last 10 amino acids of Mls1 will be targeted to peroxisomes by Ymr018w. Interestingly the C’ of Mls1 was sufficient to mediate targeting by Ymr018w (Fig.3F).
Together, our results show that Ymr018w targets a subset of PTS1-containing matrix proteins, and that targeting specificity is defined by the context of the PTS1 sequence. In a parallel and independent approach Ymr018w was found to have the same function (Effelsberg et al., 2016), hence we jointly named Ymr018w, Pex9.
Discussion
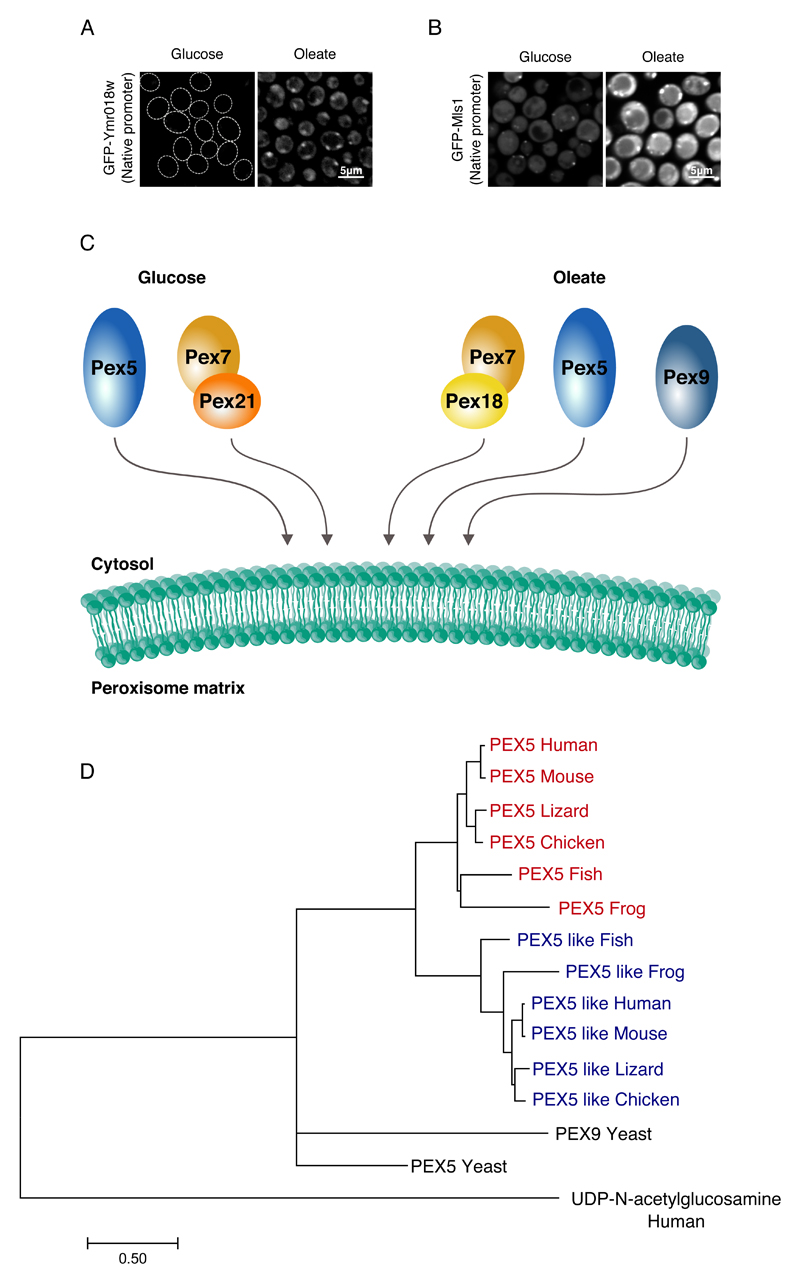
Our work demonstrates that two PTS1 targeting receptors exist in yeast - the well characterized receptor, Pex5, that targets all PTS1 proteins, and Pex9 that targets a specific subset of PTS1 proteins. Why would another targeting receptor be required for only a few proteins? Interestingly the three proteins that we found to be substrates for Pex9 targeting, Mls1, Mls2 and Gto1, are highly regulated and their expression is induced dramatically in oleate (Barreto et al., 2006; Kunze et al., 2002; Kunze et al., 2006). Correspondingly, we could see that just like GFP-Mls1, GFP-Pex9, when driven under its native promoter, was up regulated upon growth in oleate (Fig.4A,B). We therefore suggest that Pex9 functions under specific conditions, such as oleate growth, to support and specify targeting of essential peroxisomal proteins under such conditions.
Figure 4. Pex9 is a condition specific targeting receptor.
(A) When GFP tagged Ymr018w (Pex9) was expressed under its native promoter, a punctate localization was observed only in oleate. (B) When GFP tagged Mls1 was expressed under its native promoter, a higher expression was detected in oleate compared to glucose. (C) A suggested model for targeting of matrix proteins to peroxisomes. The model consists of two machineries: The basal machinery that is constitutively expressed which includes Pex5 for targeting PTS1 proteins, and Pex7 assisted by the co-receptor Pex21 for targeting PTS2 proteins. On the other hand, there exists a condition-specific arm that we see induced in oleate that includes Pex5, Pex7 assisted by the co-receptor Pex18, and Pex9 that is activated under special conditions and regulates the import of specific PTS1 proteins. (D) A phylogenetic tree showing the human PEX5, PEX5-like and their orthologs from selected vertebrates. The analysis reveals two distinct phylogenetic branches for PEX5 and PEX5-like, which indicates that a duplication took place in early vertebrates and that two dedicated PTS1 targeting receptors may also exist in vertebrates. All images show a single focal plane.
Until now two major pathways for targeting proteins to the matrix of peroxisomes were characterized (Hasan et al., 2013) (Fig.4C). One pathway is mediated by the Pex5 receptor that mainly targets PTS1-containing proteins. The second pathway is mediated by the Pex7 receptor that targets two PTS2-containing proteins and Pnc1 (Effelsberg et al., 2015; Kumar et al., 2016). Pex7 cannot work independently and hence it uses co-receptors (Purdue et al., 1998). Our results put down a new framework for thinking of peroxisomal targeting – a basal machinery (Pex5 and Pex7 with co-receptor Pex21) that functions under normal conditions, and an environmentally regulated arm (Pex7 with co-receptor Pex18 and Pex9), that may be less robust, but gives priority to specific proteins required under these conditions or may be able to work under more adverse conditions.
The requirement for a condition-specific targeting machinery may not be unique to yeast. It was previously shown that Pex5 has a Pex5-like paralog in humans. Interestingly, the Pex5-like protein is specifically expressed in the brain, which is one of the organs most dependent on peroxisome function (Amery et al., 2001). We found that indeed, most vertebrates contain both a Pex5 protein and a Pex5-like protein (Fig.4D) implying a duplication of the PEX5 gene in early vertebrates. The existence of paralogs for Pex5 might suggest that two PTS1 targeting receptors, one which targets all PTS1 proteins, and another, that is expressed under specific conditions or in specific cells and targets preferential proteins, is not restricted to yeast. This highlights the beauty and complexity of targeting proteins to peroxisomes – a fascinating and important organelle.
Materials and methods
Yeast strains and strain construction
All strains in this study are based on the BY4741 laboratory strain (Brachmann et al., 1998). A complete list of strains can be found in supplementary Table 4. The libraries used were the yeast C'-GFP library (Huh et al., 2003) and the SWAT N'-GFP library (Yofe et al., 2016) which is a collection of ~1,800 strains tagged with GFP at their N’ and expressed under a generic, constitutive, promoter (SpNOP1pr). The pFA6a plasmid that was originally used for C’ terminal GFP tagging (Huh et al., 2003) was modified to contain the last 10 amino acids of Mls1 at the end of the GFP sequence. Using this plasmid, GFP-PTS1(Mls1) was genomically integrated downstream to MDH3 promoter (Fig.3F). The GFP-Pex9 and GFP-Mls1 expressed under their native promoter (Fig.4A,B) were picked from the seamless N’ GFP library (Yofe et al., 2016).
Yeast library preparation
To create collections of haploid strains containing GFP-tagged proteins with additional genomic modification such as a peroxisomal marker (Pex3-mCherry) or different deletions (Δpex5, Δpex7, Δpex5Δpex7 and Δymr018w) and overexpression (TEF2-mCherry-Ymr018w), different query strains were constructed on the basis of an SGA compatible strain (for further information see supplementary Table 4). Using the SGA method (Cohen and Schuldiner, 2011; Tong and Boone, 2006) the Pex3-mCherry query strain was crossed with the SWAT-GFP library and the other query strains were crossed into a collection of strains from the SWAT-GFP library containing ~90 strains including known and potential peroxisomal proteins and controls. To perform the SGA in high-density format we used a RoToR bench top colony arrayer (Singer Instruments). In short: mating was performed on rich medium plates, and selection for diploid cells was performed on SD-URA plates containing Geneticin (200μg/mL) and/or Neurseothricin (200μg/mL). Sporulation was induced by transferring cells to nitrogen starvation media plates for 7 days. Haploid cells containing the desired mutations were selected by transferring cells to SD-URA plates containing Geneticin (200μg/mL) and/or Neurseothricin (200μg/mL), alongside the toxic amino acid derivatives Canavanine and Thialysine (Sigma-Aldrich) to select against remaining diploids, and lacking Histidine to select for spores with an A mating type.
Automated high-throughput fluorescence microscopy
The collections were visualized using an automated microscopy setup as described previously (Breker et al., 2013). In short: cells were transferred from agar plates into 384-well polystyrene plates for growth in liquid media using the RoToR arrayer robot. Liquid cultures were grown in LiCONiC incubator, overnight at 30°C in SD-URA medium (for the SWAT-GFP collections) or SD-HIS medium (for the C'-GFP library). A JANUS liquid handler (PerkinElmer) connected to the incubator was used to dilute the strains to an OD600 of ~0.2 into plates containing SD medium (6.7 g/L yeast nitrogen base and 2% glucose) or S-Oleic (6.7 g/L yeast nitrogen base, 0.2% oleic acid and 0.1% Tween-40) supplemented with complete amino acids. Plates were incubated at 30°C for 4 hours in SD medium or for 20 hours in S-Oleic. The cultures in the plates were then transferred by the liquid handler into glass-bottom 384-well microscope plates (Matrical Bioscience) coated with Concanavalin A (Sigma-Aldrich). After 20 minutes, wells were washed twice with SD-Riboflavin complete medium (for screens in glucose) or with double-distilled water (for screens in oleate) to remove non-adherent cells and to obtain a cell monolayer. The plates were then transferred to the ScanR automated inverted fluorescent microscope system (Olympus) using a robotic swap arm (Hamilton). Images of cells in the 384-well plates were recorded in the same liquid as the washing step at 24°C using a 60× air lens (NA 0.9) and with an ORCA-ER charge-coupled device camera (Hamamatsu). Images were acquired in two channels: GFP (excitation filter 490/20 nm, emission filter 535/50 nm) and mCherry (excitation filter 572/35 nm, emission filter 632/60 nm). All images were taken at a single focal plane.
Data analysis
The localization of the GFP tagged proteins in oleate was obtained manually and was compared to the published localization databases of cells grown in SD medium that contains glucose. The localizations of the N'-GFP library were compared to the SWAT-GFP localization database (Yofe et al., 2016). The localizations of the C'-GFP library were first compared to the LoQAtE database (Breker et al., 2014). Whenever the localization in oleate was different from the localization observed in LoQAtE, we also compared the localization to the YeastGFP database, which is based on the same library (Huh et al., 2003). Only cases in which the localization of a protein in oleate was different to the localization observed in either of the two databases were further analyzed.
In cases where an obvious GFP signal was viewed but it was impossible to define a specific localization, the localization was described as “ambiguous” and it was not included in the analysis. Cases with technical problems such as contaminations, unfocused images and not enough cells were not included in the analysis as well. Since mitochondria were fragmented in oleate, we did not consider changes of mitochondrial proteins from mitochondria in glucose to punctate in oleate as different localization. Bud and bud neck localizations were defined as “same” localization.
The proteins that exhibited a different localization in oleate compared to glucose were subdivided into group according to the criteria described in the following table:
| Localization in glucose | Localization in oleate | Definition of change in localization |
|---|---|---|
| X | Y | Complete change in localization |
| XY | Z | Complete change in localization |
| X | YZ | Complete change in localization |
| XY | Y | Loss of subset of localizations |
| XYZ | XY | Loss of subset of localizations |
| XY | XZ | Partial change in localization |
| XYZ | XYM | Partial change in localization |
| XYZ | XM | Partial change in localization |
| X | XY | Acquired additional localization |
| XY | XYZ | Acquired additional localization |
| Below threshold | X | Acquired defined Localization |
| X | Below threshold | Loss of defined localization |
The analysis was done using Matlab software version R2015a (http://www.mathworks.com/products/matlab/). All changes in localizations are summarized in supplementary Table 1 (N’ GFP library) and 2 (C’ GFP library).
Quantification of Lipid droplet size and mitochondria shape
In order to get a quantitative information about the size of lipid droplets and the shape of mitochondria under growth in glucose and oleate (Fig.S1 A-E), a home-made script was written in Imagej (http://imagej.net/Fiji/Downloads), which includes rolling ball algorithm for subtracting the background and Otsu's threshold clustering algorithm for adjusting the threshold. After a mask was created, parameters such as area and circularity (the ratio between two perpendicular axes) were measured on the original images. The statistical analysis and the relevant plots were generated using R software (https://cran.r-project.org/bin/windows/base/old/3.2.2/). Overall, nine lipid droplets proteins and seven mitochondrial proteins were analyzed including thousands of cells per analysis.
Manual microscopy
Manual microscopy imaging was performed to the following strains: Mtc4-GFP+Erg6-mCherry (Fig.2D), GFP-Mls1 (Fig.3B), GFP-Mls2 (Fig.3C), GFP-Gto1 (Fig.3D), GFP-Pxp1 (Fig.S4C), GFP-Pex9 and GFP-Mls1 under the regulation of their native promoter (Fig.4A,B). Yeast strains were grown as described above for the high-throughput microscopy with changes in the selection required for each strain. Imaging was performed using the VisiScope Confocal Cell Explorer system, composed of a Zeiss Yokogawa spinning disk scanning unit (CSU-W1) coupled with an inverted Olympus microscope (IX83; x60 oil objective; Excitation wavelength of 488nm for GFP). Images were taken by a connected PCO-Edge sCMOS camera controlled by VisView software. All images were taken at a single focal plane.
Phylogenetic analysis
The phylogenetic analysis was conducted in MEGA7 software (Kumar S., 2015) (http://www.megasoftware.net/), with the Maximum Likelihood method (Jones et al., 1992). The tree topology was tested using 500 bootstrap iterations, which confirmed the separation of the two vertebrate branches (bootstrap value 99, Fig.4D). The scale represents the number of substitutions per site. Positions containing gaps were eliminated. The analysis involved 14 amino acid sequences: PEX5 (human (ENST00000266563), mouse (ENSMUSG00000005069), lizard (ENSACAG00000007658), chicken (ENSGALG00000014683), frog (ENSXETG00000023910), fish (ENSTRUG00000009964)), PEX5 like (human (ENST00000465751), mouse (ENSMUSG00000027674), lizard (ENSACAG00000004210), chicken (ENSGALG00000008929), frog (ENSXETG00000032445)), yeast PEX5 (YDR244W) and yeast PEX9 (YMR018W). Human UDP-N-acetylglucosamine, the closest protein to human PEX5 was used as an outgroup. The sequences were aligned using Clustalx software (http://www.clustal.org/download/current/).
Supplementary Material
Summary statement.
A high-content screen uncovered many changes in protein localization in yeast grown in oleate and highlighted a novel condition-specific peroxisomal protein, Pex9, which targets a subset of proteins to peroxisomes.
Acknowledgements
This work was supported by the European Research Council (ERC) (Consolidator grants Peroxisystem 64660). We wish to thank Yoav Peleg for constructing the GFP-PTS1(Mls1) plasmid. We wish to thank Ralf Erdmann and Wolfgang Schliebs for their collegiality.
Abbreviations List
- GFP
Green Fluorescent Protein
- PTS
Peroxisomal Targeting Signal
- PEX
Peroxin
- GID
Glucose Induced degradation Deficient
- SGA
Synthetic Genetic Array
Footnotes
Competing interests
No competing interests declared
Author contributions
EY, ND, SM, LZ and UW performed the experiments, SGC and EY performed the analysis, UW and IY constructed the SWAT GFP library, TO performed the phylogenetic analysis of the PTS1 receptors, MS and EZ supervised the work, MS, EZ and EY wrote the manuscript (with comments from the other authors).
References
- Amery L, Sano H, Mannaerts GP, Snider J, Van Looy J, Fransen M, Van Veldhoven PP. Identification of PEX5p-related novel peroxisome-targeting signal 1 (PTS1)-binding proteins in mammals. Biochem J. 2001;357:635–646. doi: 10.1042/0264-6021:3570635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto L, Garcera A, Jansson K, Sunnerhagen P, Herrero E. A peroxisomal glutathione transferase of Saccharomyces cerevisiae is functionally related to sulfur amino acid metabolism. Eukaryot Cell. 2006;5:1748–1759. doi: 10.1128/EC.00216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Braun B, Pfirrmann T, Menssen R, Hofmann K, Scheel H, Wolf DH. Gid9, a second RING finger protein contributes to the ubiquitin ligase activity of the Gid complex required for catabolite degradation. FEBS Lett. 2011;585:3856–3861. doi: 10.1016/j.febslet.2011.10.038. [DOI] [PubMed] [Google Scholar]
- Breker M, Gymrek M, Moldavski O, Schuldiner M. LoQAtE--Localization and Quantitation ATlas of the yeast proteomE. A new tool for multiparametric dissection of single-protein behavior in response to biological perturbations in yeast. Nucleic Acids Res. 2014;42:D726–730. doi: 10.1093/nar/gkt933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker M, Gymrek M, Schuldiner M. A novel single-cell screening platform reveals proteome plasticity during yeast stress responses. J Cell Biol. 2013;200:839–850. doi: 10.1083/jcb.201301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breker M, Schuldiner M. The emergence of proteome-wide technologies: systematic analysis of proteins comes of age. Nat Rev Mol Cell Biol. 2014;15:453–464. doi: 10.1038/nrm3821. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Klug YA, Dimitrov L, Erez Z, Chuartzman SG, Elinger D, Yofe I, Soliman K, Gartner J, Thoms S, Schekman R, et al. Peroxisomes are juxtaposed to strategic sites on mitochondria. Mol Biosyst. 2014;10:1742–1748. doi: 10.1039/c4mb00001c. [DOI] [PubMed] [Google Scholar]
- Cohen Y, Schuldiner M. Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods Mol Biol. 2011;781:127–159. doi: 10.1007/978-1-61779-276-2_8. [DOI] [PubMed] [Google Scholar]
- Effelsberg D, Cruz-Zaragoza LD, Schliebs W, Erdmann R. Pex9p is a novel import receptor for a subset of peroxisomal enzymes with a type1 peroxisomal targeting signal in Saccharomyces cerevisiae. J Cell Sci. 2016 doi: 10.1242/jcs.195271. submitted. [DOI] [PubMed] [Google Scholar]
- Effelsberg D, Cruz-Zaragoza LD, Tonillo J, Schliebs W, Erdmann R. Role of Pex21p for Piggyback Import of Gpd1p and Pnc1p into Peroxisomes of Saccharomyces cerevisiae. J Biol Chem. 2015;290:25333–25342. doi: 10.1074/jbc.M115.653451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurvitz A, Hiltunen JK, Erdmann R, Hamilton B, Hartig A, Ruis H, Rottensteiner H. Saccharomyces cerevisiae Adr1p governs fatty acid beta-oxidation and peroxisome proliferation by regulating POX1 and PEX11. J Biol Chem. 2001;276:31825–31830. doi: 10.1074/jbc.M105989200. [DOI] [PubMed] [Google Scholar]
- Hasan S, Platta HW, Erdmann R. Import of proteins into the peroxisomal matrix. Front Physiol. 2013;4:261. doi: 10.3389/fphys.2013.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Islinger M, Cardoso MJ, Schrader M. Be different--the diversity of peroxisomes in the animal kingdom. Biochim Biophys Acta. 2010;1803:881–897. doi: 10.1016/j.bbamcr.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jung S, Smith JJ, von Haller PD, Dilworth DJ, Sitko KA, Miller LR, Saleem RA, Goodlett DR, Aitchison JD. Global analysis of condition-specific subcellular protein distribution and abundance. Mol Cell Proteomics. 2013;12:1421–1435. doi: 10.1074/mcp.O112.019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpichev IV, Durand-Heredia JM, Luo Y, Small GM. Binding characteristics and regulatory mechanisms of the transcription factors controlling oleate-responsive genes in Saccharomyces cerevisiae. J Biol Chem. 2008;283:10264–10275. doi: 10.1074/jbc.M708215200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpichev IV, Small GM. Global regulatory functions of Oaf1p and Pip2p (Oaf2p), transcription factors that regulate genes encoding peroxisomal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6560–6570. doi: 10.1128/mcb.18.11.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel JA, Veenhuis M, van der Klei IJ. PEX genes in fungal genomes: common, rare or redundant. Traffic. 2006;7:1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- Kumar S, Singh R, Williams CP, van der Klei IJ. Stress exposure results in increased peroxisomal levels of yeast Pnc1 and Gpd1, which are imported via a piggy-backing mechanism. Biochim Biophys Acta. 2016;1863:148–156. doi: 10.1016/j.bbamcr.2015.10.017. [DOI] [PubMed] [Google Scholar]
- Kumar S, S G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution. 2015 doi: 10.1093/molbev/msw054. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze M, Kragler F, Binder M, Hartig A, Gurvitz A. Targeting of malate synthase 1 to the peroxisomes of Saccharomyces cerevisiae cells depends on growth on oleic acid medium. Eur J Biochem. 2002;269:915–922. doi: 10.1046/j.0014-2956.2001.02727.x. [DOI] [PubMed] [Google Scholar]
- Kunze M, Pracharoenwattana I, Smith SM, Hartig A. A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim Biophys Acta. 2006;1763:1441–1452. doi: 10.1016/j.bbamcr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Purdue PE, Yang X, Lazarow PB. Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol. 1998;143:1859–1869. doi: 10.1083/jcb.143.7.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliebs W, Kunau WH. PTS2 co-receptors: diverse proteins with common features. Biochim Biophys Acta. 2006;1763:1605–1612. doi: 10.1016/j.bbamcr.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Shai N, Schuldiner M, Zalckvar E. No peroxisome is an island - Peroxisome contact sites. Biochim Biophys Acta. 2016;1863:1061–1069. doi: 10.1016/j.bbamcr.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Aitchison JD. Peroxisomes take shape. Nat Rev Mol Cell Biol. 2013;14:803–817. doi: 10.1038/nrm3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Marelli M, Christmas RH, Vizeacoumar FJ, Dilworth DJ, Ideker T, Galitski T, Dimitrov K, Rachubinski RA, Aitchison JD. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J Cell Biol. 2002;158:259–271. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Ramsey SA, Marelli M, Marzolf B, Hwang D, Saleem RA, Rachubinski RA, Aitchison JD. Transcriptional responses to fatty acid are coordinated by combinatorial control. Mol Syst Biol. 2007;3:115. doi: 10.1038/msb4100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol Biol. 2006;313:171–192. doi: 10.1385/1-59259-958-3:171. [DOI] [PubMed] [Google Scholar]
- Trotter PJ. The genetics of fatty acid metabolism in Saccharomyces cerevisiae. Annu Rev Nutr. 2001;21:97–119. doi: 10.1146/annurev.nutr.21.1.97. [DOI] [PubMed] [Google Scholar]
- Veenhuis M, Mateblowski M, Kunau WH, Harder W. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 1987;3:77–84. doi: 10.1002/yea.320030204. [DOI] [PubMed] [Google Scholar]
- Velazquez AP, Tatsuta T, Ghillebert R, Drescher I, Graef M. Lipid droplet-mediated ER homeostasis regulates autophagy and cell survival during starvation. J Cell Biol. 2016;212:621–631. doi: 10.1083/jcb.201508102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Ferdinandusse S, Wanders RJ. Human disorders of peroxisome metabolism and biogenesis. Biochim Biophys Acta. 2016;1863:922–933. doi: 10.1016/j.bbamcr.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Yofe I, Weill U, Meurer M, Chuartzman S, Zalckvar E, Goldman O, Ben-Dor S, Schutze C, Wiedemann N, Knop M, Khmelinskii A, et al. One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nat Methods. 2016;13:371–378. doi: 10.1038/nmeth.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.