Extended Data Figure 1: Parametric design: workflow and shortcomings.
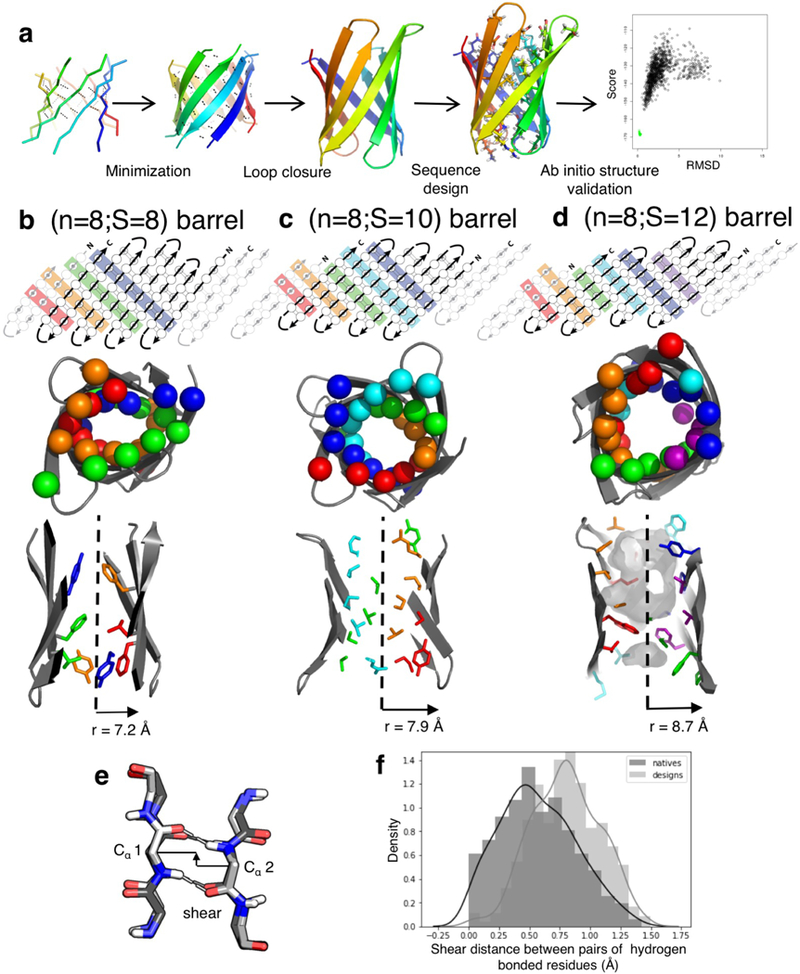
a, Schematic representation of the parametric approach to generate β-barrel designs. b-d, Comparison between β-barrels of type (n=8;S=8, b), type (n=8;S=10, c) and type (n=8;S=12, d); showing an example of 2D map with residue connectivity (top), the arrangement of the Cβs in the Cβ-strips (middle) and the packing pattern of the core side-chains (bottom). The difference in shear number translates into different overall strand staggering and barrel radii. The number of core Cβ-strips (top, middle) results in different arrangements of side-chains in the core of the barrel. e&f, The parametric designs exhibited distorted hydrogen bonds, reflected by the shear distance (defined in e) between paired antiparallel β-strands residues. The shear distance in the design deviate from the distribution observed in native β-sheet proteins (f).
