Abstract
Nucleotide sequences representing nine genes and five presumptive genetic loci were used to infer phylogenetic relationships among seven Baylisascaris species, including one species with no previously available molecular data. These genes were used to test the species status of B. procyonis and B. columnaris using a coalescent approach. Phylogenetic analysis based on combined analysis of sequence data strongly supported monophyly of the genus and separated the species into two main clades. Clade 1 included B. procyonis, B. columnaris, and B. devosi, species hosted by musteloid carnivores. Clade 2 included B. transfuga and B. schroederi from ursids, B. ailuri, a species from the red panda (a musteloid), and B. tasmaniensis from a marsupial. Within clade 2, geographic isolates of B. transfuga, B. schroederi (from giant panda), and B. ailuri formed a strongly supported clade. In certain analyses (e.g., some single genes), B. tasmaniensis was sister to all other Baylisascaris species rather than sister to the species from ursids and red panda. Using one combination of priors corresponding to moderate population size and shallow genetic divergence, the multispecies coalescent analysis of B. procyonis and B. columnaris yielded moderate support (posterior probability 0.91) for these taxa as separate species. However, other prior combinations yielded weak or no support for delimiting these taxa as separate species. Similarly, tree topologies constrained to represent reciprocal monophyly of B. columnaris and B. procyonis individuals (topologies consistent with separate species) were significantly worse in some cases, but not others, depending on the dataset analyzed. An expanded analysis of SNPs and other genetic markers that were previously suggested to distinguish between individuals of B. procyonis and B. columnaris was made by characterization of additional individual nematodes. The results suggest that many of these SNPs do not represent fixed differences between nematodes derived from raccoon and skunk hosts.
Keywords: Baylisascaris, Raccoon roundworm, Phylogenetics, Molecular systematics, Species delimitation, Arctoidea, Tasmanian devil
Graphical abstract
Highlights
-
•
A phylogenetic hypothesis for Baylisascaris species was produced using nine genes.
-
•
Genetic data was generated for two new species- B. devosi and B. tasmaniensis.
-
•
Baylisascaris devosi and B. tasmaniensis were part of a monophyletic Baylisascaris.
-
•
B. procyonis (raccoon) and B. columnaris (skunk) could not be reliably distinguished.
-
•
Established SNPs may not be diagnostic for Baylisascaris from raccoons and skunks.
1. Introduction
Baylisascaris species are intestinal nematode parasites that primarily infect hosts in Arctoidea (Mammalia: Carnivora). Sprent (1968) created the genus to account for distinct morphological features of seven species previously classified as Ascaris or Toxascaris. Baylisascaris currently contains 11 recognized species (Table 1). Baylisascaris procyonis, commonly known as the raccoon roundworm, is the most studied species because it is widespread in North America, and causes severe pathogenicity in paratenic and accidental human hosts due to extensive larval migration through host tissues (Kazacos, 2001; Graeff-Teixeira et al., 2016). Less research has been done on the other ten Baylisascaris species, due to reduced availability of specimens and because they are considered less important than B. procyonis from a human health perspective.
Table 1.
Recognized species of Baylisascaris.
| Species | Primary Host | Original description |
|---|---|---|
| B. transfuga | Ursine species | Rudolphi, 1819 |
| B. columnaris | Mephitis mephitis | Leidy, 1856 |
| B. laevis | Marmota monax | Leidy, 1856 |
| B. melis | Meles meles | Gedoelst, 1920 |
| B. schroederi | Ailuropoda melanoleuca | McIntosh, 1939 |
| B. procyonis | Procyon lotor | Stefanski and Zarnowski, 1951 |
| B. devosi | Pekania pennanti, M. americana, Gulo gulo, Martes zibellina | Sprent, 1952 |
| B. tasmaniensis | Sarcophilus harrisii | Sprent, 1970 |
| B. ailuri | Ailurus fulgens | Wu et al., 1987 |
| B. potosis | Potus flavus | Tokiwa et al., 2014 |
| B. venezuelensis | Tremarctos ornatus | Pérez Mata et al., 2016 |
Genetic analyses of Baylisascaris species have been limited, but sequence data from certain species has been used to investigate: 1) phylogenetic relationships among species in order Ascaridida (Nadler, 1992; Nadler and Hudspeth, 1998, 2000; Xie et al., 2011a, 2011b, 2013; Li et al., 2012; Liu et al., 2014); 2) detection of species from fecal samples (B. procyonis, Dangoudoubiyam et al., 2009, Gatcombe et al., 2010; B. transfuga, De Ambrogi et al., 2011; B. schroederi, Zhou et al., 2013); 3) identification of adults from different hosts (B. devosi, Tranbenkova and Spiridonov, 2017; B. schroederi, Lin et al., 2012; Zhao et al., 2012; B. transfuga, Testini et al., 2011) or geographic areas (Davidson et al., 2013); 4) species delimitation of B. procyonis and B. columnaris (Franssen et al., 2013; Choi et al., 2017); and 5) descriptions of new species (B. ailuri, He et al., 2008, cited in Xie et al., 2011a; B. potosis, Taira et al., 2013 and Tokiwa et al., 2014; B. venezuelensis, Pérez Mata et al., 2016).
The first phylogenetic analysis of Baylisascaris involved comparison of B. procyonis and B. transfuga among superfamily Ascaridoidea and was based on small (18S) and large subunit (28S) nuclear ribosomal DNA (rDNA) (Nadler, 1992). These analyses were expanded by adding more rDNA sequence (Nadler and Hudspeth, 1998), mitochondrial cytochrome c oxidase subunit 2 (cox2) sequences, and morphological characters (Nadler and Hudspeth, 2000). Baylisascaris procyonis and B. transfuga were monophyletic (with moderate to low support) based on cox2 alone and based on cox2 combined with rDNA and morphological data (Nadler and Hudspeth, 2000). He et al. (2008) examined evolutionary relationships within Baylisascaris based on sequences of internal transcribed spacer 2 (rDNA, ITS2), confirming that Baylisascaris specimens from giant panda and multiple bear species formed a clade, along with an ascarid from red panda. Based on this result, they concluded that the red panda parasite belonged to Baylisascaris rather than Toxascaris (cited in Xie et al., 2011a). Phylogenetic analyses based on amino acid sequences of 12 mitochondrial protein-coding genes from ten ascarid species yielded a monophyletic group of Baylisascaris (B. transfuga, B. schroederi, and B. ailuri) with absolute support; B. transfuga and B. ailuri were sister taxa with varying support (Xie et al., 2011a, 2011b). Other recent phylogenetic analyses have been based on nuclear rDNA and mitochondrial cox1 and cox2 sequences and included five to six Baylisascaris species (e.g. Franssen et al., 2013; Tokiwa et al., 2014).
These phylogenetic analyses represent preliminary assessments of species relationships within Baylisascaris. Monophyly of the genus was supported for most inference methods and genes (Taira et al., 2013; Xie et al., 2013; Tokiwa et al., 2014), and two well-supported clades have consistently been resolved: clade 1 consisting of B. procyonis, B. columnaris; and clade 2 consisting of B. ailuri, B. schroederi, B. transfuga. A recent analysis placed B. potosis as monophyletic with B. procyonis and B. columnaris (Tokiwa et al., 2014) based on 28S rDNA and cox1. For both nuclear rDNA and cox1 data, B. potosis and B. devosi resolve as members of clade 1 (Tranbenkova and Spiridonov, 2017). Baylisascaris venezuelensis has been resolved as part of clade 2 based on ITS1 and ITS2 (Pérez Mata et al., 2016). Despite these advances, our understanding of Baylisascaris relationships is incomplete for four reasons: 1) three of the eleven species (B. laevis, B. melis, and B. tasmaniensis) have no published sequence data; 2) the genes used to infer phylogenies represent only two loci and do not provide strong support for all relationships; 3) there are no genetic markers that reliably distinguish B. procyonis from B. columnaris; and 4) the number of Baylisascaris species present in bear hosts has not been evaluated.
The goals of the present study are to: 1) infer relationships among Baylisascaris species and evaluate monophyly of the genus after including newly sampled species (B. tasmaniensis) and genes; 2) use multiple loci to test the validity of species status for B. procyonis and B. columnaris; and 3) compare Baylisascaris specimens isolated from different species of bears (including giant pandas) representing different geographic areas. The molecular objectives were accomplished by obtaining sequence data from three variable nuclear loci not previously used for Baylisascaris systematics, along with nuclear ribosomal and mitochondrial gene sequences.
2. Materials and methods
2.1. Specimen collection and DNA extraction
All nematodes were collected from host intestines at necropsy. Parasite species, host species, and collection locations are listed in Table 2. Specimens were preserved in 95% ethanol and then stored at −20 °C. Prior to dissection, specimens were treated in a 62 °C water bath for 4 min to inactivate potentially infective eggs (Shafir et al., 2011), including eggs ingested by, or on the cuticle of, males. Nematodes were dissected to obtain muscle tissue, which was placed in digestion buffer containing 10% Sarkosyl, 100 mM Tris HCl (pH 7.6), 200 mM NaCl, and 0.5 M EDTA. Two μL of proteinase K (10 mg/mL) was added to each sample, and specimens were incubated at 56 °C until tissues were fully digested. DNA was extracted from each specimen using DNAzol reagent (Molecular Research Center, Cincinnati, Ohio) following the manufacturer's protocol. DNA was precipitated with 2-propanol and the resulting pellet was dried overnight. Extracts were resuspended in 35 μL of TE buffer and stored at −20 °C. Prior to PCR, all extracts were diluted 1:10 with TE buffer to reduce the effects of potential PCR inhibitors.
Table 2.
List of Baylisascaris species and outgroups included in analyses. Note: no hars1 sequence for B. devosi.
| Species | Abbreviation | Host | Collection location | GenBank accession #s: 12S, cox1, cox2, 28S, ITS, ard1, hars1, msp |
|---|---|---|---|---|
| B. columnaris CT | BcCT | Mephitis mephitis | Connecticut, USA | MG937785, MH795147, MH469662, MG937772, MH030594, MH900134, MH900147, MH891571 |
| B. columnaris IL | BcIL | M. mephitis | Illinois, USA | MG937786, MH795148, MH469663, MG937773, MH030595, MH900135, MH900148, MH891572 |
| B. procyonis CT | BpCT | Procyon lotor | Connecticut, USA | MG937787, MH795149, MH469664, MG937774, MH030596, MH900136, MH900149, MH891573 |
| B. procyonis CA | BpCA | P. lotor | California, USA | MG937788, MH795150, MH469665, MG937775, MH030597, MH900137, MH900150, MH891574 |
| B. devosi | Bd | Pekania pennanti | Ontario, Canada | MG937789, MH795151, MH469666, MG937776, MH030598, MH900138, MH891575 |
| B. schroederi | Bs | Ailuropoda melanoleuca | Sichuan, China | MG937790, MH795152, MH469667, MG937777, MH030599, MH900139, MH900151, MH891576 |
| B. ailuri | Ba | Ailurus fulgens | Sichuan, China | MG937791, MH795153, MH469668, MG937778, MH030600, MH900140, MH900152, MH891577 |
| B. transfuga ALB | BtrALB | U. arctos | Alberta, Canada | MG937792, MH795154, MH469669, MG937779, MH030601, MH900141, MH900153, MH891578 |
| B. transfuga WV | BtrWV | Ursus americanus | West Virginia, USA | MG937793, MH795155, MH469670, MG937780, MH030602, MH900142, MH900154, MH891579 |
| B. tasmaniensis | Btas | Sarcophilus harrisii | Tasmania, Australia | MG937794, MH795156, MH469671, MG937781, MH030603, MH900143, MH900155, MH891580 |
| A. suum | As | Sus scrofa domesticus | Louisiana or Michigan, USA | MG937795, MH795157, MH469672, MG937782, MH030604, MH900144, MH900156, MH891581 |
| P. equorum | Pe | Equus ferus caballus | Louisiana, USA | MG937796, MH795158, MH469673, MG937783, MH030605, MH900145, MH900157, MH891582 |
| T. leonina | Tl | Vulpes vulpes | South Dakota, USA | MG937797, MH795159, MH469674, MG937784, MH030606, MH900146, MH900158, MH891583 |
2.2. Genes and primer design
To provide variable sequences for resolving relationships among closely related Baylisascaris species, primers were successfully designed for two exon-primed, intron-crossing loci (EPICs; Lessa, 1992; Slade et al., 1993; Palumbi and Baker, 1994). Initial candidate genes for designing primers for EPIC loci came from Regier et al. (2008); we targeted the ten fastest evolving genes identified by Regier et al. (2008), and used protein sequences to query BLAST databases (WormBase v. 240) of Ascaris suum (ftp://ftp.wormbase.org/pub/wormbase/releases/WS240/species/a_suum/) and Brugia malayi (ftp://ftp.wormbase.org/pub/wormbase/releases/WS240/species/b_malayi/) genomes. EPIC primers were designed for six loci based on amino acid alignments of B. malayi and A. suum. These primers were tested using A. suum DNA, and those yielding positive PCRs were tested in Baylisascaris species and outgroups. Primer design for Baylisascaris was successful for two EPIC loci – alcohol/ribitol dehydrogenase (ard-1) and histidyl tRNA synthetase (hars-1) (Table 3). Alternative primers for these genes were designed and used as needed if initial amplification of these loci was suboptimal for particular Baylisascaris species. Amplicon lengths for ard-1 ranged from 617 to 900 bp, and for hars-1 from 678 to 737 bp.
Table 3.
Unique primers used in this study.
| Gene | Primer name | Direction | Sequence (5′ to 3′) | PCR (P) or Sequencing (S) |
|---|---|---|---|---|
| cox1 | dp508 | Forward | ATAATTTTTTTTATRGTTATRCC | P and S |
| dp509 | Reverse | AATCTCAGACTGRTATCTRTGACCAAATACTRA | P and S | |
| dp615 | Forward | GTTCTGGCGGGGGCTATTAC | P and S | |
| dp616 | Reverse | CCCAGTAATAAAACGCCACC | P and S | |
| ITS | dp617 | Forward | CTCCGAACGTGCATAAGCACC | S |
| msp | dp818 | Forward | GTTCCTCCTGGCGATATCAACACC | P and S |
| dp819 | Reverse | CATGCCATCACCCTGGAACCATTC | P and S | |
| ard1 | dp828 | Forward | TTCGCTGAGAACGAGAAAGACG | P and S |
| dp833 | Reverse | CTTATCGGGGAAGGATGCCATC | P and S | |
| dp835 | Forward | GTGTCATTATCAACACGGGCTC | P and S | |
| hars1 | dp824 | Forward | GAGAGGGCGTTATCGAGAGTTC | P and S |
| dp836 | Reverse | GACCTCATCCCATGAAACCTTGTC | P and S | |
| dp853 | Reverse | TGAAACCTTGTCCAGTTTATCG | P and S |
In addition to ard-1 and hars-1, regions of three mitochondrial genes (12S ribosomal DNA, cytochrome oxidase subunits 1 and 2 (cox1, cox2)), three nuclear rDNA genes (large-subunit (28S) and both internal transcribed spacers (ITS-1, ITS-2)), and one nuclear gene (Major Sperm Protein (msp)) were amplified by PCR. Primer sequences unique to this study are in Table 3. For 12S, primers 505 and 506 (Nadler et al., 2006) were used to amplify a region of 535–542 bp. For cox1, a region of 1163–1211 bp was amplified using one of two forward primers (508 or 615) with one of two reverse primers (509 or 616). Primers 211 and 210 (Nadler and Hudspeth, 2000) were used to amplify cox2 sequences of 629 bp for all species. The 5′ end of 28S rDNA (domains D1-D3, 1106–1110 bp) was amplified using primers 391 (Nadler and Hudspeth, 1998) and 501 (Thomas et al., 1997). Full-length ITS-1, 5.8S, ITS-2 (889–975 bp) was amplified using primers 521 (Gasser et al., 1996) and 94 (Gasser et al., 1993). ITS sequences were truncated prior to analysis due to poor quality sequence for A. suum at the 5′ end (ITS-1) and B. transfuga ALB and B. schroederi at the 3’ end (ITS-2). Finally, msp was amplified using forward primer 566 (Anderson and Jaenike, 1997) or 818 with reverse primer 567 (Anderson and Jaenike, 1997) or 819, yielding amplicons of 529–615 bp.
2.3. PCR amplification, sequencing, and cloning
For all primer combinations, polymerase chain reactions contained 3 mM MgCl2, 200 μM deoxynucleoside triphosphates, 1 unit of AmpliTaq polymerase, 0.5 μM of each primer, and 1.5–3 μl of DNA template. To enhance PCR yield, the KOD XL polymerase kit (EMD Millipore, Merck KGaA, Darmstadt, Germany) was used for some amplifications. Cycling parameters differed for each gene. For 12S and LSU, cycling parameters followed Nadler et al. (2006). For cox1, cycling parameters started with denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 50 °C or 55 °C for 30 s, 72 °C for 1 min, and a final extension of 72 °C for 7 min. For cox2, cycling parameters followed Nadler and Hudspeth (2000). For ITS, cycling parameters started with a 4 min denaturation at 94 °C, then 35 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min, and a final extension of 7 min at 72 °C. For msp, cycling parameters started with denaturation at 94 °C for 3 min, and then 35 cycles of 94 °C for 30 s, 55 °C or 60 °C for 45 s, 72 °C for 1.5 min, followed by a final extension at 72 °C for 7 min. For ard1, cycling parameters included an initial denaturation at 94 °C for 4 min, then 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 49 s, and a final extension of 7 min at 72 °C. For hars1, cycling parameters began with an initial denaturation at 94 °C for 4 min, then 35 cycles of 94 °C for 30 s, 57 °C for 30 s, 72 °C for 45 s, and a final extension at 72 °C for 7 min. Annealing temperature and number of PCR cycles were adjusted empirically for optimal amplification of these genes in each species.
PCR products were enzymatically treated for direct sequencing with exonuclease I and shrimp alkaline phosphatase (US Biochemical, Affymetrix Pre-sequencing kit, USA). Products were sequenced using an ABI 3730 DNA Sequencer (Applied Biosystems, Thermo Fisher Scientific) with PCR primers, internal primers, or both (Table 3). Cloning was done for PCR products that could not be sequenced directly, generally due to repetitive sequence regions, double PCR bands, or weak amplification. Cloning was needed for ITS (both B. procyonis isolates), ard1 (A. suum), hars1 (all species), and msp (B. devosi). Prior to cloning PCR products with double-bands, the product of interest was gel isolated using centrifugal gel extraction devices (EMD Millipore, Merck KGaA, Darmstadt, Germany). PCR products were cloned using pGEM-T vector (Promega, Madison, Wisconsin) using methods described previously (Nadler and Hudspeth, 1998) except JM109 Escherichia coli was transformed. Putative clones were checked by PCR amplification with modified pGEM-T vector primers (Nadler and Hudspeth, 1998; 156 5′-GGCCAGTGAATTGTAATACGACTC; 157 5′-GACACTATAGAATACTCAAGCTATGC), and for positive clones the resulting PCR products were directly sequenced. Clones that yielded low quality (direct) sequence were prepared as purified plasmids using a commercial kit (Qiaprep, Qiagen) and sequenced. Clones were sequenced using primers 156 and 157 and with PCR or internal primers as needed.
Contigs were assembled for directly sequenced and cloned products in CodonCode Aligner (version 5.1.5, CodonCode Corporation, Centerville, Massachusetts) using Phred base calling. All sequences were double-stranded for verification. Sequences corresponding to PCR primers were removed. Polymorphisms were only recorded if the following conditions were met: both possible peaks were present in DNA strands sequenced from both directions; the shorter peak was significantly higher than background peaks; and the shorter peak was at least 25% of the height of the taller peak. GenBank accession numbers for all sequences are provided in Table 2.
2.4. Phylogenetic analysis
For non-protein-coding genes (12S, 28S, ITS), loci that contained large intron regions (ard1, and hars1), and msp, sequences were aligned using ProAlign (Löytynoja and Milinkovitch, 2003). Unreliably aligned sites were detected based on the minimum posterior probability (PP) of sites. Alignment ambiguous regions were removed (filtered) for all non-coding genes and those with introns based on a 60% minimum PP threshold, to avoid excluding sites that were aligned correctly (Löytynoja and Milinkovitch, 2003). Thus unfiltered (FULL) and filtered (FILT) datasets were obtained for these six genes.
Nucleotide sequences of protein-coding genes cox1 and cox2 were translated using the web-based program ExPASY (http://web.expasy.org/translate/). Amino acid sequences for each gene were aligned with default options in CLUSTAL_X (Larkin et al., 2007). The online RevTrans Server (v1.4; Wernersson and Pedersen, 2003) was used to align nucleotide sequences of cox1 and cox2 based on their amino acid alignments. Following alignment, Gblocks (v 0.91b; Castresana, 2000) with default options was used to test for ambiguously aligned sites; cox1 and cox2 lacked such sites.
Maximum parsimony (MP) analyses were done for the FULL and FILT dataset for each gene using PAUP* (Swofford, 2003). Branch-and-bound searches were conducted to generate MP trees; strict consensus trees were created if there was more than one MP tree. To assess clade support values, bootstrap searches with character resampling were done using 10,000 pseudoreplicates, with 1000 replicates of random-taxon addition, and tree-bisection-reconnection branch swapping. PAUP* was also used to obtain matrices of uncorrected “p” pairwise distance for each gene.
Bayesian inference (BI) analyses were conducted on the FULL and FILT datasets for each gene using MrBayes v3.2.6 (Ronquist et al., 2012) executed on the Cyberinfrastructure for Phylogenetic Research (CIPRES) web portal (Miller et al., 2010; http://www.phylo.org). For non-coding genes, best-fit evolutionary models (Table S1) were chosen prior to Bayesian analysis based on the Akaike Information Criterion (AIC) using MrModelTest v2.3 (Nylander, 2004). For cox1 and cox2, partitioning schemes and evolutionary models (Table S1) were selected based on the AIC using PartitionFinder v1.1.1 (Lanfear et al., 2012). These models were applied to each gene or partition for Bayesian analyses. Two independent Bayesian runs were conducted for each gene with four Markov Chain Monte Carlo chains for 4 or 8 million generations, with chains sampled every 4000 or 8000 generations (respectively), and the initial 25% of trees were discarded as burn-in. In the post burn-in samples, stationarity was assessed based on the following: an average standard deviation of split frequencies below 0.01; an average potential scale reduction factor of 1.000; and similar mean marginal likelihoods for both runs. Convergence of the remaining parameters was assessed using Tracer v1.6 (Rambaut et al., 2014; http://tree.bio.ed.ac.uk/software/tracer/). The trees remaining after burn-in were used to create 50% majority-rule consensus trees with posterior probability distributions for each clade. Consensus trees from MP and BI runs were visualized in FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and edited using InkScape v0.91 (http://inkscape.org/) or GravitDesigner (https://designer.io).
Additional datasets of cox1, 28S, and ITS were created to incorporate published sequences from Baylisascaris species including B. devosi (cox1, KX682028, KM216978-85; 28S, KY465564; and ITS1 and ITS2, KY465505), B. potosis (cox1, AB893609; 28S, AB893608; and ITS2, AB901104), and B. venezuelensis (ITS1 and ITS2, KX151725-27). Published sequences were aligned to original alignments of single genes using the Profile Alignment function in CLUSTAL_X (Larkin et al., 2007). Trees for each gene were inferred using Bayesian analyses following the same procedures described in Section 2.4 for single genes. Sequences from single genes were concatenated to form datasets containing mitochondrial genes (12S, cox1, and cox2), nuclear genes (28S, ITS, msp, ard1), and combined mitochondrial and nuclear genes (Table S2). Three datasets contained unfiltered sequence data (Mitochondrial FULL, Nuclear FULL, and Combined FULL) and three datasets contained sequence data where alignment ambiguous characters were removed (filtered; Mitochondrial FILT, Nuclear FILT, and Combined FILT). Four additional datasets (Table S2) were necessary to analyze hars1 sequences because the B. devosi hars1 was a putative paralog (see Results). Incongruence length difference (ILD) tests (Farris et al., 1994, 1995) were performed using PAUP* (“partition homogeneity test”; Swofford, 2003) for the following genes or datasets (FULL data only) to test for incongruence: combined mitochondrial genes vs. combined nuclear genes; nuclear rDNA vs. each low or single copy nuclear locus (msp, ard1, hars1); msp vs. ard1; and msp vs. hars1. Trees were inferred for all combined datasets in PAUP* for MP and MrBayes for BI, using the procedures followed for single genes. Alternative topology tests were conducted on trees obtained from MP analyses of all combined datasets: mitochondrial, nuclear (with and without hars1), and combined data (with and without hars1) using Templeton's modified parsimony test (Templeton, 1983) implemented in PAUP*. For Bayesian analyses, datasets were partitioned based on genes or codon positions (cox1 and cox2), and models of evolution were applied separately to each partition. Parameters were unlinked across all partitions and were estimated as part of the analyses. Bayesian analyses of combined datasets were run for 20–40 million generations with chain sampling every 20,000 or 40,000 generations, respectively. All other procedures for Bayesian runs followed those used for single genes.
2.5. Species delimitation of B. procyonis and B. columnaris with BP&P
The program BP&P v3.3 (Bayesian Phylogenetics and Phylogeography; Yang, 2015) was used for species delimitation analysis of B. procyonis and B. columnaris. BP&P implements the multispecies coalescent model (MSC) to account for possible conflicts between gene trees and species trees as coalescent models accommodate independent evolutionary histories of different loci (Caviedes-Solis et al., 2015). Two parameters are included in the MSC: θs are the population size parameters for ancestral and modern species, and τs are species divergence times. In BP&P the population size parameter is assigned a gamma prior of G θ (α, β) with a mean of α/β. The root age (τ0) also has a gamma prior G τ0 (α, β), while the other species divergence times have a Dirichlet prior. The shape parameter α determines how informative each prior is – values of α ∼2 represent diffuse (non-informative) priors; diffuse priors were used for this analysis. However, for B. procyonis and B. columnaris we predict a moderate effective population size and a shallow divergence time (the latter based on previously observed low sequence divergence), and therefore initial priors were chosen to reflect those conditions. Multiple combinations of parameters for the gamma priors were run in BP&P module A00 to test the sensitivity of posterior estimates for θ and τ to the priors before running the species delimitation module A11 (Yang, 2015). We chose three priors each for θ and τ. For θ, the priors represented a large population (G θ (1.95, 30)), a moderate population (G θ (1.95, 300)), and a small population (G θ (1.95, 3000)). For τ, the priors represented a deep divergence (G τ0 (2, 200)), a shallow divergence (G τ0 (2, 2000)), and a very shallow divergence (G τ0 (2, 20000)).
The A11 module in BP&P was used to test the hypothesis of separate species for specimens from raccoon (B. procyonis) and skunk (B. columnaris). This module allows species delimitation and species tree inference to be conducted jointly. Specimens from grizzly bears (B. transfuga) collected in Alberta, Canada were used as an outgroup. Sequence files used in the analyses contained five loci: mitochondrial (12S + cox2), ITS, msp, ard1, and hars1. In BP&P, two sequences from each potential species are used to estimate θs. Sequence data from two individuals of Baylisascaris from grizzly bears were not available for msp or ard1, so these loci only included sequence data for specimens from skunk and raccoon hosts. The prior for the number of species was three, with the assumption that specimens derived from a given host were the same species. All nine combinations of θ and τ0 priors were run for 5 × 105 generations, with a burn-in of 1 × 104, and a sample frequency of 5. Two independent runs were conducted for each combination of priors to assess convergence. Similar results from both runs demonstrate adequate chain mixing.
3. Results
3.1. Sequence characteristics and datasets
Sequences of hars1 were excluded from the largest combined datasets containing nuclear genes and combined mitochondrial and nuclear genes because the sequence obtained for B. devosi appeared to be a paralog. In order to analyze hars1 sequences alone, and in combination with other genes, additional datasets were created that included hars1 and excluded B. devosi (Table S2). There was little indication of incongruence among genes as significant partition homogeneity tests occurred only if msp was included as a separate partition. Notably, msp sequences did not have the expected open reading frame or translation, suggesting they might be msp pseudogenes. The numbers of characters for all genes and datasets (e.g. FULL and FILT) are in Table S2. The percentage of characters filtered (removed) based on the ProAlign result was highest for ard1 at 47.1% and lowest for 28S at 0.4%.
Among Baylisascaris species, average pairwise percent sequence divergence was highest for msp at 15.5%, and lowest for 28S at 1.5% (Tables S3-S6). Among B. columnaris and B. procyonis individuals, values of pairwise divergence only exceeded 1% for cox1 (Table S3), ard1 (Table S4), and hars1 (Table S4). Pairwise divergence values varied between 0.15% and 4.9% within clade 2 (B. ailuri, B. schroederi, and B. transfuga; Tables S3-S6). For this group, relatively high divergence values were obtained when comparing B. schroederi with B. ailuri for cox1 (4.9%, Table S3) and B. ailuri with B. transfuga WV for cox2 (4.6%, Table S3).
In the following sections, descriptions of results and trees are based on FULL, combined datasets. Support values for monophyly of selected clades based on single genes are summarized in Tables S7 and S8. Bootstrap values below 70% and Bayesian posterior probabilities (BPP) less than 0.90 are not provided or shown. Trees for combined datasets (Fig. 1, Fig. 2, Fig. 3, Figs. S1, and S2) are only shown for Bayesian inference of FULL datasets. Trees based on combined datasets for parsimony analysis were very similar to trees from Bayesian analyses, with the only ingroup difference involving the position of B. tasmaniensis. Due to these similarities, parsimony trees are only described for single genes (section 3.5).
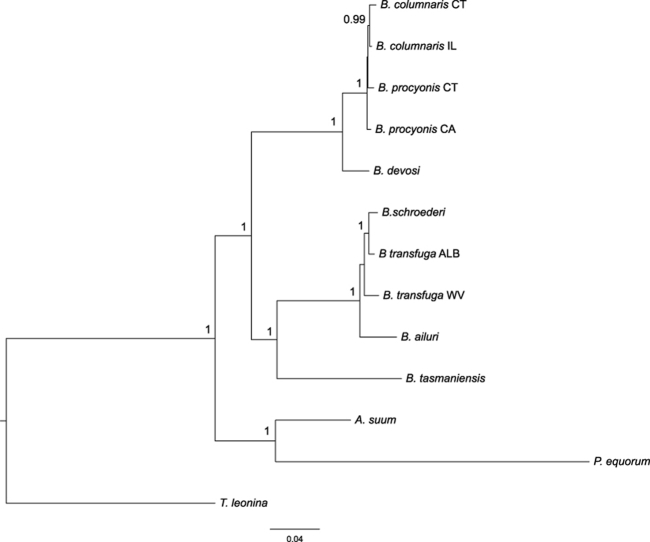
Fig. 1.
Bayesian consensus tree based on combined FULL data (8 genes; not including hars1). Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes are Bayesian posterior probabilities, shown when 0.90 and greater.
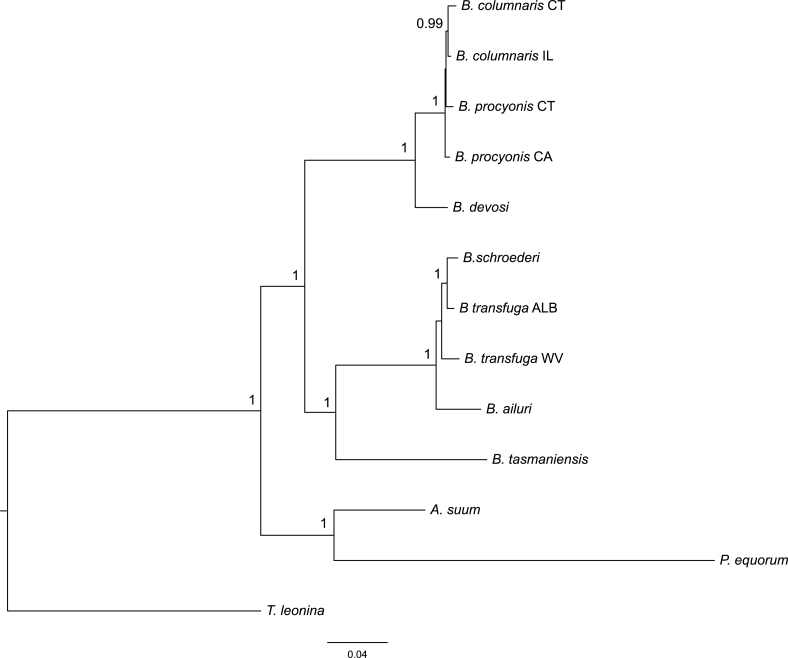
Fig. 2.
Bayesian consensus tree based on FULL data from nuclear genes (5 genes; not including hars1). Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes represent Bayesian posterior probabilities, shown when 0.90 and greater.
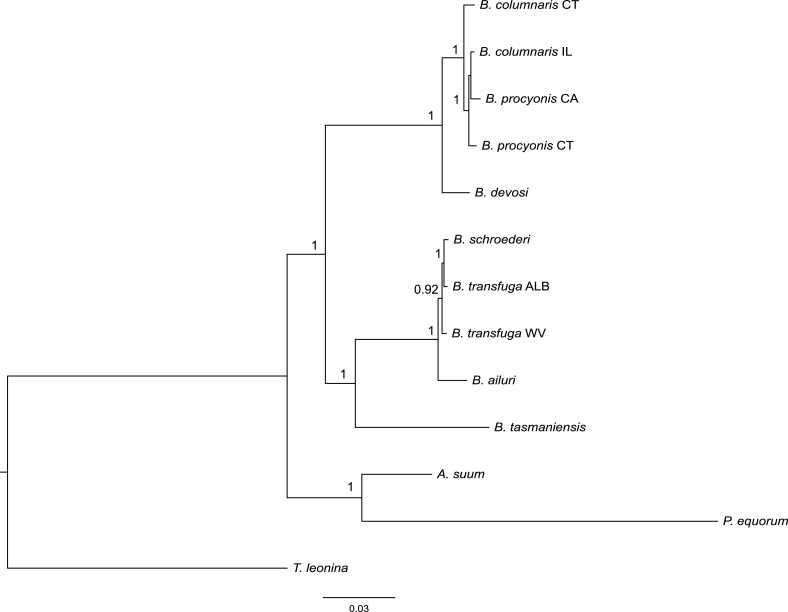
Fig. 3.
Bayesian consensus tree based on FULL mitochondrial gene sequences (3 genes). Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes represent Bayesian posterior probabilities, shown when 0.90 and greater.
3.2. Combined mitochondrial and nuclear genes
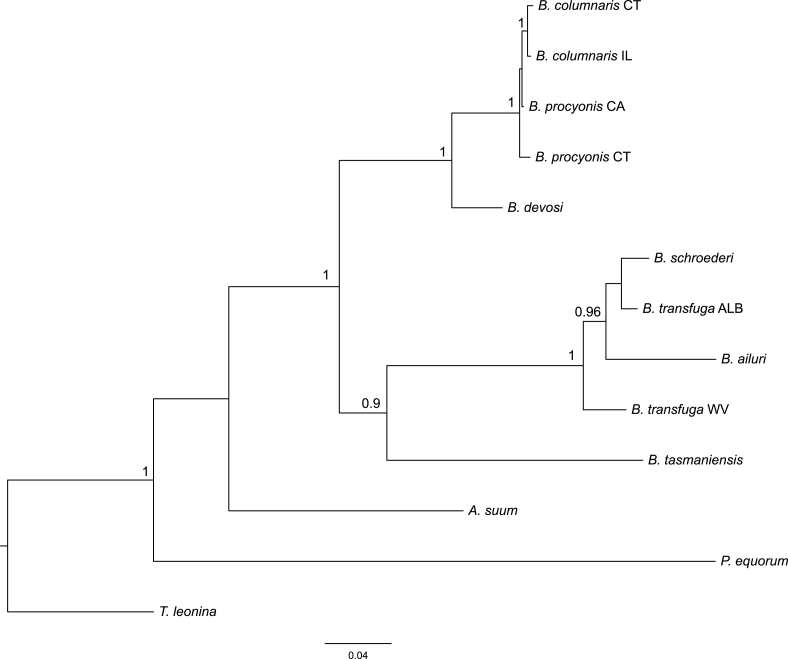
Two well-supported Baylisascaris clades were resolved: clade 1 including B. columnaris, B. procyonis, B. devosi; and clade 2 including B. ailuri, B. schroederi, B. transfuga, B. tasmaniensis. Both of these main clades were monophyletic for combined data with absolute BPP support for datasets with (Fig. 1) and without hars1 (Fig. S1). Analyses of combined data without hars1 resolved a clade of B. devosi and geographic isolates of B. columnaris and B. procyonis with absolute BPP support (clade 1, Fig. 1). Baylisascaris columnaris isolates were monophyletic for combined data without hars1 (Fig. 1), with BPP support of 0.99 (Fig. 1). Baylisascaris procyonis CT was resolved as sister to the B. columnaris group, but this relationship was not supported by BPP (<0.90). Reciprocal monophyly of B. columnaris and B. procyonis isolates was resolved for the dataset including hars1, but these clades were poorly supported by BPP (Fig. S1, BPP B. columnaris clade 0.64, BPP B. procyonis clade 0.63).
Within clade 2, which contained parasites from ursids (B. schroederi and the geographic isolates of B. transfuga) and the red panda (B. ailuri), B. schroederi and B. transfuga ALB were monophyletic with absolute BPP support (Fig. 1 and Fig. S1). Baylisascaris transfuga WV was sister to B. schroederi + B. transfuga ALB in trees with or without hars1, but this clade was not supported by BPP (Fig. 1 and Fig. S1).
Baylisascaris tasmaniensis was part of clade 2 including B. ailuri, B. schroederi, and both B. transfuga specimens based on combined data without hars1 (Fig. 1) with absolute BPP support. In the tree based on combined data including hars1, the two main Baylisascaris clades (clades 1 and 2) were sister groups (unsupported by BPP, Fig. S1), and B. tasmaniensis was sister to that clade. The seven species of Baylisascaris were resolved as a clade with absolute support for datasets with and without hars1.
3.3. Combined nuclear genes
Baylisascaris clades 1 and 2 were supported in trees inferred from combined nuclear genes with and without hars1 sequences (Fig. 2 and Fig. S2). Within clade 1, monophyly of B. devosi with geographic isolates of B. columnaris and B. procyonis had absolute BPP support without hars1 data (Fig. 2). Baylisascaris columnaris and B. procyonis isolates were resolved as a clade with absolute support without hars1 (Fig. 2) and 0.99 BPP support with hars1 (Fig. S2). Baylisascaris columnaris IL grouped with both B. procyonis specimens with absolute support without hars1 (Fig. 2) and BPP support of 0.97 with hars1 (Fig. S2). Baylisascaris procyonis CA and B. columnaris IL were resolved as sister taxa with and without hars1, but this relationship was only supported in the dataset with hars1 (BPP 0.93, Fig. S2).
In clade 2, Baylisascaris schroederi and B. transfuga ALB were monophyletic with absolute support in trees based on data with and without hars1 (Fig. 2 and Fig. S2). Baylisascaris transfuga WV was sister to (B. schroederi, B. transfuga ALB) in both trees with BPP support of 0.92 without hars1 (Fig. 2) and absolute support with hars1 (Fig. S2).
Baylisascaris tasmaniensis was resolved as part of clade 2 containing B. ailuri, B. schroederi, and the B. transfuga isolates for the dataset without hars1 sequences (Fig. 2); this relationship had absolute support. For the tree with hars1 sequences, B. tasmaniensis was sister to both Baylisascaris clades 1 and 2 (Fig. S2). Support for Baylisascaris monophyly was absolute in both trees.
3.4. Combined mitochondrial genes
Baylisascaris clades 1 and 2 had absolute support based on combined mitochondrial genes (Fig. 3). Monophyly of B. devosi and isolates of B. columnaris and B. procyonis had absolute BPP support. Within this clade, the B. columnaris specimens were monophyletic with absolute BPP support. Baylisascaris procyonis CA and the B. columnaris isolates were resolved as a group, but this relationship was not supported by BPP (Fig. 3).
Monophyly of B. ailuri, B. schroederi, and both B. transfuga isolates had absolute support (Fig. 3). Within clade 2, B. schroederi and B. transfuga ALB were monophyletic, but this relationship was not supported by BPP. Baylisascaris ailuri was sister to B. schroederi + B. transfuga ALB, and this relationship had BPP support of 0.96 (Fig. 3).
Baylisascaris tasmaniensis was part of a monophyletic clade 2 with B. ailuri, B. schroederi, and B. transfuga, and this relationship had BPP support of 0.90 (Fig. 3). Baylisascaris was monophyletic for combined mitochondrial genes with absolute BPP support (Fig. 3).
3.5. Single genes
Monophyly of selected clades based on parsimony and Bayesian analyses of single genes are shown in Tables S7 and S8. Baylisascaris clades 1 and 2 were present in all single gene trees, but bootstrap and BPP support varied by gene. Baylisascaris devosi and geographic isolates of B. columnaris and B. procyonis were monophyletic in all trees (except for trees with hars1, which do not include sequence data from B. devosi). This clade was not reliably supported for cox1 but received moderate to absolute support for all other genes. Isolates of B. columnaris and B. procyonis were monophyletic in all trees, and this clade had moderate to absolute support for all genes except cox2 and msp. Monophyly of B. columnaris isolates had absolute support for cox1 (Table S8).
Monophyly of B. ailuri, B. schroederi, and B. transfuga isolates (B. ailuri + Baylisascaris from ursid hosts in Tables S7 and S8) had absolute support for all individual nuclear genes, and high to absolute support for all individual mitochondrial genes. Monophyly of B. schroederi and B. transfuga ALB was supported by bootstrap for cox1 and cox2 (Table S7) and by BPP for 12S and 28S (Table S8). Parasites from ursids were monophyletic with varying support based on trees from four genes (Tables S7 and S8): 12S (bootstrap 95%, BPP 0.91), msp (bootstrap 76%, BPP 0.93), ard1 (bootstrap 85%, BPP 0.78), and hars1 (bootstrap 97%, BPP 0.95).
Major relationships did not change in single gene trees that incorporated published sequences of cox1, 28S, and ITS (compare Tables S7 and S8 with Figures S3-S5) though BPP values and the position of B. tasmaniensis varied. Clade 1 included B. potosis, a parasite of kinkajous. However, for cox1 and 28S, the analysis including B. potosis and additional B. devosi sequences from wolverines (cox1 only) and Kamchatka sables did not yield a monophyletic clade 1 (BPP <0.90). Clade 2 included B. venezuelensis (ITS analysis only) and was monophyletic for all three genes, but B. tasmaniensis was only part of clade 2 in the cox1 tree.
3.6. Alternative topology tests
Combined datasets were used to test alternative phylogenetic hypotheses of reciprocal monophyly for B. columnaris and B. procyonis isolates (Table S9) and of monophyly for B. transfuga ALB and B. transfuga WV (Table S10). Reciprocal monophyly of B. columnaris and B. procyonis isolates was not significantly worse (P-values >0.05) than the original topologies for mitochondrial genes, nuclear genes without hars1, or combined genes with hars1. However, reciprocal monophyly of B. columnaris and B. procyonis isolates was significantly worse than the original topologies for nuclear genes with hars1 and combined genes without hars1. For all datasets, monophyly of B. transfuga isolates was not significantly worse (P-values >0.05) than the original topology (monophyly of B. schroederi + B. transfuga ALB in all datasets).
3.7. Species delimitation of B. procyonis and B. columnaris using BP&P
The results of species delimitation using BP&P were dependent on the priors used for θ and τ0. Posterior probabilities (PP) for each species and the total number of species are summarized for each prior combination in Table 4. The strongest support for B. columnaris and B. procyonis as separate species occurred with a moderate population size (G θ (1.95, 300)) and a shallow divergence time (G τ0 (2, 2000)). At this prior combination, both species had PP support of 0.91, and B. transfuga had a PP of 1; the PP for three species was also 0.91. Priors for a large population size (G θ (1.95, 30)) and a shallow divergence time (G τ0 (2, 2000)) resolved lower support for B. procyonis and B. columnaris as separate species (0.72 for both). No other prior combinations supported B. columnaris and B. procyonis as separate species (Table 4).
Table 4.
Results from Bayesian Phylogenetics and Phylogeography analyses for each combination of priors. The posterior probability (PP) values are averages from two independent runs of module A11. Each species is represented by a single letter abbreviation corresponding to the species epithet, e.g. T is B. transfuga.
| Priors |
PP of each species |
PP for number of species |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| θ | τCPT | P [T] | P [C] | P [P] | P [CP] | P [CPT] | P [3] | P [2] | P [1] |
| G θ (1.95, 30) | G τCPT (2, 200) | 1 | 0.30 | 0.30 | 0.70 | 0 | 0.30 | 0.70 | 0 |
| G θ (1.95, 30) | G τCPT (2, 2000) | 0.93 | 0.72 | 0.72 | 0.22 | 0.04 | 0.70 | 0.26 | 0.039 |
| G θ (1.95, 30) | G τCPT (2, 20000) | 0.48 | 0.48 | 0.48 | 0.12 | 0.29 | 0.36 | 0.35 | 0.291 |
| G θ (1.95, 300) | G τCPT (2, 200) | 1 | 0.34 | 0.34 | 0.66 | 0 | 0.34 | 0.66 | 0 |
| G θ (1.95, 300) | G τCPT (2, 2000) | 1 | 0.91 | 0.91 | 0.09 | 0 | 0.91 | 0.09 | 0 |
| G θ (1.95, 300) | G τCPT (2, 20000) | 0.53 | 0.50 | 0.50 | 0.12 | 0.28 | 0.41 | 0.62 | 0.28 |
| G θ (1.95, 3000) | G τCPT (2, 200) | 1 | 0.02 | 0.02 | 0.98 | 0 | 0.02 | 0.98 | 0 |
| G θ (1.95, 3000) | G τCPT (2, 2000) | 0.03 | 0.01 | 0.01 | 0 | 0.98 | 0.01 | 0.01 | 0.98 |
| G θ (1.95, 3000) | G τCPT (2, 20000) | 0.15 | 0.15 | 0.15 | 0.03 | 0.80 | 0.12 | 0.08 | 0.80 |
4. Discussion
4.1. Phylogeny of Baylisascaris
In the present study, relationships among seven species of Baylisascaris were analyzed using multiple loci, including one species with no previously published sequence data – B. tasmaniensis. Monophyly of the genus Baylisascaris was supported in all trees based on combined sequence data (Fig. 1, Fig. 2, Fig. 3, Figs. S1, and S2) and in most single gene trees (Tables S7 and S8). Baylisascaris devosi was sister to isolates of B. columnaris and B. procyonis in all trees based on combined sequence data with very high or absolute support (Fig. 1, Fig. 2, Fig. 3). The inclusion of B. devosi in clade 1 is not unexpected given a previous analysis of rDNA data that resolved B. devosi and B. potosis as sister species (Tranbenkova and Spiridonov, 2017) and considering that Sprent (1968) originally grouped B. devosi with B. columnaris, B. procyonis, and B. laevis based on reduced size of cervical alae and overall body size. In addition, hosts of three of these five species (not B. laevis) are from the superfamily Musteloidea: B. potosis infects Potos flavus (kinkajou) and B. devosi infects Martes americana (American marten), Martes zibellina (sable), Pekania pennanti (fisher), and Gulo gulo (wolverine). Several of these B. devosi hosts co-occur in parts of the northern and western extents of raccoon (B. procyonis) and striped skunk (B. columnaris) ranges. In addition, analyses of previously published sequences (limited data, often single genes) for B. potosis and B. venezuelensis confirmed their placement in clade 1 and clade 2, respectively (Fig. S3-S5).
Baylisascaris tasmaniensis was also part of a monophyletic Baylisascaris, and although the position of this species was inconsistent in trees based on single genes (Tables S7 and S8), the combined analyses often resolved this species as part of clade 2, sister to the parasites from ursids and red panda (Fig. 1, Fig. 2, Fig. 3). This relationship had absolute BPP support for combined nuclear genes and the combined analysis of all genes, and lower support for mitochondrial genes (0.90), but this relationship was not obtained for datasets that included hars1 (Figs. S1, S2).
Limited research has been done on B. tasmaniensis since the work of Sprent (1970) and colleagues (Sprent et al., 1973). Sprent (1970) suggested that B. tasmaniensis was morphologically similar to B. melis of European badgers and B. transfuga of bears, because all three species have noticeable cervical alae. However, Sprent et al. (1973) also noted similarities between larvae of B. tasmaniensis and B. devosi in terms of development, behavior, and morphology. Sequence data from B. melis was not available, but sequences of B. devosi and two Baylisascaris species from bears were analyzed. Baylisascaris tasmaniensis was part of a monophyletic group containing bear parasites in eight of 10 trees based on combined sequence data with moderate to absolute support (Fig. 1, Fig. 2, Fig. 3). More complete sampling of Baylisascaris species has the potential to increase resolution in molecular phylogenetic trees, and for described species this would require adding B. melis and B. laevis. However, recent descriptions of two new species (Tokiwa et al., 2014; Pérez Mata et al., 2016) suggests that Baylisascaris biodiversity is incompletely known and requires additional investigation.
As Sprent et al. (1973) noted, it is not clear how B. tasmaniensis initially infected Tasmanian devils (Sarcophilus harrisii) and quolls (Dasyurus species) because the other definitive hosts of Baylisascaris spp. are arctoid carnivorans, which are not found in Australia or Tasmania. Sprent (1970) provided two potential explanations: 1) marsupials originally occurring in Australia had a phylogenetic relationship with arctoid carnivorans and ascaridoid nematodes were shared between these host groups; and 2) convergent evolution of ascaridoids, due to infecting hosts that occupy similar niches, led to the morphological similarity between B. tasmaniensis and other Baylisascaris species (Sprent, 1970). A recent phylogenetic analysis of mammals provides no support for a close phylogenetic relationship between arctoid carnivorans and marsupials (Tarver et al., 2016). Sprent's second hypothesis of convergent evolution from a non-Baylisascaris ascaridoid ancestor is contradicted by the inclusion of B. tasmaniensis as part of a monophyletic Baylisascaris in the present study. Sprent et al. (1973) also noted the possibility of B. transfuga occurring in bears at the far southern extent of South East Asia but did not directly connect this idea to the colonization of marsupial hosts in the Australian region. Sun bears (Helarctos malayanus) and the Asiatic black bear (Ursus thibetanus, syn. U. torquatus) occur in SE Asia, and both species have been recorded as hosts of Baylisascaris (Sprent, 1968). The potential colonization of marsupials by Baylisascaris from SE Asian bears would need to be explained relative to the apparent restricted host range of Baylisascaris species in Australian marsupials. Additional information on the phylogenetic relationships of Baylisascaris species in bears and B. tasmaniensis may be key to determining whether the origin of Baylisascaris in dasyurids was due to a host-colonization (switching) event.
According to Kazacos (2001, 2016), baylisascariasis in humans can also be caused by B. columnaris, B. melis, B. devosi, B. transfuga, and B. tasmaniensis. Baylisascaris procyonis is believed to be the primary cause of baylisascariasis in paratenic hosts and humans, but the lack of a clear, rapid molecular diagnostic test that can specifically identify B. procyonis means that we do not know the health risk of other Baylisascaris species. Clinical diagnosis of baylisascariasis is primarily based on serological tests, but these tests cannot discriminate among Baylisascaris species (Graeff-Teixeira et al., 2016). In such cases, it would be useful to diagnose specimens or potential environmental sources of Baylisascaris using a DNA sequence-based method. One approach is to use phylogenetic analysis of gene sequence data to place unknown Baylisascaris samples in an evolutionary tree (Hoberg et al., 2018). This approach does not depend upon predefined species-specific sequence signatures and can accommodate previously unknown variation in the sequences during the analysis. For example, phylogenetic analysis of the three mitochondrial genes used herein (12S, cox-1, cox-2) provides a well-resolved tree and sequencing these genes does not normally require cloning. In addition, size (electrophoretic) comparisons of ard1 amplicons can distinguish between taxa in the two main Baylisascaris clades as B. ailuri, B. schroederi, and both B. transfuga isolates have a continuous 222 bp gap in ard-1 that is not present in B. columnaris, B. procyonis, B. devosi, or B. tasmaniensis.
4.2. Discrimination of raccoon and skunk Baylisascaris
Baylisascaris procyonis and B. columnaris are believed to be closely related, but distinct, species. However, discriminating between B. procyonis and B. columnaris is difficult. Morphologically, they are almost indistinguishable, and identifying the species of larval or adult B. procyonis or B. columnaris based on morphology alone is not always possible even for specialists (Kazacos, 2001; Graeff-Teixeira et al., 2016). Previous authors have also attempted to discriminate between B. columnaris and B. procyonis based on pathogenicity in paratenic hosts, protein electrophoresis (Berry, 1985), larval excretory-secretory antigens (Dangoudoubiyam et al., 2010), and DNA sequence data (Dangoudoubiyam et al., 2009; Gatcombe et al., 2010; Franssen et al., 2013; Choi et al., 2017). The most pronounced difference between skunk- and raccoon-derived worms is in pathogenicity for paratenic hosts. Clinical baylisascariasis can occur when paratenic hosts ingest infective eggs of either B. procyonis or B. columnaris, but fewer B. procyonis eggs are needed to cause disease (Kazacos, 2001).
Morphological characters proposed to discriminate between B. columnaris and B. procyonis include shape of the tail tip in males, rough areas near the cloaca in males, and lip denticle shape. Subsets of these characters have recently been used in research focused on distinguishing B. columnaris from B. procyonis (e.g. Franssen et al., 2013) or describing new species (B. potosis, Tokiwa et al., 2014). However, when Berry (1985) analyzed these characters for multiple individuals of B. columnaris, B. procyonis, and B. laevis, he determined that they were too variable to be useful for discrimination. Therefore, most contemporary studies have turned to molecular data in attempts to discriminate between B. procyonis and B. columnaris.
Phylogenetic analyses based on molecular data have consistently resolved skunk- and raccoon-derived Baylisascaris specimens as members of the same clade (e.g., Tranbenkova and Spiridonov, 2017) but they have been unclear with regard to further delimiting B. procyonis and B. columnaris. The inclusion of variable nuclear genes (msp, ard1, and hars1) in the current phylogenetic analysis resolved reciprocal monophyly of specimens derived from raccoon and skunk hosts in a single tree: the BI tree for combined genes including hars1 (Fig. S1). However, both clades lacked strong support (BPP 0.64 for B. columnaris specimens and 0.63 for B. procyonis specimens). In some analyses, the B. columnaris specimens were monophyletic, but the B. procyonis specimens were not (cox1, cox2, mitochondrial genes, and certain combined data, Tables S7 and S8, Fig. 1, Fig. 3). In other cases, there was either no resolution or the resolved clades included individuals of both species (Tables S7 and S8, Fig. 2 and Fig. S2). The alternative topology depicting reciprocal monophyly for B. columnaris and B. procyonis was significantly worse for two datasets (nuclear genes with hars1, and combined data without hars1) but was not significantly worse than the original topologies for the other three datasets (Table S9).
In order to more fully utilize the data collected from multiple loci, we employed a multispecies coalescent approach using the program BP&P (Yang, 2015) to test the hypothesis that B. procyonis and B. columnaris are separate species. Given a prior of three species (skunk-derived worms, raccoon-derived worms, and B. transfuga ALB as the test taxa), support for B. columnaris and B. procyonis as separate species was resolved by two combinations of θ and τ0 priors that we tested. These priors correspond to a shallow divergence time with either a large or moderate effective population size (Table 4). With large population size and shallow divergence time, support for B. procyonis and B. columnaris as separate species was PP 0.72, and support for B. transfuga ALB as a species was higher (PP 0.93; Table 4). With priors for a moderate population size and shallow divergence, support for separate species was stronger (PP 0.91), and support for B. transfuga ALB was absolute (Table 4). Effective population sizes for Baylisascaris species are probably similar to Ascaris in that they are large, but smaller than trichostrongylids in domesticated ruminants (Anderson and Jaenike, 1997), which tend to have thousands of worms per host. Raccoons have been documented to have hundreds of worms in their small intestines (Kazacos, 2001, 2016; Weinstein, 2016), although average infrapopulations are smaller. For the divergence time prior, it is unlikely that the split between B. procyonis and B. columnaris was deep given their low sequence divergence across genes included in this and other studies. Prior combinations corresponding to a shallow divergence were the most reasonable based on the posterior distributions obtained using the BP&P program (module A00). Future analyses using BP&P would benefit from including more nuclear loci and additional geographic isolates and individuals of skunk- and raccoon-derived worms.
Recent attempts at using molecular data to distinguish B. columnaris and B. procyonis were conducted by Franssen et al. (2013) and Choi et al. (2017). Franssen et al. (2013) analyzed specimens obtained from skunk hosts in the Netherlands, and from raccoon hosts in Indiana, USA and Norway, and reported a GA repeat region in ITS-2 with unique patterns for individuals of B. procyonis (nine repeats) and B. columnaris (six or seven repeats). In addition, the authors reported 14 single nucleotide polymorphisms (SNPs) among several genes. Five of these SNPs were reported to be identical in isolates from skunk hosts, but different in isolates from raccoons: three SNPs in cox1 and one SNP each in cox2 and ITS-1. These comparisons were based on 12 B. procyonis and 15 B. columnaris for ITS-1; 19 B. procyonis and 43 B. columnaris for cox1; and 10 B. procyonis and 38 B. columnaris for cox2.
Choi et al. (2017) obtained mitochondrial sequence data for 10 skunk-derived Baylisascaris in Salt Lake County, Utah, USA. They compared complete sequences of 11 mitochondrial genes – cox1, cox2, nd2, and 8 tRNA genes – with mitochondrial genome sequences of one B. procyonis specimen (Xie et al., 2011b) and with cox1 and cox2 sequences obtained by Franssen et al. (2013). In total, Choi et al. (2017) reported 11 SNPs that were presumed diagnostic for skunk- and raccoon-derived Baylisascaris: six SNPs in cox1; three SNPs in nd2; one SNP in tRNA-Leu; and one SNP in tRNA-Ser. In contrast to Franssen et al. (2013) no diagnostic SNPs were identified in cox2. The SNPs identified by Choi et al. (2017) were based on one B. procyonis and 34 B. columnaris for cox1; one B. procyonis and 9 B. columnaris for cox2; and one B. procyonis and 10 B. columnaris for nd2 and the tRNA genes.
In order to assess the reliability of putatively diagnostic SNPs reported by Franssen et al. (2013) and Choi et al. (2017), we compared their sequences with cox1, cox2, ITS-1, and ITS-2 sequences from additional isolates of raccoon- and skunk-derived worms generated in our lab. For ITS, cox1, and cox2, we compared additional sequences of B. columnaris (two, six, and two, individuals, respectively) and B. procyonis (two, 44, and two, respectively). We did not have additional sequences for nd2 or tRNA and could not further assess SNPs in those genes (Choi et al., 2017).
When more individuals from the United States were compared (based on RFLP screening of individuals followed by confirmation by sequencing) with the individuals sequenced by Franssen et al. (2013) and Choi et al. (2017), intraindividual polymorphism at the SNP from ITS-1 (position 201 from Franssen et al., Table 4 – T in B. columnaris and C in B. procyonis) was revealed. The pattern of GA repeats in ITS-2 was the same in our specimens as in those of Franssen and colleagues (six for B. columnaris isolates and nine for B. procyonis isolates, Table 5). For cox1 SNPs, none of those identified as diagnostic by Franssen et al. (2013) were unique to skunk- or raccoon-derived worms. Choi and colleagues identified five additional putatively diagnostic SNPs in full-length cox1 (Fig. 1, Choi et al. (2017)) at the following positions: 231; 1266; 1315; 1491; and 1506. Partial cox1 sequences from skunk- and raccoon-derived worms generated in our lab were used to assess cox1 SNPs at positions 1266 and 1315 of Choi et al. (2017), but SNPs at other positions could not be assessed due to lack of sequence overlap. The SNPs at positions 1266 and 1315 of cox1 were not specific for skunk- and raccoon-derived worms. For the cox2 SNP, B. procyonis CA had the same sequence as the skunk isolates sequenced by Franssen et al. (position 66 from Table 3 in Franssen et al.). Considering all three studies, these comparisons are based on 14 B. procyonis and 17 B. columnaris for full length ITS, 13 B. procyonis and 49 B. columnaris for cox2, and 64 B. procyonis and 83 B. columnaris for cox1. Further testing of the ITS-2 GA repeat diagnostic region is needed, however, this is made more difficult by the need to clone PCR products to obtain high quality sequence from this region. These results emphasize the importance of having sufficient sample sizes and appropriate sampling from all hosts and geographic regions for testing SNPs as species diagnostic markers. For example, in comparison to Baylisascaris species, Ascaris from humans and pigs have been broadly geographically sampled for comparative genetics (Betson et al., 2013), and geographic variation in the cox1 gene of Ascaris (e.g., Betson et al., 2011) has revealed more than 50 haplotypes.
Table 5.
Repeat pattern for ITS-2 in Baylisascaris species. For B. columnaris, B. procyonis, B. transfuga and B. schroederi, the region included corresponds to the highlighted region in Fig. 2 from Franssen et al. (2013).
| Species | Repeat pattern |
|---|---|
| B. columnaris isolates | GAGAGAGAGAGAGAAAGAGAAA |
| B. procyonis isolates | GAGAGAGAGAGAGAGAGAGAAAGAGAAA |
| B. devosi | CAGAGAGAGAGAGAGAGAAAGAGAAAGAAAGAAAGAGAAAGAA |
| B. ailuri | GAGGGAGAGA |
| B. schroederi | GAGAAGAGA |
| B. transfuga isolates | GAGAAGAGA |
| B. tasmaniensis | GAGAGAGAAAA |
One potential problem with using only molecular methods to distinguish B. columnaris and B. procyonis is the possibility that these species lack strict host specificity for skunks and raccoons. Nadler (2010, unpublished) used comparisons of ITS sequences for skunk- and raccoon-derived worms to develop a restriction fragment length polymorphism test based on a SNP difference in their ITS-1 sequences; this is the same SNP reported by Franssen et al. (2013). The restriction enzyme Apo I recognizes the sequence 5′-RAATTY; at the SNP site, sequences derived from raccoon Baylisascaris have the sequence AAACTT, whereas sequences from skunks have AAATTT. Raccoon Baylisascaris ITS lacks the Apo I recognition site relative to skunk Baylisascaris specimens. Using this RFLP test, 148 individuals from raccoons were assignable to B. procyonis, 34 individuals from skunks were assignable to B. columnaris, but 12 individuals from raccoon hosts had the polymorphic pattern, and a single individual from a raccoon was assignable to B. columnaris. The polymorphic ITS pattern found in some raccoon Baylisascaris could reflect an ancestral polymorphism maintained in B. procyonis rDNA despite concerted evolution of multicopy rDNA, or perhaps even past hybridization event(s) with subsequent backcrosses to B. procyonis individuals (Camp et al., 2011). These polymorphic individuals call into question the species-specific diagnostic value of this ITS-1 SNP.
4.3. Baylisascaris in bears
The first Baylisascaris species was described by Rudolphi in 1819 as Ascaris transfuga. Rudolphi's description was based on specimens from polar bear (Ursus maritimus) and brown bear (U. arctos). When Sprent (1968) created Baylisascaris, he redescribed A. transfuga, and reported this parasite from six species in Ursinae – U. maritimus, U. arctos, U. thibetanus (syn. U. torquatus), U. americanus, Helarctos malayanus, and Melursus ursinus. Ascarids from the six ursines (and their numerous subspecies) are typically assumed to be B. transfuga. Spectacled bears (Tremarctos ornatus, Tremarctinae) and giant panda (Ailuropoda melanoleuca, Ailuropodinae) were not part of this host list for B. transfuga. Giant pandas are consistently resolved as outside the clade containing other Ursidae (Yu et al., 2007; Pagès et al., 2008; Nyakatura and Bininda-Emonds, 2012; Kutschera et al., 2014), and are hosts for a distinct Baylisascaris species (B. schroederi). Spectacled bears are also commonly resolved as sister to the six ursine species and a new Baylisascaris species specific to T. ornatus was recently described (B. venezuelensis; Pérez Mata et al., 2016).
Specimens of Baylisascaris from these six bear species, or their subspecies, have been included in recent molecular phylogenies (Li et al., 2012; Franssen et al., 2013; He et al., 2013; Tokiwa et al., 2014; Pérez Mata et al., 2016). Importantly, the distributions of these bear species and their subspecies do not usually overlap. For a variety of genes and inference methods, Baylisascaris specimens from ursine hosts are monophyletic, but with clade support ranging from absent (Pérez Mata et al., 2016) to high (Li et al., 2012; He et al., 2013; Tokiwa et al., 2014). Based on mitochondrial ND1, He et al. (2013) concluded that Baylisascaris from polar bear, an Asian black bear subspecies (U. t. mupinensis), and two subspecies of the brown bear (Ussuri and Tibetan blue) were all B. transfuga. These analyses provide a start to assessing species validity of Baylisascaris from bears, but none of them explicitly tested the hypothesis of multiple species (including cryptic species) of Baylisascaris in ursine hosts. The historical biogeography of certain bear species is consistent with the isolation and allopatric speciation of their nematode parasites (Catalano et al., 2015). For example, the hookworm species Uncinaria rauschi and U. yukonensis are believed to have originated, respectively, in the Nearctic (with black bears) and Palearctic (with grizzly bears), during millions of years of geographic isolation of their hosts (Rausch et al., 1979). Such biogeographic histories could likewise influence the speciation of Baylisascaris in bears.
Our analysis included B. transfuga from two North American bear species – grizzly bear (U. arctos horribilis, Alberta, Canada) and American black bear (U. americanus, West Virginia, USA). Unlike previous studies, specimens of B. transfuga from these hosts were not monophyletic in any trees based on single genes or combined sequence data. Pairwise distance values between B. transfuga ALB and B. transfuga WV were higher than values between B. transfuga ALB and B. schroederi for all genes except msp and ard1 (Tables S4 and S6). Baylisascaris transfuga ALB was usually sister to B. schroederi, which suggests that the Baylisascaris specimens from grizzly and black bears may be different species. However, the alternative topology test depicting monophyly of B. transfuga isolates was not significantly worse than monophyly of B. schroederi and B. transfuga ALB for any combined dataset (Table S10). These results indicate that more research to test the hypothesis of multiple species of Baylisascaris from these and other bears is warranted. When choosing a sample size, investigators should consider the possibility that a single host species might be infected with multiple cryptic species of Baylisascaris, and sample from wild hosts throughout their ranges, rather than captive or zoo-kept hosts, which could host aberrant species.
4.4. Relationships of Baylisascaris species with their hosts in Arctoidea (Carnivora)
Nine Baylisascaris species infect arctoid carnivorans as definitive hosts – B. transfuga, B. columnaris, B. melis, B. schroederi, B. procyonis, B. devosi, B. ailuri, B. potosis, and B. venezuelensis. Six of these species were included in the multigene phylogenetic analyses conducted herein, which resolved two strongly supported Baylisascaris clades, consistent with previous analyses. Additional single gene analyses included sequences from B. potosis (cox1, 28S, and ITS2; Tokiwa et al., 2014) and B. venezuelensis (ITS; Pérez Mata et al., 2016) as well as additional sequences of B. devosi (cox1, Nemeth and Tannis, 2014; cox1, 28S, and ITS, Tranbenkova and Spiridonov, 2017). Clade 1 includes parasites from musteloid hosts native to North America, Central America, and Eurasia (B. columnaris, B. procyonis, and B. devosi), and one species (Fig. S1-3, B. potosis) from a musteloid host (kinkajou) occurring in both North and South America. Baylisascaris ailuri, a member of clade 2, also infects a musteloid, the red panda (Ailurus fulgens), but this host species is native to China. Baylisascaris ailuri was part of Baylisascaris clade 2 that includes parasites from ursid hosts. Support for monophyly of B. ailuri + ursid Baylisascaris spp. was strong among datasets (Fig. 1, Fig. 2, Fig. 3, Figs. S1, and S2, Tables S7 and S8). Previous phylogenetic analyses have not provided strong support for relationships between B. ailuri and other ursid Baylisascaris spp. (Xie et al., 2011a; Franssen et al., 2013; Tokiwa et al., 2014). Cophylogeny would predict monophyly of B. ailuri and the other Baylisascaris species from musteloid hosts, but this is falsified by the phylogeny. The red panda range in China has overlapped that of two ursid Baylisascaris hosts – A. melanoleuca (giant panda) and Ursus thibetanus (Asian black bear) – and is close to the range of Melursus ursinus (sloth bear) (IUCN, 2016). Biogeography and host colonization (switching) have likely both been important factors in determining host-parasite associations in this assemblage. The general absence of cospeciation suggests important roles for factors such as ecological fitting, taxon pulses, and oscillation (Hoberg and Brooks, 2008, 2010) in determining host-parasite associations of Baylisascaris species.
In the clade of Baylisascaris from North American musteloids, cophylogeny would predict a sister group relationship between B. procyonis from raccoons and B. devosi from fishers. Instead, B. devosi is sister to B. potosis (Tranbenkova and Spiridonov, 2017) and B. procyonis and B. columnaris are strongly supported as a clade in phylogenetic analyses. Interpretation of this result depends on the status of B. procyonis and B. columnaris as separate species. Biogeography and host colonization may also play a role in the close relationship of these species. For example, raccoons and skunks currently overlap for most of their North American distributions but overlap less with hosts of B. devosi (fishers, martens, and wolverines) (IUCN, 2016).
Conflicts of interest
The authors have no conflicts of interest to report.
Acknowledgements
This research was supported by National Science Foundation grants DEB-0228692 and DEB-0731516 to S. Nadler. Host animals were collected and necropsied in accordance with the guidelines of animal ethics committees at cooperating institutions. We thank Ian Beveridge for providing specimens of B. tasmaniensis. We also thank Lance Truong and Stefanie Frank for assistance with laboratory work.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijppaw.2018.09.010.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Bayesian consensus tree based on FULL combined data including hars1 sequences (9 genes). Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes represent Bayesian posterior probabilities, shown when 0.90 and greater.
Bayesian consensus tree based on FULL nuclear data including hars1 sequences (6 genes). Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes represent Bayesian posterior probabilities, shown when 0.90 and greater.
Bayesian consensus tree for cox1 including previously published sequences of Baylisascaris. Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes are Bayesian posterior probabilities, shown when 0.90 and greater.
Bayesian consensus tree for 28S including previously published sequences of Baylisascaris. Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes are Bayesian posterior probabilities, shown when 0.90 and greater.
Bayesian consensus tree for ITS including previously published sequences of Baylisascaris. Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes are Bayesian posterior probabilities, shown when 0.90 and greater.
References
- Anderson T., Jaenike J. Host specificity, evolutionary relationships and macrogeographic differentiation among Ascaris populations from humans and pigs. Parasitology. 1997;115:325–342. doi: 10.1017/s0031182097001339. [DOI] [PubMed] [Google Scholar]
- Berry J.F. University of Guelph; 1985. Phylogenetic Relationship between Baylisascaris Spp. Sprent, 1968 (Nematoda: Ascarididae) from Skunks, Raccoons and Groundhogs in Southern Ontario; p. 86. [Google Scholar]
- Betson M., Nejsum P., Stothard J.R. Elsevier; 2013. From the Twig Tips to the Deeper Branches: New Insights into Evolutionary History and Phylogeography of Ascaris, Ascaris: the Neglected Parasite; pp. 265–285. [Google Scholar]
- Betson M., Halstead F.D., Nejsum P., Imison E., Khamis I.S., Sousa-Figueiredo J.C., Rollinson D., Stothard J.R. A molecular epidemiological investigation of Ascaris on Unguja, Zanzibar using isoenyzme analysis, DNA barcoding and microsatellite DNA profiling. Trans. R. Soc. Trop. Med. Hyg. 2011;105:370–379. doi: 10.1016/j.trstmh.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Camp L.E., Pagan C., Nadler S.A. American Society of Parasitologists 86th Annual Meeting; Anchorage, AK: 2011. Diagnosis of Baylisascaris Hybrid Individuals. [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Catalano S., Lejeune M., van Paridon B., Pagan C.A., Wasmuth J.D., Tizzani P., Duignan P.J., Nadler S.A. Morphological variability and molecular identification of Uncinaria spp. (Nematoda: ancylostomatidae) from grizzly and black bears: new species or phenotypic plasticity? J. Parasitol. 2015;101:182–192. doi: 10.1645/14-621.1. [DOI] [PubMed] [Google Scholar]
- Caviedes-Solis I.W., Bouzid N.M., Banbury B.L., Leaché A.D. Uprooting phylogenetic uncertainty in coalescent species delimitation: a meta-analysis of empirical studies. Current Zoology. 2015;61:866–873. [Google Scholar]
- Choi Y., Mason S., Ahlborn M., Zscheile B., Wilson E. Partial molecular characterization of the mitochondrial genome of Baylisascaris columnaris and prevalence of infection in a wild population of Striped skunks. Int. J. Parasitol. Parasites and Wildl. 2017;6:70–75. doi: 10.1016/j.ijppaw.2017.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangoudoubiyam S., Vemulapalli R., Kazacos K.R. PCR assays for detections of Baylisascaris procyonis eggs and larvae. J. Parasitol. 2009;95:571–577. doi: 10.1645/GE-1905.1. [DOI] [PubMed] [Google Scholar]
- Dangoudoubiyam S., Vemulapalli R., Hancock K., Kazacos K.R. Molecular cloning of an immunogenic protein of Baylisascaris procyonis and expression in Escherichia coli for use in developing improved serodiagnostic assays. Clin. Vaccine Immunol. 2010;17:1933–1939. doi: 10.1128/CVI.00404-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R.K., Øines Ø., Hamnes I.S., Schulze J.E. Illegal wildlife imports more than just animals-Baylisascaris procyonis in raccoons (Procyon lotor) in Norway. J. Wildl. Dis. 2013;49:986–990. doi: 10.7589/2012-06-154. [DOI] [PubMed] [Google Scholar]
- De Ambrogi M., Aghazadeh M., Hermosilla C., Huber D., Majnaric D., Reljic S., Elson-Riggins J. Occurrence of Baylisascaris transfuga in wild populations of European brown bears (Ursus arctos) as identified by a new PCR method. Vet. Parasitol. 2011;179:272–276. doi: 10.1016/j.vetpar.2011.02.025. [DOI] [PubMed] [Google Scholar]
- Farris J.S., Källersjö M., Kluge A.G., Bult C. Testing significance of incongruence. Cladistics. 1994;10:315–319. [Google Scholar]
- Farris J.S., Källersjö M., Kluge A.G., Bult C. Constructing a significance test for incongruence. Syst. Biol. 1995;44:570–572. [Google Scholar]
- Franssen F., Xie K., Sprong H., van der Giessen J. Molecular analysis of Baylisascaris columnaris revealed mitochondrial and nuclear polymorphisms. Parasites Vectors. 2013;6:1. doi: 10.1186/1756-3305-6-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R., Stevenson L., Chilton N., Nansen P., Bucknell D., Beveridge I. Species markers for equine strongyles detected in intergenic rDNA by PCR-RFLP. Mol. Cell. Probes. 1996;10:371–378. doi: 10.1006/mcpr.1996.0050. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., Chilton N.B., Hoste H., Beveridge I. Rapid sequencing of rDNA from single worms and eggs of parasitic helminths. Nucleic Acids Res. 1993;21:2525. doi: 10.1093/nar/21.10.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatcombe R.R., Jothikumar N., Dangoudoubiyam S., Kazacos K.R., Hill V.R. Evaluation of a molecular beacon real-time PCR assay for detection of Baylisascaris procyonis in different soil types and water samples. Parasitol. Res. 2010;106:499–504. doi: 10.1007/s00436-009-1692-6. [DOI] [PubMed] [Google Scholar]
- Gedoelst L. Sur une espece nouvelle d'Ascaride, parasite du blaireau. CR Soc. Biol. 1920;83:1291–1292. [Google Scholar]
- Graeff-Teixeira C., Morassutti A.L., Kazacos K.R. Update on baylisascariasis, a highly pathogenic zoonotic infection. Clin. Microbiol. Rev. 2016;29:375–399. doi: 10.1128/CMR.00044-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Niu L., Wang T., Wang Q., Chen S., Yan Y., Zhang Z., Yu H., Deng J., Wang S. 2013. Molecular Phylogenetic Studies on Ascarid Nematodes from Ailuropoda Melanoleuca and Seven Other Species of Captive Wild Mammals Based on ND1 Genes. [Google Scholar]
- He G.Z., Niu L.L., Yang G.Y., Deng J.P., Wang S., Yu X.M., Wang T., Gu X.B., Chen W.G. Sequence analysis of ITS-2 rDNA of roundworms from Ailuropoda melanoleuca and seven rare wild animals. Chin. J. Vet. Sci. 2008;11:933–938. [Google Scholar]
- Hoberg E.P., Brooks D.R. A macroevolutionary mosaic: episodic host-switching, geographical colonization and diversification in complex host–parasite systems. J. Biogeogr. 2008;35:1533–1550. [Google Scholar]
- Hoberg E.P., Brooks D.R. 2010. Beyond Vicariance: Integrating Taxon Pulses, Ecological Fitting, and Oscillation in Evolution and Historical Biogeography, the Biogeography of Host-parasite Interactions; pp. 7–20. [Google Scholar]
- Hoberg E.P., Burek-Huntington K., Beckmen K., Camp L.E., Nadler S.A. Transuterine infection by Baylisascaris transfuga: neurological migration and fatal debilitation in sibling moose calves (Alces alces gigas) from Alaska. Int. J. Parasitol.: Parasites Wildlife. 2018;7:280–288. doi: 10.1016/j.ijppaw.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN . 2016. The IUCN Red List of Threatened Species. Version 2016-1. [Google Scholar]
- Kazacos K.R. Baylisascaris procyonis and related species. In: Samuel W.M., Pybus M.J., Kocan A.A., editors. Parasitic Diseases of Wild Mammals. second ed. Iowa State University Press; Ames, Iowa: 2001. pp. 301–341. [Google Scholar]
- Kazacos K.R. In: Abbott R.C., Van Riper C. III, editors. vol. 1412. U.S. Geological Survey Circular; 2016. p. 122. (Baylisascaris Larva Migrans). Reston, vol. A. [Google Scholar]
- Kutschera V.E., Bidon T., Hailer F., Rodi J.L., Fain S.R., Janke A. Bears in a forest of gene trees: phylogenetic inference is complicated by incomplete lineage sorting and gene flow. Mol. Biol. Evol. 2014;31:2004–2017. doi: 10.1093/molbev/msu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R., Calcott B., Ho S.Y., Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., Thompson J.D. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leidy J. A synopsis of entozoa and some of their ectocongeners observed by the author. Proc. Acad. Natl. Sci. Phila. 1856;8:42–58. [Google Scholar]
- Lessa E.P. Rapid surveying of DNA sequence variation in natural populations. Mol. Biol. Evol. 1992;9:323–330. doi: 10.1093/oxfordjournals.molbev.a040723. [DOI] [PubMed] [Google Scholar]
- Li Y., Niu L., Wang Q., Zhang Z., Chen Z., Gu X., Xie Y., Yan N., Wang S., Peng X. Molecular characterization and phylogenetic analysis of ascarid nematodes from twenty-one species of captive wild mammals based on mitochondrial and nuclear sequences. Parasitology. 2012;139:1329–1338. doi: 10.1017/S003118201200056X. [DOI] [PubMed] [Google Scholar]
- Lin Q., Li H.M., Gao M., Wang X.Y., Ren W.X., Cong M.M., Tan X.C., Chen C.X., Yu S.K., Zhao G.H. Characterization of Baylisascaris schroederi from Qinling subspecies of giant panda in China by the first internal transcribed spacer (ITS-1) of nuclear ribosomal DNA. Parasitol. Res. 2012;110:1297–1303. doi: 10.1007/s00436-011-2618-7. [DOI] [PubMed] [Google Scholar]
- Liu G.-H., Zhou D.-H., Zhao L., Xiong R.-C., Liang J.-Y., Zhu X.-Q. The complete mitochondrial genome of Toxascaris leonina: comparison with other closely related species and phylogenetic implications. Infect. Genet. Evol. 2014;21:329–333. doi: 10.1016/j.meegid.2013.11.022. [DOI] [PubMed] [Google Scholar]
- Löytynoja A., Milinkovitch M.C. A hidden Markov model for progressive multiple alignment. Bioinformatics. 2003;19:1505–1513. doi: 10.1093/bioinformatics/btg193. [DOI] [PubMed] [Google Scholar]
- McIntosh A. A new nematode, Ascaris schroederi, from a giant panda, Ailuropoda melanoleuca. Zoologica. 1939;24:355–357. [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. Gateway Computing Environments Workshop (GCE), 2010. IEEE; 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Nadler S.A. Phylogeny of some ascaridoid nematodes, inferred from comparison of 18S and 28S ribosomal-RNA sequences. Mol. Biol. Evol. 1992;9:932–944. doi: 10.1093/oxfordjournals.molbev.a040768. [DOI] [PubMed] [Google Scholar]
- Nadler S.A., Bolotin E., Stock S.P. Phylogenetic relationships of Steinernema Travassos, 1927 (Nematoda : cephalobina : Steinernematidae) based on nuclear, mitochondrial and morphological data. Syst. Parasitol. 2006;63:161–181. doi: 10.1007/s11230-005-9009-3. [DOI] [PubMed] [Google Scholar]
- Nadler S.A., Hudspeth D.S.S. Ribosomal DNA and phylogeny of the ascaridoidea (nemata : secernentea): implications for morphological evolution and classification. Mol. Phylogenet. Evol. 1998;10:221–236. doi: 10.1006/mpev.1998.0514. [DOI] [PubMed] [Google Scholar]
- Nadler S.A., Hudspeth D.S.S. Phylogeny of the ascaridoidea (Nematoda : Ascaridida) based on three genes and morphology: hypotheses of structural and sequence evolution. J. Parasitol. 2000;86:380–393. doi: 10.1645/0022-3395(2000)086[0380:POTANA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Nemeth C., Tannis M. The Identification of Baylisascaris Devosi Parasite. 2014. Wacky work on worms in wolverines.http://words.usask.ca/wcvm/files/2014/07/wolverine-research-poster.pdf Retrieved August 2018 from. [Google Scholar]
- Nyakatura K., Bininda-Emonds O.R. Updating the evolutionary history of Carnivora (Mammalia): a new species-level supertree complete with divergence time estimates. BMC Biol. 2012;10:1. doi: 10.1186/1741-7007-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylander J.A.A. Evolutionary Biology Centre. Uppsala University; 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- Pagès M., Calvignac S., Klein C., Paris M., Hughes S., Hänni C. Combined analysis of fourteen nuclear genes refines the Ursidae phylogeny. Mol. Phylogenet. Evol. 2008;47:73–83. doi: 10.1016/j.ympev.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Palumbi S.R., Baker C.S. Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. Mol. Biol. Evol. 1994;11:426–435. doi: 10.1093/oxfordjournals.molbev.a040115. [DOI] [PubMed] [Google Scholar]
- Pérez Mata A., García Pérez H., Gauta Parra J. Morphological and molecular description of Baylisascaris venezuelensis, n. sp. from a natural infection in the South American spectacled bear Tremarctos ornatus Cuvier, 1825 in Venezuela. Neotropical Helminthol. 2016;10:85–103. [Google Scholar]
- Rambaut A., Suchard M., Drummond A. Tracer v1.6, available from rausch, R., krechmar, A., rausch, V., 1979. New records of helminths from the brown bear, Ursus arctos L., in the soviet far East. Can. J. Zool. 2014;57:1238–1243. http://beast.bio.ed.ac.uk/Tracer [Google Scholar]
- Rausch R., Krechmar A., Rausch V. New records of helminths from the brown bear, Ursus arctos L., in the soviet far East. Can. J. Zool. 1979;57:1238–1243. [Google Scholar]
- Regier J.C., Shultz J.W., Ganley A.R., Hussey A., Shi D., Ball B., Zwick A., Stajich J.E., Cummings M.P., Martin J.W. Resolving arthropod phylogeny: exploring phylogenetic signal within 41 kb of protein-coding nuclear gene sequence. Syst. Biol. 2008;57:920–938. doi: 10.1080/10635150802570791. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolphi C.A. Rücker; 1819. Entozoorum synopsis, cui accedunt mantissa duplex et indices locupletissimi. Cum tab. III aeneis. [Google Scholar]
- Shafir S.C., Sorvillo F.J., Sorvillo T., Eberhard M.L. Viability of Baylisascaris procyonis eggs. Emerg. Infect. Dis. 2011;17:1293–1295. doi: 10.3201/eid1707.101774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slade R., Moritz C., Heideman A., Hale P. Rapid assessment of single-copy nuclear DNA variation in diverse species. Mol. Ecol. 1993;2:359–373. doi: 10.1111/j.1365-294x.1993.tb00029.x. [DOI] [PubMed] [Google Scholar]
- Sprent J.F.A. On an Ascaris parasite of the fisher and marten, Ascaris devosi sp. nov. Proc. Helm. Soc. Wash. 1952;19:27–37. [Google Scholar]
- Sprent J.F.A. Notes on Ascaris and Tosascaris with a definition of Baylisascaris gen. nov. Parasitology. 1968;58:185–196. doi: 10.1017/s0031182000073534. [DOI] [PubMed] [Google Scholar]
- Sprent J.F.A. Baylisascaris tasmaniensis sp. nov. in marsupial carnivores - heirloom or souvenir. Parasitology. 1970;61:75–86. [Google Scholar]
- Sprent J.F.A., Lamina J., McKeown A. Observations on migratory behaviour and development of Baylisascaris tasmaniensis. Parasitology. 1973;67:67–83. doi: 10.1017/s0031182000046291. [DOI] [PubMed] [Google Scholar]
- Stefanski W., Zarnowski E. Ascaris procyonis n. sp. from the intestine of Procyon lotor L. Ann. Musei Zool. Pol. 1951;14:199–203. [Google Scholar]
- Swofford D.L. 2003. PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods) [Google Scholar]
- Taira K., Une Y., Šnábel V., Sugiyama H. Baylisascaris sp. infection in a pet kinkajou Potos flavus. Helminthologia. 2013;50:238–243. [Google Scholar]
- Tarver J.E., dos Reis M., Mirarab S., Moran R.J., Parker S., O'Reilly J.E., King B.L., O'Connell M.J., Asher R.J., Warnow T. The interrelationships of placental mammals and the limits of phylogenetic inference. Genome Biol. Evolut. 2016;8:330–344. doi: 10.1093/gbe/evv261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A.R. Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution. 1983:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- Testini G., Papini R., Lia R.P., Parisi A., Dantas-Torres F., Traversa D., Otranto D. New insights into the morphology, molecular characterization and identification of Baylisascaris transfuga (Ascaridida, Ascarididae) Vet. Parasitol. 2011;175:97–102. doi: 10.1016/j.vetpar.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Thomas W.K., Vida J., Frisse L.M., Mundo M., Baldwin J.G. DNA sequences from formalin-fixed nematodes: integrating molecular and morphological approaches to taxonomy. J. Nematol. 1997;29:250. [PMC free article] [PubMed] [Google Scholar]
- Tokiwa T., Nakamura S., Taira K., Une Y. Baylisascaris potosis n. sp., a new ascarid nematode isolated from captive kinkajou, Potos flavus, from the Cooperative Republic of Guyana. Parasitol. Int. 2014;63:591–596. doi: 10.1016/j.parint.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Tranbenkova N.A., Spiridonov S.E. Molecular characterization of Baylisascaris devosi sprent, 1952 (ascaridoidea, nematoda) from Kamchatka sables. Helminthologia. 2017;54:105–112. [Google Scholar]
- Weinstein S.B. Baylisascaris procyonis demography and egg production in a California raccoon population. J. Parasitol. 2016;102:622–628. doi: 10.1645/15-747. [DOI] [PubMed] [Google Scholar]
- Wernersson R., Pedersen A.G. RevTrans: multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., He G.Z., Hu H.G. Study on a new species, Toxascaris ailuri sp.n. (Nematoda: Ascaridae) from red panda. Sichuan J. Zool. 1987;6:1–3. [Google Scholar]
- Xie Y., Niu L., Zhao B., Wang Q., Nong X., Chen L., Zhou X., Gu X., Wang S., Peng X. Complete mitochondrial genomes of chimpanzee-and gibbon-derived Ascaris isolated from a zoological garden in southwest China. PloS One. 2013;8 doi: 10.1371/journal.pone.0082795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhang Z., Niu L., Wang Q., Wang C., Lan J., Deng J., Fu Y., Nie H., Yan N. The mitochondrial genome of Baylisascaris procyonis. PloS One. 2011;6 doi: 10.1371/journal.pone.0027066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhang Z., Wang C., Lan J., Li Y., Chen Z., Fu Y., Nie H., Yan N., Gu X., Wang S., Peng X., Yang G. Complete mitochondrial genomes of Baylisascaris schroederi, Baylisascaris ailuri and Baylisascaris transfuga from giant panda, red panda and polar bear. Gene. 2011;482:59–67. doi: 10.1016/j.gene.2011.05.004. [DOI] [PubMed] [Google Scholar]
- Yang Z. The BPP program for species tree estimation and species delimitation. Curr. Zool. 2015;61:854–865. [Google Scholar]
- Yu L., Li Y.-W., Ryder O.A., Zhang Y.-P. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol. Biol. 2007;7:1. doi: 10.1186/1471-2148-7-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G.-H., Li H.-M., Ryan U.M., Cong M.-M., Hu B., Gao M., Ren W.-X., Wang X.-Y., Zhang S.-P., Lin Q. Phylogenetic study of Baylisascaris schroederi isolated from Qinling subspecies of giant panda in China based on combined nuclear 5.8 S and the second internal transcribed spacer (ITS-2) ribosomal DNA sequences. Parasitol. Int. 2012;61:497–500. doi: 10.1016/j.parint.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Zhou X., Yu H., Wang N., Xie Y., Liang Y.-n., Li D.-s., Wang C.-d., Chen S.-j., Yan Y.-b., Gu X.-b. Molecular diagnosis of Baylisascaris schroederi infections in giant panda (Ailuropoda melanoleuca) feces using PCR. J. Wildl. Dis. 2013;49:1052–1055. doi: 10.7589/2012-07-175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian consensus tree based on FULL combined data including hars1 sequences (9 genes). Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes represent Bayesian posterior probabilities, shown when 0.90 and greater.
Bayesian consensus tree based on FULL nuclear data including hars1 sequences (6 genes). Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes represent Bayesian posterior probabilities, shown when 0.90 and greater.
Bayesian consensus tree for cox1 including previously published sequences of Baylisascaris. Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes are Bayesian posterior probabilities, shown when 0.90 and greater.
Bayesian consensus tree for 28S including previously published sequences of Baylisascaris. Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes are Bayesian posterior probabilities, shown when 0.90 and greater.
Bayesian consensus tree for ITS including previously published sequences of Baylisascaris. Branch lengths are scaled to the expected number of substitutions per site. Numbers above nodes are Bayesian posterior probabilities, shown when 0.90 and greater.