Microbes play critical roles in determining the quality of freshwater ecosystems, which are vital to human civilization. Because acI Actinobacteria are ubiquitous and abundant in freshwater lakes, clarifying their ecophysiology is a major step in determining the contributions that they make to nitrogen and carbon cycling. Without accurate knowledge of these cycles, freshwater systems cannot be incorporated into climate change models, ecosystem imbalances cannot be predicted, and policy for service disruption cannot be planned. Our work fills major gaps in microbial light utilization, secondary metabolite production, and energy cycling in freshwater habitats.
KEYWORDS: blh, carotenoid, freshwater, opsin, photoheterotrophy, proton pump, rhodopsin, xanthorhodopsin
ABSTRACT
Freshwater lakes harbor complex microbial communities, but these ecosystems are often dominated by acI Actinobacteria. Members of this cosmopolitan lineage are proposed to bolster heterotrophic growth using phototrophy because their genomes encode actino-opsins (actR). This model has been difficult to validate experimentally because acI Actinobacteria are not consistently culturable. Based primarily on genomes from single cells and metagenomes, we provide a detailed biosynthetic route for members of acI clades A and B to synthesize retinal and its carotenoid precursors. Consequently, acI cells should be able to natively assemble light-driven actinorhodopsins (holo-ActR) to pump protons, unlike many bacteria that encode opsins but may need to exogenously obtain retinal because they lack retinal machinery. Moreover, we show that all acI clades contain genes for a secondary branch of the carotenoid pathway, implying synthesis of a complex carotenoid. Transcription analysis of acI Actinobacteria in a eutrophic lake shows that all retinal and carotenoid pathway operons are transcribed and that actR is among the most highly transcribed of all acI genes. Furthermore, heterologous expression of acI retinal pathway genes showed that lycopene, retinal, and ActR can be made using the genes encoded in these organisms. Model cells producing ActR and the key acI retinal-producing β-carotene oxygenase formed holo-ActR and acidified solution during illumination. Taken together, our results prove that acI Actinobacteria containing both ActR and acI retinal production machinery have the capacity to natively synthesize a green light-dependent outward proton-pumping rhodopsin.
IMPORTANCE Microbes play critical roles in determining the quality of freshwater ecosystems, which are vital to human civilization. Because acI Actinobacteria are ubiquitous and abundant in freshwater lakes, clarifying their ecophysiology is a major step in determining the contributions that they make to nitrogen and carbon cycling. Without accurate knowledge of these cycles, freshwater systems cannot be incorporated into climate change models, ecosystem imbalances cannot be predicted, and policy for service disruption cannot be planned. Our work fills major gaps in microbial light utilization, secondary metabolite production, and energy cycling in freshwater habitats.
INTRODUCTION
Freshwater lakes contain complex and dynamic microbial communities that influence water quality by mediating biogeochemical cycling. These ecosystems are often numerically dominated by three clades of acI Actinobacteria (acI-A, acI-B, and acI-C), ultramicrobacteria with low-GC-content genomes that are streamlined to approximately 1.2 to 1.5 MBp (1–5). The designation acI was adopted to denote the first named branch of the ubiquitous freshwater Actinobacteria (6, 7). Because acI are so abundant, the following question arises: what are the factors that enable their success? Major proposed advantages include scavenging resources with membrane transporters (3, 8), evading protist feeding by virtue of their small size (9, 10), and supplementing energy needs via photoheterotrophy. Light utilization is feasible because each clade encodes actino-opsin (ActR), the putative platform protein for actinorhodopsin (holo-ActR) (3, 11–13). Validating this hypothesis not only requires a demonstration that acI Actinobacteria can synthesize or acquire sufficient retinal, the chromophore needed to complete holo-ActR, but also biochemical evidence that the holo-ActR is functional. Additionally, the relevant genes must be shown to be expressed in the native freshwater environments where acI Actinobacteria are dominant. That these requirements are satisfied by acI Actinobacteria is not guaranteed; Rhodoluna lacicola holo-ActR was shown to be active if, and only if, exogenous retinal was added (14). Without an encoded, retinal-producing enzyme or a solution for obtaining dilute environmental chromophores, it is unclear what the physiological role and ecosystem implications of ActR in such organisms are. On the other hand, the Luna1 bacterium “Candidatus Rhodoluna planktonica” encodes a putative retinal-synthesizing enzyme (15). This family of Actinobacteria is radically different from acI and is much less abundant and prevalent in freshwater lakes (2). Work in marine environments indicates that microorganisms encoding opsins contain genes for confirmed retinal-production enzymes, leading to functional proteorhodopsin assembly (16). However, even with extensive marine work, it is not known whether rhodopsins are truly energy-positive adaptations (17). This question provides motivation to determine if acI truly use rhodopsins for photoheterotrophy.
Rhodopsins are seven-transmembrane-helix bundles that bind retinal at an internal lysine via a Schiff base (18). The retinal pocket amino acid sidechains, specifically a tuning residue (19), determine the maximal absorption wavelength of the holo-protein. The absorbed photons trigger the geometric isomerization of retinal, which completes a transfer pathway through the interior of the bundle and thus, across the membrane. Most commonly, protons are shuttled out of the cytoplasm to form a gradient that can be harnessed for activities like ATP synthesis (20). The retinal that ultimately enables the gradient is derived from carotenoids, colorful molecules that act as photoprotectants, cofactors, pigments, and membrane stabilizers throughout nature (21, 22). The initial building blocks of carotenoids are the five-carbon monomers isopentenyl diphosphate and dimethylallyl diphosphate. These units can be polymerized and modified to form simple carotenoids, like lycopene, β-carotene, and various intermediates (23), or more complex ones, like myxoxanthophyll (24). Moreover, the ketocarotenoids, salinixanthin and echinenone, can boost rhodopsin activity by resonance energy transfer to retinal in rare xanthorhodopsin-like systems (25–27). Microorganisms with the machinery to produce functional, chromophore-loaded rhodopsins may have a competitive advantage in their native ecosystems (28, 29).
Here, we provide experimental evidence that acI Actinobacteria have photoheterotrophic potential. A significant challenge when these studies were initiated was the inability to axenically culture acI (5, 30), although it appears this hurdle may soon be overcome (31). Consequently, we pursued a multidisciplinary approach that did not rely on large amounts of acI biomass, but instead focused on bioinformatic, transcriptomic, and biochemical data. Specifically, we characterize a biosynthetic pathway for retinal and its carotenoid precursors using heterologous expression. We then demonstrate that acI-actR is highly transcribed in the freshwater biome, Lake Mendota, and an acI ActR can function as a retinylidene holoprotein to produce a light-driven proton gradient. Most importantly, the nanomachine is active in cells that produce retinal using the native acI β-carotene oxygenase. Finally, we provide evidence for a complex carotenoid that may screen cells from oxidative damage and/or boost the light harvesting ability of actinorhodopsin.
RESULTS
All acI ActR contain key amino acids for function.
We first determined whether acI ActR sequences were consistent with holo-ActR formation. Indeed, all contain the features for proper rhodopsin structure and function (Fig. 1A). Seven predicted helices (α1 to α7) match those found in xantho-opsin, a close homolog of ActR from Salinibacter ruber (25). A conserved lysine for Schiff base formation and acidic residues for proton-shuttling across the retinylidene gate are present (32), and a leucine is predicted to tune the absorbance of all acI holo-ActRs to the green region (33). We also found novel features. A proline in the middle of α4 differentiates ActRs from xantho-opsins, and the residue may serve as a means for better phylogenetic classification (Fig. S1). Additionally, 3D structure predictions closely position two cysteines on α1 and α7 in clades A and B, which may allow a disulfide bond to covalently staple the protein together (Fig. 1B). acI ActRs also contain glycine near the top of α5, a required feature for binding ketolated antenna carotenoids (26, 27). This glycine replaces a bulky residue, usually tryptophan, found in most rhodopsins, including the well-characterized bacteriorhodopsin.
FIG 1.
Features of ActRs from acI clades A, B, and C. (A) ActRs from clade A (ActRO22), clade B (ActRL06), and clade C (ActRMEE578) are aligned to a homolog, xantho-opsin (XR) from Salinibacter ruber. Proteins are subscripted with genome shorthand (Table S1). Features are highlighted: predicted helices (gray, numbered), cysteines (yellow), main proton shuttles (orange), main absorbance tuner (green), Schiff base lysine (magenta), possible antenna carotenoid residues (cyan), and proline indicative of ActR versus xantho-opsins (red). Antenna carotenoid residues are based on amino acids in proximity to salinixanthin in the crystal structure of xanthorhodopsin (PDB 3DDL). (B) Three-dimensional (3D) structure prediction for ActR with key residue side chains displayed according to the color coding in panel A. The α-carbon of glycine near the top of α5 that would allow antenna binding is shown as a sphere, the Schiff base retinal (lime green) and predicted disulfide bond in clades A and B are modeled, and the N and C termini are labeled.
Seven gene products form an actinorhodopsin pathway.
Retinal is almost certainly the chromophore needed to form holo-ActR in acI Actinobacteria. We find that in addition to encoding ActR, acI single-cell amplified genomes (SAGs), metagenome-assembled genomes (MAGs), and complete genomes from dilution-to-extinction cultures (Table S1) appear to encode genes for enzymes that produce retinal and its carotenoid precursors (Fig. 2).
FIG 2.
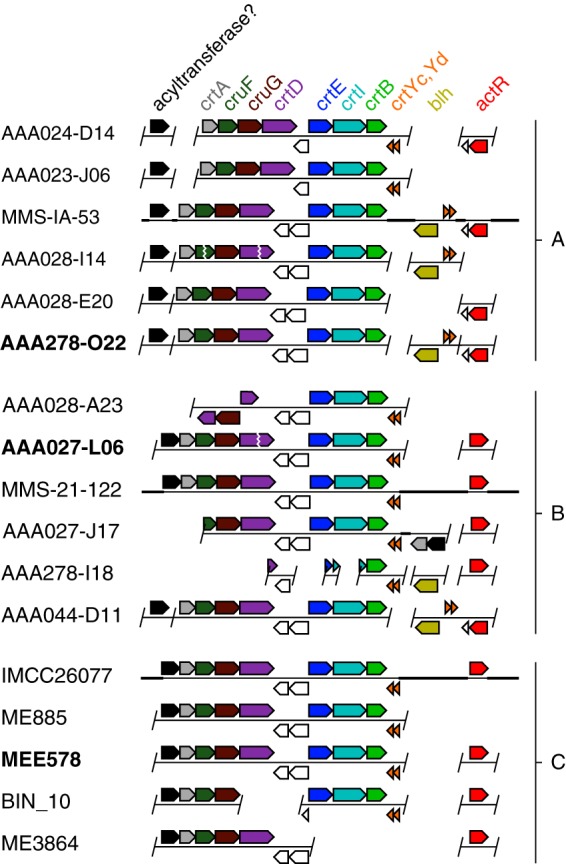
acI carotenoid-related genes in the genomic context. acI genomic contigs or genomes from clades A, B, and C are primarily labeled by shorthand notation (i.e., designation after “actinobacterium SCGC” or “int.metabat.”). Genes are arrows pointing in the direction of transcription. A slash indicates a contig boundary, which may or may not end immediately after the pictured gene, a vertical zig-zag indicates contig ends that have been manually paired, and a thick horizontal bar indicates a longer region of DNA not represented. Uncolored genes are neighboring genes that may or may not be functionally associated with the carotenoid-related genes. Boldface labels indicate a gene source for this study. Relevant Integrated Microbial Genomes (IMG) database locus tags for this study from AAA278-O22 are A278O22DRAFT_00003540 (proposed acyltransferase), A278O22DRAFT_00010590-10560 (crtA to crtD), A278O22DRAFT_00010530-10510 (crtE to crtB), A278O22DRAFT_00007000-6980 (crtYd to blh), and A278O22DRAFT_00001360 (actR).
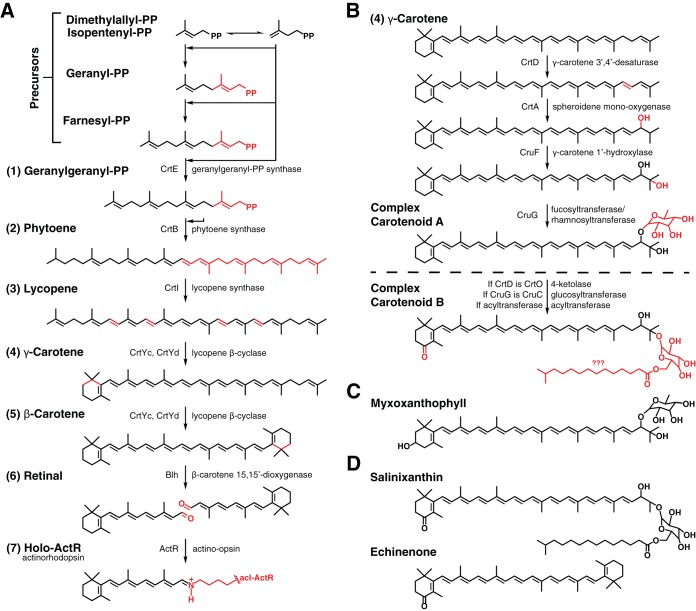
We assembled a plausible, complete pathway with protein assignments for forming lycopene, β-carotene, retinal, and subsequently, actinorhodopsin (Fig. 3A). Lycopene synthesis in acI Actinobacteria requires three major steps, as follows: synthesis of geranylgeranyl-PP (step 1), linkage of two geranylgeranyl-PP molecules to phytoene (step 2), and tetra-desaturation of phytoene to lycopene (step 3), predicted to be carried out by CrtE, CrtB, and CrtI, respectively. Subsequent β-cyclization of lycopene would first produce γ-carotene (step 4) and then β-carotene (step 5). A heterodimeric enzyme composed of CrtYc and CrtYd likely carries out these serial cyclizations, much like Myxococcus xanthus, β-cyclase genes [36% and 33% identity, trihelical transmembrane topology, PxE(E/D) catalytic motif] (Fig. S2) (34–37). The symmetric cleavage of β-carotene by a β-carotene oxygenase, Blh, would then form retinal (step 6). Blh is a putative dioxygenase, based on 27% sequence identity, predicted helical topology, and four histidines for nonheme iron coordination that are shared with the characterized β-carotene dioxygenase from an uncultured marine bacterium (Fig. S3) (35, 36, 38). Functional holo-ActR requires retinal to autocatalytically form a Schiff base with the side chain of a conserved lysine in ActR (step 7).
FIG 3.
Predicted carotenoid-related pathways in acI. (A) The actinorhodopsin synthesis pathway requires isopentenyl precursors to be assembled into carotenoids (1 to 3), which are modified (4 to 5) and cleaved to produce retinal (6). A Schiff base forms between retinal and a lysine of acI-ActR to form acI-holo-ActR (7). Chemical changes are shown in red, proteins from this study with their predicted functions are shown to the sides of progress arrows, and product names are shown to left of their chemical structures. Step 6 cannot be performed in acI Actinobacteria that lack Blh; retinal must be exogenously sourced. (B) A complex carotenoid synthesis pathway is predicted to require γ-carotene produced in the actinorhodopsin pathway, with further desaturation, hydroxylation, and glycosylation for a myxoxanthophyll-like carotenoid. A carotenoid with different stucture can result if enzyme identities are different and a proposed acyltransferase is involved in the pathway. Question marks indicate uncertainty in naming and/or chemical structure. (C) A complex carotenoid used by many cyanobacteria for photosynthesis. (D) Complex carotenoids from other organisms that function as carotenoid antennae on proton-pumping rhodopsins.
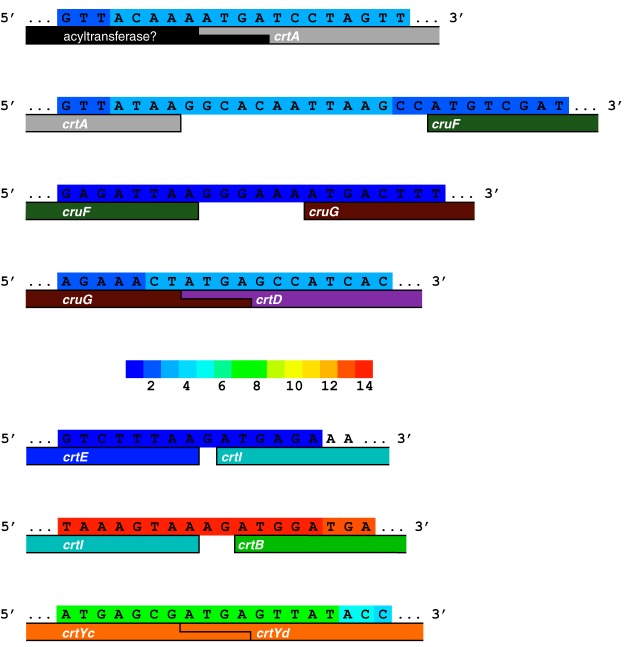
In the genomic context, genes encoding the enzymes for chromophore production are grouped into functional regions (Fig. 2). Lycopene production genes (crtE, crtB, and crtI) are found as a neighborhood in the order of steps 1, 3, and 2 in all acI clades. Genes for steps 4 to 6 (crtYc, crtYd, and blh) are found at various distances from the lycopene synthesis cluster. In many cases, the 3′ ends of crtB and crtYd are adjacent but encoded in the opposite sense, thus forming a β-carotene synthesis neighborhood. In other cases, the cyclase genes are instead found near blh. In most cases, actR is widely separated from other pathway genes. An interesting exception is AAA044-D11, where actR, crtYc, crtYd, and blh are contiguous (Fig. 2).
A pathway for a complex carotenoid contains at least nine genes.
We previously noted that genes for production of a predicted carotenoid glycoside ester may be encoded adjacent to the lycopene gene neighborhood (3) (Fig. 2). In this study, we used bioinformatics, sequence homology, topology predictions, and known carotenoid structures to assign functions to gene products and assemble two plausible pathways (Fig. 3B and Fig. S4 to S7) (24, 35, 36, 39–44).
The crucial branch point compound for initiating the pathway is γ-carotene (step 4), which is predicted to be produced in the retinal pathway after synthesis of phytoene and lycopene. Modifications of γ-carotene are expected to include desaturation of a carbon-carbon bond at the noncyclized end by CrtD, introduction of two hydroxyl groups in separate steps by CrtA and CruF, and transfer of a monosaccharide, like fucose or rhamnose, by CruG onto the CrtA-inserted hydroxyl group. The carotenoid resulting from CrtD and CruG would be similar to the known cyanobacterial carotenoid myxol 2′-fucoside, although lacking a ring hydroxyl (Fig. 3C) (24). For the sake of discussion, we will refer to this molecule as complex carotenoid A, with the caveat that the carotenoid could have a different structure than predicted, especially with regard to substituent modifications, and it may be a previously characterized molecule.
Genes encoding enzymes for complex carotenoid synthesis (crtA, cruF, cruG, and crtD) are found upstream of the lycopene neighborhood as a contiguous group, which most often includes a putative acyltransferase (Fig. 2 and 3). The proposed acyltransferase identity and chemistry are also not well defined, and it is not homologous to the acyltransferase CruD (45), yet the involvement of the gene in carotenoid synthesis is supported by database annotation as a coenzyme A (CoA) methyl esterase. Because many carotenoid modification proteins are related in protein sequence (i.e., lycopene desaturases and carotene ketolases) (46), functional validation will be required for all of the carotenoid pathway enzymes. Specifically, the crtD gene could encode a ketolase, CrtO, which would introduce a carbonyl onto the β-ionone ring rather than desaturating an additional bond, and the cruG gene product may instead be CruC, which attaches a glucose onto a CruF-inserted hydroxyl group (45). CrtO, CruC, and a proposed acyltransferase combination may produce a rhodopsin antenna, like salinixanthin or echinenone (Fig. 3D) (25–27). This carotenoid will be referred to as complex carotenoid B.
Metatranscriptomic analysis of pathway gene transcripts in environmental acI populations.
For actinorhodopsin assembly in acI cells, actR and retinal synthesis genes must be expressed. To measure gene expression in environmental acI Actinobacteria, four metatranscriptome samples were collected across multiple time points from the surface of eutrophic Lake Mendota (Dane County, WI, USA), and RNA was isolated and sequenced. The resulting transcripts were mapped to available acI SAGs and MAGs (Table S1) to quantify relative gene expression levels in acI cells. Notably, actR is the most highly expressed acI-A gene and the second most highly expressed acI-B gene (13), as well as the most highly expressed gene from either pathway in each of the three acI clades (Table 1). Transcription is also observed for other genes in both the retinal and putative complex carotenoid pathways, but at levels several hundredfold lower than for actR (Table 1). No transcripts were mapped for blh from acI-C because the gene is not present in any acI-C SAG or MAG or in the first complete acI-C genome obtained by dilution-to-extinction cultures (4).
TABLE 1.
Log2 RPKM values for pooled carotenoid-related gene transcripts within each clade
| Gene name | Log2 RPKM values |
||
|---|---|---|---|
| acI-A | acI-B | acI-C | |
| crtE | 3.69 | 4.93 | 6.48 |
| crtB | 2.74 | 3.45 | 6.62 |
| crtI | 4.33 | 5.93 | 6.90 |
| crtYc | 1.57 | 5.62 | 7.07 |
| crtYd | NDa | 3.29 | 4.02 |
| blh | 0.07 | 2.06 | NAb |
| actR | 14.36 | 13.86 | 12.52 |
| crtD | 3.30 | 3.93 | 4.03 |
| crtA | 2.11 | 3.39 | 5.24 |
| cruF | 2.63 | 3.85 | 5.59 |
| cruG | 2.90 | 3.62 | 4.51 |
| Putative acyltransferase | 2.64 | 4.04 | 5.37 |
| recA | 7.85 | 9.16 | 10.09 |
ND, not determined; gene is present in clade member assemblies but no transcript was mapped.
NA, not applicable; gene is not present in any clade member assembly.
This mapping of Lake Mendota metatranscriptome samples provides direct evidence for complex carotenoid, lycopene, and lycopene cyclase operons (Fig. 4). Specifically, reads that mapped to the most populous genome, ME885, overlap the intergenic regions within each of the three operons (Fig. 3 and 4). Synteny across acI clades, especially for the lycopene and complex carotenoid operons, similarly supports assignment of the three neighborhoods as genuine operons. The reading frame overlap in acI-C Actinobacteria of crtA with the putative acyltransferase supports a lipid attachment step in complex carotenoid B synthesis.
FIG 4.
Intergenic transcripts that map to carotenoid-related genes in genome ME885. Operons are evident for complex carotenoid (putative acyltransferase through crtD), lycopene (crtE through crtB), and lycopene cyclase (crtYc and crtYd) biosynthesis in an acI-C member. The color key in the center indicates of the number of times a base was covered. Transcripts and genes may continue beyond the edges of the DNA window.
CrtE, CrtB, and CrtI produce lycopene.
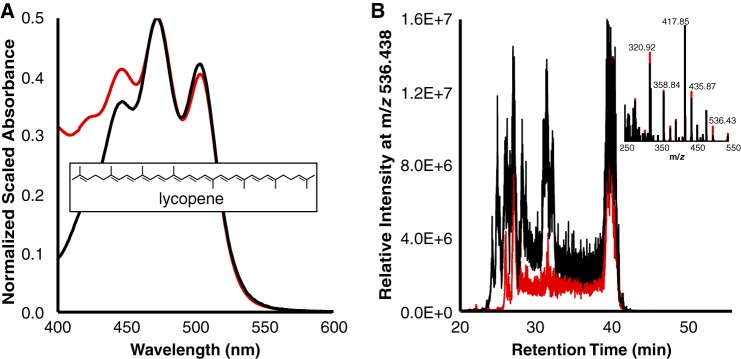
We sought to demonstrate whether CrtE, CrtB, and CrtI enzymes from acI can form lycopene as predicted (Fig. 3A, steps 1 to 3). Therefore, a fast, reproducible method for pigment extraction from whole Escherichia coli cells and identification by high-performance liquid chromatography (HPLC)-mass spectrometry (MS) analysis was developed. Relevant genes (crtE, crtB, and crtI) from assembly AAA278-O22 were cloned into pCDFDuet1 and expressed from the resulting acI-CrtEBI/− cassette (Tables 2 and 3). Assembly AAA278-O22 was chosen for carotenoid production work because it contained all relevant genes and source DNA was available (3). The extracted compounds of these cells displayed absorbance maxima which exactly matched those of a lycopene standard at expected positions of 447, 472, and 504 nm (Fig. 5A) and displayed an intense red color (data not shown). HPLC-MS elution profiles indicated the presence of cis and all-trans isomer peaks at m/z 536.438 (Fig. 5B). The retention time of the all-trans species (39.90 min) coincides for sample and standard as the peak with highest intensity. The lycopene assignment was further confirmed by tandem mass spectrometry (MS/MS) fragmentation of the parent ion.
TABLE 2.
DNA sources for this study
| Gene vector | Gene(s)a | E. coli codon-optimized? |
|---|---|---|
| Single-cell genome | acI-crtE, acI-crtB, acI-crtI | No |
| acI-blh | No | |
| Plasmid | ||
| pJExpress404 | acI-actRL06 | Yes |
| Bba_K274210 | Pa-crtE, Pa-crtB, Pa-crtI, Pa-crtY | Yes |
Pa, Pantoea ananatis.
TABLE 3.
Plasmids utilized for this study
| Backbone | Cassette shorthandc | Cloning site contentsd |
|---|---|---|
| pCDFDuet1a | –/– | NA/NA |
| acI-CrtEBI/− | acI-crtE, acI-crtB, acI-crtI/NA | |
| acI-CrtEBI/Pa-CrtY | acI-crtE, acI-crtB, acI-crtI/Pa-crtY | |
| Pa-CrtEBI/− | Pa-crtE, Pa-crtB, Pa-crtI/NA | |
| Pa-CrtEBIY/− | Pa-crtE, Pa-crtB, Pa-crtI, Pa-crtY/NA | |
| Pa-CrtEBIY/acI-Blh | Pa-crtE, Pa-crtB, Pa-crtI, Pa-crtY/acI-blh | |
| pET21b+b | – | NA |
| acI-ActRL06 | acI-actRL06 |
Plasmid contains two cloning sites with contents separated by a slash.
Plasmid contains one cloning site in-frame with a C-terminal hexahistidine tag.
–, empty.
NA, not applicable because the cloning site is unmodified.
FIG 5.
CrtE, CrtB, and CrtI from acI members catalyze lycopene formation. (A) Spectra of a control extract subtracted from an extract from cells containing acI enzymes (CrtE, CrtB, CrtI) (black) and standard lycopene (red). All extracts are in acetone. The inset depicts lycopene, and maxima for both curves are at 447, 472, and 504 nm. (B) HPLC-MS data for the same samples. The inset depicts MS/MS data from all-trans peak retention times.
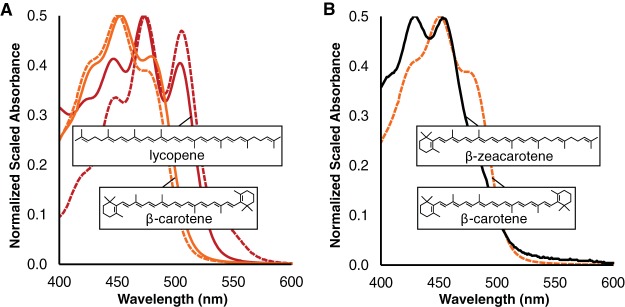
To further characterize the lycopene product of acI CrtE, CrtB, and CrtI enzymes, we tested whether it serves as the substrate in a lycopene cyclization reaction. Pantoea ananatis genes crtE, crtB, crtI, and crtY were expressed in E. coli as positive controls (Table 2, Table 3, and Fig. 6A). When the crtY cyclase gene and acI lycopene pathway genes were coexpressed from pCDFDuet1 acI-CrtEBI/Pa-CrtY, the cellular extract absorbance maxima were 407, 429, and 453 nm (Fig. 6B). These dramatically blue-shifted maxima, compared to those of β-carotene, combined with the defined cyclase activity of CrtY, identify the major extract product as β-zeacarotene, γ-carotene saturated between C-7′ and C-8′ (Fig. 6B). Thus, CrtE, CrtB, and CrtI from acI produce a chromophore, which serves as a substrate for a lycopene cyclase. Presumably, more β-carotene would be produced after complete desaturation and a second cyclization, which were inefficiently carried out in our heterologous expression system.
FIG 6.
A lycopene-like carotenoid produced by CrtE, CrtB, and CrtI from acI members is cyclized by CrtY from Pantoea ananatis. (A) Spectra of a control extract subtracted from an extract from cells containing Pantoea ananatis enzymes (CrtE, CrtB, and CrtI; red dash), standard lycopene (red), a control extract subtracted from an extract from cells containing Pantoea ananatis enzymes (CrtE, CrtB, CrtI, and CrtY; orange dash), and standard β-carotene (orange). All extracts are in acetone. The insets depict lycopene and β-carotene, and the maxima are 472/474 nm and 453/451 nm. (B) The spectrum for synthesized β-carotene as in panel A (orange dash), and the spectrum of a control extract subtracted from an extract from cells containing acI enzymes (CrtE, CrtB, and acI-CrtI) and CrtY from Pantoea ananatis. All extracts are in acetone. The insets depict β-zeacarotene and β-carotene, and the shifted maxima are at 429 and 453 nm.
Blh produces retinal from β-carotene.
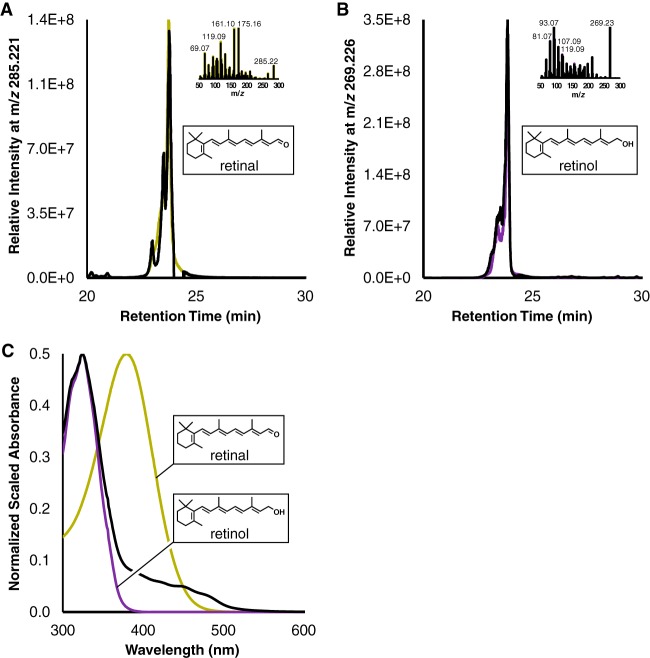
The final enzymatic step in retinal biosynthesis is the symmetric cleavage of a cyclized carotene (Fig. 3A, step 6). This enzyme is not encoded in any acI-C genomes (Fig. 2). To show that acI can natively perform retinal synthesis, we tested the enzymatic activity of AAA278-O22 Blh. This blh gene was expressed from pCDFDuet1 Pa-CrtEBIY/acI-Blh (Table 3) to ensure a large β-carotene substrate pool. Introduction of Blh yielded yellow cells instead of the intensely orange-colored cells observed when β-carotene is abundant (data not shown). HPLC-MS confirmed the presence of retinal at m/z 285.221 (Fig. 7A). Retinol was also identified via its dehydrated species at m/z 269.226 (Fig. 7B), as described by Breeman and Huang (47). As is the case for lycopene, geometric isomer peak patterning was observed. Maximal all-trans species retention times match those of retinal and retinol standards at 23.76 min and 23.88 min, respectively. Additionally, MS/MS fragmentation of the appropriate m/z species confirmed retinoid identities (Fig. 7A and B). Retinol was roughly 7× more abundant than retinal in the extract, as judged by MS intensities. Accordingly, the UV-visible (UV-Vis) absorbance profile appears more like that of retinol (Fig. 7C). Retinal produced by Blh is likely serving as a potent electron acceptor for the E. coli alcohol dehydrogenase, ybbO (48, 49).
FIG 7.
acI-Blh is a β-carotene oxygenase that catalyzes retinal formation. (A) HPLC-MS data for Pa-EBIY/acI-Blh extract (black) and a retinal standard (yellow). The inset depicts MS/MS data from all-trans peak retention times. (B) HPLC-MS and MS/MS (inset) data for the same experimental extract but with standard retinol spectra (purple). Retinol forms a dehydrated species in positive mode, and the species was used as a proxy for retinol detection (C) Spectra of a control extract subtracted from an extract from cells containing Pantoea ananatis enzymes (CrtE, CrtB, CrtI, and CrtY) and Blh from acI (black), standard retinal (yellow), and standard retinol (purple). All extracts are in ethanol. Insets depict retinal and retinol to highlight conjugation differences, and important maxima are at 379, 324, and 325 nm.
ActRL06 with retinal forms an active, green light-dependent proton pump.
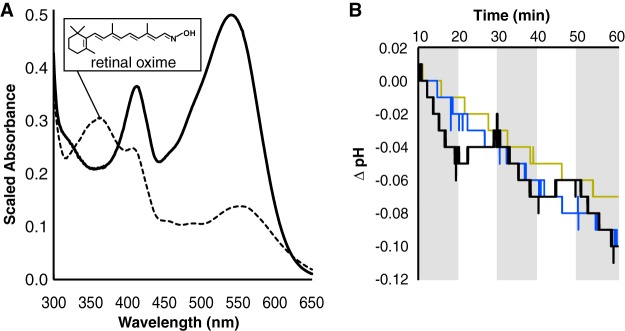
The gold-standard test of a functional rhodopsin is light-dependent activity. Given that acI ActR proteins contain the residues for chromophore binding and proton movement, we tested an opsin from each clade (AAA278-O22, AAA027-L06, and MEE578) for production and chromophore binding in E. coli. Each opsin was expressed from pET21b+ alongside the plasmid confirmed for retinal production. ActRL06 was selected for further characterization due to its high expression and efficient retinal binding, as judged from samples captured by metal affinity from detergent-solubilized membranes. holo-ActRL06 maximally absorbed in the green region at 541 nm (Fig. 8A). The covalent attachment of retinal was confirmed by incubation with hydroxylamine hydrochloride, which frees the retinal and leads to production of retinal oxime.
FIG 8.
holo-acI-ActRL06 is a light-driven proton pump. (A) Spectra for a holo-ActRL06 enrichment purification from cells also producing retinal using Pantoea ananatis CrtE, CrtB, and CrtI, and Blh from acI members without (solid) and with (dashed) hydroxylamine hydrochloride. The retinal oxime that results from hydroxylamine incubation is shown, and the maximum absorbance is at 358 nm. (B) Microelectrode pH traces for retinal-producing cells expressing as in panel A with (black, blue) or without (yellow) acI-ActRL06. CCCP is present in the blue trace. The room was illuminated by a white light with a red plastic overlay for the entire assay, and shaded regions represent extra direct illumination by white light. Curves are from one of three independent experiments.
For conclusive demonstration that holo-ActRL06 outwardly pumps protons in the presence of light when in a membrane environment, microelectrode pH measurements were performed. During exposure to white light, the pH of a nonbuffered assay solution sharply decreases compared to that in periods of dim red light (Fig. 8B). To confirm protons as the source of the steep pH drop, carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to discharge the proton gradient. CCCP trials did not display light-dependent proton pumping but rather the constant downward drift of control cells. Thus, the acI holo-ActR is a retinal-bound, green light-dependent proton-pumping rhodopsin.
DISCUSSION
Prior to this study, there was no biochemical evidence to support the hypothesis that acI Actinobacteria use opsin-based phototrophy in freshwater. We have provided experimental verification of the advantages that allow the acI lineage to be so abundant. We describe two favorable environmental adaptations, carotenoid biosynthesis and light utilization. Transformation of simple isoprenoid precursors into carotenoids and retinal allow ActR to function as a green light-absorbing, outward proton-pumping holo-ActR. An additional pathway is predicted to branch from a carotenoid intermediate in the holo-ActR pathway to synthesize a chromophore, like complex carotenoid A or B, with a yet unconfirmed structure and function.
The pathway for retinal and rhodopsin synthesis has been experimentally confirmed using acI actinobacterial proteins. The machinery to start retinal production consists of three enzymes, CrtE, CrtB, and CrtI (steps 1 to 3). The enzymes produce lycopene (Fig. 3A and Fig. 5) and cluster into the crtEIB operon (Fig. 2 and Fig. 4). The reactions producing γ-carotene and β-carotene (steps 4 to 5) that follow lycopene synthesis are likely carried out by a heterodimeric membrane protein expressed from the crtYc and crtYd operon (Fig. 2 to 4). Homologs of these gene products are found in Myxococcus xanthus, where they were shown to synthesize a mixture of γ-carotene and β-carotene (34). Therefore, we propose that acI can also synthesize these compounds. Although the ratio of these cyclized products in acI is unknown, it may be intrinsically linked to intracellular concentrations of retinal and complex carotenoid. Unsaturated but cyclized intermediates like β-zeacarotene (Fig. 6) may result from differing levels of cyclase compared to those of CrtE, CrtB, and CrtI. We note that a system for stable β-zeacarotene production could prove a fruitful biotechnology tool because monocyclized carotenoids are not readily available for purchase.
A key finding of our work is the presence, expression, and robust activity of a retinal-producing oxygenase from the acI gene, blh. The enzyme symmetrically cleaves β-carotene to two retinal molecules (Step 6) (Fig. 3A and Fig. 7) and might additionally use alternative substrates, such as a β-zeacarotene or other γ-carotene-like carotenoids, to yield a single retinal. Notably, the blh gene is not found in acI-C genomes; in five acI-C genomes ranging in completeness from 25 to 100%, blh has never been recovered (Table S1) (4). This absence points toward clade-level differentiation with respect to retinal production and subsequent holo-ActR assembly in acI Actinobacteria. Specifically, we propose that populations of acI-A and acI-B maintain a high relative abundance in the community throughout the year (50, 51) because they can synthesize their own retinal to support phototrophy; therefore, phototrophy could support heterotrophy. In contrast, acI-C Actinobacteria exhibit “bloom and bust” cycles coincident with cyanobacterial blooms (52). It will require further study to determine if this acI-C variability is due to an inability to harvest light or a dependence on exogenous retinal, potentially from lysed cyanobacteria. Indeed, many species of cyanobacteria encode functional β-carotene oxygenases (38, 53, 54), and retinoid concentrations in eutrophic lakes during cyanobacterial blooms are measurable (55). This proposal is consistent with actinorhodopsin studies in the culturable freshwater organisms Rhodoluna lacicola and “Candidatus Rhodoluna planktonica” (14, 15). While both carry actR, only the latter contains blh and thus exhibits self-sufficient rhodopsin activity in the laboratory. R. lacicola is dependent on retinal supplementation. As such, it may be analogous to blh-lacking organisms. Indeed, recently reported complete genomes (5) indicate that even some acI-A and acI-B Actinobacteria would require an alternative retinal source. This heterogeneity in gene content has major implications for niche filling by acI-A and acI-B members and may dictate community-related growth patterns.
Regardless of whether acI cells synthesize or scavenge retinal, all acI ActR proteins contain the necessary features for proper activity as retinal holoproteins with green maximal absorbance, and we have demonstrated that an example acI ActR functions as a retinylidene protein (Fig. 1 and Fig. 8). Green absorption correlates with light penetration depth for many freshwater bodies where acI Actinobacteria thrive and strengthens the case that holo-ActRs in acI are light-activated proteins in the environment. Native production of holo-ActR would enable acI cells to produce a proton gradient even when central metabolism intermediates are scarce. In addition to showing the proton-pumping activity of holo-ActR, we discovered interesting qualities of ActRs (Fig. 1 and Fig. S1). A proline in helix four was determined to be a phylogenetically differentiating residue between actino-opsins and other xantho-opsins, like the one in Salinibacter ruber. Prolines kink helices, and these residues may have broad structural effects on photocycling times and/or intermediate photocycle structures. Similarly, a disulfide bond joining α1 to α7 in acI-A and acI-B Actinobacteria could also impact activity and/or protein stability.
In addition to retinal and ActR synthesis, members of all acI clades contain an operon for other carotenoid biosynthetic machinery (a putative acyltransferase, crtA, cruF, cruG, and crtD) (Fig. 2 and Fig. 4). The gene products can be predicted to produce complex carotenoids with glycosyl or glycosyl and acyl modifications, complex carotenoid A or B, respectively. (Fig. 3B). Although the structure and presence of the carotenoid remain to be experimentally validated, the glycine void near the top of α5 is a necessary (although not sufficient) condition for binding of an antenna ketocarotenoid (56, 57). acI ActR would then be another example of a special class of rhodopsins that we call antenna rhodopsins (25–27), but others are predicted (57). We are left with the following questions: can acI synthesize a complex carotenoid using the genes found upstream from the lycopene cluster, and does it interact with acI ActR as an antenna?
Two biological scenarios for carotenoid-related light utilization in acI include (i) holo-ActR is an antenna rhodopsin that augments proton pumping via carotenoid energy transfer, or (ii) complex carotenoids protect the cell from oxidative species and excess light while the proton pump functions (58). Both situations favor production of a proton gradient to power numerous nutrient uptake transporters, particularly when nutrients are limited (13). Such adaptations go far toward explaining how acI Actinobacteria can reach nearly 50% of the microbial cell number in freshwater lake epilimnia, and why they exist in virtually all freshwater ecosystems (2, 50). Further defining the physiology of a dominant ecological force within the lake biome will expand our conceptual models by including heterotrophs that perform phototrophy. Ultimately, this addition will better explain freshwater carbon and energy cycling and improve ecosystem behavior forecasting.
MATERIALS AND METHODS
acI gene identification and pathway assembly.
Annotations for multiple acI SAGs (3) and MAGs were analyzed for genes relating to carotenoids using the Joint Genome Institute’s Integrated Microbial Genomes Viewer (59). Translated candidate protein sequences were used to identify homologs in other acI genomes, and those with consistent gene neighborhoods and known carotenoid-related functions were prioritized. After function assignments, two pathways for carotenoid biosynthesis and use in acI Actinobacteria were assembled.
acI biomass collection and transcriptomics.
Environmental sampling, metatranscriptome sequencing, and gene expression calculations were all performed as previously described (13). Briefly, four samples were collected from within the top 12 m of the Lake Mendota (Dane County, WI) water column and filtered through cheesecloth and onto 0.2 µm mixed cellulose filters (Whatman). Filters were immediately frozen in liquid nitrogen and stored at −80°C. Samples were subjected to TRIzol-based RNA extraction, phenol-chloroform separation, and RNA precipitation. RNA was further purified using the RNeasy minikit (Qiagen) with on-column digestion of DNA via the RNase-free DNase set (Qiagen). RNA was then sent to the University of Wisconsin—Madison Biotechnology center for rRNA depletion, cDNA synthesis, and sequencing. rRNA was depleted using the RiboZero rRNA removal kit (bacteria) (Illumina). Samples were then prepared for sequencing using the TruSeq RNA library prep kit v2 (Illumina), pooled in an equimolar ratio, and sequenced on an Illumina HiSeq 2500 platform using 2 × 100-bp paired-end sequencing.
After sequencing, metatranscriptomic reads were trimmed, merged, subjected to in silico rRNA removal, mapped to carotenoid-related genes, and counted. Raw paired-end reads were trimmed using Sickle (60), merged using FLASH (61), and subjected to in silico rRNA removal using SortMeRNA (62). Sickle was run using default parameters; FLASH was run with a maximum overlap of 100 nucleotides; and SortMeRNA was run using databases for bacterial, archaeal, and eukaryotic rRNA, derived from SILVA v119 (63) and RFAM v12.0 (64). Trimmed and merged reads from all four samples were then pooled and mapped to a single reference FASTA file containing 36 high-quality acI genomes from a larger freshwater genome collection (11). Reads were competitively mapped to genes using BBMap (https://sourceforge.net/projects/bbmap/) with the ambig = random and minid = 0.95 options. Next, reads mapping to each carotenoid-related gene were counted using hts-count (65) with the intersection_strict option. Within each clade, reads mapping to each carotenoid-related gene were pooled, and gene expression was computed on a reads per kilobase per million (RPKM) basis (66), while also accounting for different gene lengths and total mapped reads for each acI genome. All scripts are found on GitHub (https://github.com/joshamilton/Hamilton_acI_2017/tree/actR).
Plasmid construction.
All cloning was performed using Phusion high-fidelity (HF) GC master mix (Thermo Fisher) and the listed primers (Table 4). A collection of stable DNA sources for amplification and subcloning was first created. A plasmid containing E. coli-codon-optimized crtE, crtB, crtI, and crtY from Pantoea ananatis (Pa) was obtained from the International Genetically Engineered Machine (iGEM) organization catalog (http://parts.igem.org/Main_Page ) (Tables 2 and 3). The genes were amplified by PCR from the iGEM plasmid as a block. acI biosynthetic genes were amplified by PCR from AAA278-O22 genomic DNA as gene clusters. The L06 opsin sequence was obtained as an E. coli-optimized gene from DNA2.0.
TABLE 4.
Primers for this studya
| Category | Name | Sequence (5′–3′) |
|---|---|---|
| Genomic | F_EIB | ATGCAGAGCGCATCAGAGGT |
| R_EIB | TTAGCCAATTCGTGCTGCAA | |
| F_YcYdBlh | TTAGTGTCTTTGCAGCCCT | |
| R_YcYdBlh | CTAACGAAGGTGCGATTTAT | |
| CrtEBI/− and CrtEBI/PaY | F_E_NcoI | aaataattttgtttaactttaataaggagatataccatggATGCAGAGCGCATCAGAGGT |
| R_E_AflII | tgtacaatacgattactttctgttcgacttaagTTAGTAGCTCCTACGGATTGCTGTCTC | |
| F_B_Erbs | agcaatccgtaggagctactaacaagaaggagatatacttATGGATGCAGAACTTGCAGC | |
| R_B_EcoAfl | cgtgtacaatacgattactttctgttcgacttaagaattcttaGCCAATTCGTGCTGCAA | |
| F_E/−_BEcoAfl | attgcagcacgaattggctaagaattcTTAAGTCGAACAGAAAGTAATCGTATTGTACAC | |
| R_E/−_ErbsB | caagttctgcatccataagtatatctccttcttGTTAGTAGCTCCTACGGATTGCTGTCT | |
| F_I_Brbs | gcagcacgaattggctaagaagaaggagatatacttATGACAAGAAAAGTTAAGGGACCC | |
| R_I_EcoRIAflII | tgtacaatacgattactttctgttcgacttaagaattttaCTTGACCGGACCCACAATAC | |
| F_EB/−_IEcoAfl | cgtattgtgggtccggtcaagtaagAATTCTTAAGTCGAACAGAAAGTAATCGTATTGTA | |
| R_EB/−_BrbsI | ggtcccttaacttttcttgtcataagtatatctccttcttCTTAGCCAATTCGTGCTGCA | |
| F_PaY_NdeI | tcttagtatattagttaagtataagaaggagatatacatATGCAACCGCATTATGATCTG | |
| R_PaY_AvrII | atgctagttattgctcagcggtggcagcagcctaggTTAACGATGAGTCGTCATAATGGC | |
| PaCrtEBIY/−, PaCrtEBIY/Blh, and PaCrt EBI/− | F_PaEBIY_NcoI | ataattttgtttaactttaagaaggagatataccatggATGACGGTCTGCGCAAAAAAAC |
| R_PaEBIY_AfIII | acaatacgattactttctgttcgacttaagttatTTAACGATGAGTCGTCATAATGGCTTG | |
| F_PaI_H246H | GCCAGAGTCAGCCAcATGGAAACGACAGGAAAC | |
| R_PaI_H246H | GTTTCCTGTCGTTTCCATgTGGCTGACTCTGGC | |
| F_PaEBIY_PaY | TAATAACTTAAGTCGAACAGAAAGTAATCGTATTGTAC | |
| R_PaEBIY_PaI | agatatacttaagTTATTATATCAGATCCTCCAGCATCAAAC | |
| F_blh_NdeI | tattagttaagtataagaaggagatatacatATGGAGATGGCAAAGTTAAAGACATTTTC | |
| R_blh_AvrII | tattgctcagcggtggcagcagcctaggttaCTAACGAAGGTGCGATTTATTCTTGAGAG | |
| ActR | F_ActRB_NdeI | aaataattttgtttaactttaagaaggagatatacatATGTCGAGCACCATCG |
| R_ActRB_XhoI | atctcagtggtggtggtggtggtgctcgagATGGTCGTCTTTCATGCC |
Annealing regions are indicated with capital letters, while mismatches or overhangs are lowercase.
All genes were individually amplified by PCR, if needed, and added to a cloning pipeline. Primer design included up to 40 bp of cloning site flanking regions from pCDFDuet-1 (Novagen). Genes were placed into the first site between NcoI and AflII restriction sites and between NdeI and AvrII sites of the second site by ligation-free recombination in E. cloni 10G cells (Lucigen). The insert molarity was up to 10-fold more abundant than that of appropriately digested and purified backbone. After selection on 100 µg/ml spectinomycin sulfate LB-Miller agar plates, insert presence was validated by colony PCR using GoTaq Green master mix (Thermo Fisher). Restriction enzyme digestion in lab and Sanger sequencing at the UW Biotechnology Center further confirmed plasmid correctness. pET21b+ (Novagen) was used in the same pipeline, ensuring that genes were in-frame with a C-terminal hexahistidine tag with selection by 100 µg/ml disodium carbenicillin.
Chromophore production and extraction.
For chromophore production, multiple colonies of freshly transformed BL21(DE3) Tuner E. coli (EMD Millipore) were picked into half-full 2-liter flasks of LB-Miller broth plus 100 µg/ml spectinomycin sulfate and shaken in 37°C darkness at 250 rpm for 24 h. ODml (ODml = optical density at 600 nm [OD600] × dilution × volume in milliliters) cells (500 or 1,500) were centrifuged at 3,300 × g, washed in 100 mM Tris (pH 6.8), centrifuged again, and frozen in liquid nitrogen. For chromophore extraction, cells stored at −80°C were thawed at 23°C for up to 30 min. Cells were suspended at 500 ODml cells/3 ml acetone (HPLC grade; Sigma), intensely vortexed for 1 min, and incubated on ice for 5 min. After clarification by centrifugation at 8,000 × g for 10 min, the chromophore-containing supernatant was added to 1 ml/500 ODml of 23°C NaCl-saturated water (American Chemical Society [ACS] grade; Fisher) and 1 ml/500 ODml 23°C dichloromethane (ACS-grade; ACROS) and vortexed for 1 min. Further centrifugation resulted in a colorless aqueous bottom layer and a colorful organic top layer. The organic layer was evaporated under nitrogen, resulting solids were suspended in acetone for carotenoids or ethanol for retinoids, and the resulting solution was filtered through a compatible 4-mm nylon 0.22-µm filter.
UV-Vis and LC-MS/MS analysis of chromophores.
Absorbance spectra were acquired on a Beckman Coulter DU640B spectrophotometer. A Dionex Ultimate 3000 ultra HPLC (UHPLC) coupled by electrospray ionization (ESI; positive mode) to a hybrid quadrupole–high-resolution mass spectrometer (Q Exactive Orbitrap, Thermo Scientific) was used for detection of target compounds based on their accurate masses, mass spectra, and retention times (all matched to purified standards). Liquid chromatography (LC) was based on a published protocol (47). Separation was achieved using a C30 reversed-phase, 150 mm × 2.5 mm, 3-µm particle column (YMC Carotenoid) at a flow rate of 0.2 ml/min. Solvent A consisted of equal parts methanol and water with 0.5% vol/vol acetic acid; solvent B was equal parts methanol and methyl tert-butyl ether with 0.5% vol/vol acetic acid (47). Total run time was 58 min, with the following two gradients: 5 min at 30% B, followed by a 20-min ramp to 100% B and 100% B (retinoids); or 5 min at 30% B, followed by a 25-min ramp to 100% B, and 100% B (carotenoids). MS scans consisted of full positive mode scanning for m/z 200 to 600 from 5 min onwards. In addition, MS/MS scans were obtained by isolating fractions with m/z values of 537.44548 (lycopene), 285.22129 (retinal), and 269.22639 (retinol). MS/MS fragmentations were performed at a normalized-collision energy (NCE) of 30 with an isolation window of m/z 1.4 and a postfragmentation window of m/z 50 to approximately 25 above the isolation mass. For all scans, mass resolution was set at m/z 35,000 ppm, AGC target was 1 × 106 ions, and injection time was 40 ms. Settings for the ion source were as follows: auxiliary gas flow rate, 50; sheath gas flow rate, 10; sweep gas flow rate, 2; spray voltage, 3.5 kV; capillary temperature, 350°C; heater temperature, 250°C; and S-lens RF level, 55.0. Nitrogen was used as the nebulizing gas by the ion trap source. Standards for lycopene (L487500, lot 7ANR-20-1; Toronto Research Chemicals, Inc.) β-carotene (PHR1239, 3 × 100 mg, lot LRAA6761; Sigma-Aldrich), retinal (R2500-25MG, lot SLBN4199V; Sigma-Aldrich), and retinol (R7632-25MG, lot BCBP8066V; Sigma-Aldrich) were analyzed along with experimental samples. Isomer peaks within the mass window result most often from environmentally induced changes during sample preparation. Control cells did not display any significant sustained signal within the time-mass window. Data analysis was performed using Thermo Xcalibur and visualized on MAVEN (67, 68) and Thermo Xcalibur (Thermo Scientific) software.
Proposed gene product and actino-opsin analysis.
Clustal Omega was used to align proposed biosynthetic enzymes and opsin sequences for identification of protein characteristics (69–71). Opsin sequences were submitted to the I-TASSER server for structure prediction using xantho-opsin (PDB 3DDL:A) as the template (72–74). The Basic Local Alignment Search Tool (BLAST) and TM/HMM v2.0 were used for identity percentage calculation and transmembrane helix estimates, respectively (35, 36). PyMOL was used to visualize the resulting protein structures (75).
The actino-opsin phylogeny was reconstructed using opsin protein sequences from bacterial isolate references, SAGs, and MAGs. Opsin protein sequences were aligned with the PROMALS3D multiple sequence and structure alignment tool (76). The alignment was trimmed to exclude positions that contained gaps for more than 30% of the included sequences. Poorly aligned positions and divergent regions were further eliminated by using Gblocks (77). A maximum-likelihood phylogenetic tree was constructed using PhyML v3.0 (78), with the LG substitution model, the gamma distribution parameter estimated by PhyML, and a bootstrap value of 100 replicates. The phylogenetic tree was visualized with Dendroscope v3.2.10 with midpoint root (79). The ActR and other XR sequence subtree was extracted and displayed.
Actinorhodopsin enrichment and analysis.
BL21(DE3) Tuner E. coli cells cotransformed with a plasmid expressing Pa-CrtEBIY/acI-Blh and acI-ActRL06 were grown as during carotenoid production, except that disodium carbenicillin was also added to 100 µg/ml. All further steps were done under red light or darkness at 4°C and/or on ice using 4°C buffers. A sample of 2,000 ODml cells was harvested and suspended in 5 g/ml lysis buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 1 50-ml EDTA-free protease inhibitor tablet [Roche], 1 mg/ml lysozyme, 20 µg/ml DNase I, 5 mM MgCl2, 130 µM CaCl2, and 4 mM phenylmethylsulfonyl fluoride [PMSF]). Cells were lysed at 16,000 lb/in2 by five passes through a French pressure cell. Sequential centrifugation at 10,000 × g for 15 min and 100,000 × g for 45 min cleared debris and pelleted membranes. Membranes were loosened with lysis buffer and transferred to a Potter-Evelhjem homogenizer. After homogenization, membranes were diluted with 4°C lysis buffer to 25 ml and recentrifuged at 100,000 × g for 45 min. Suspension and homogenization were repeated with solubilization buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 2% mass/volume [m/v] dodecyl-maltoside, 10 mM imidazole). Membranes were diluted to 15 ml with solubilization buffer and rocked for 18 h in darkness. Centrifugation at 20,000 × g for 20 min clarified the material before chromatography. The soluble fraction was loaded at 0.5 ml/min onto an equilibrated 1 ml Ni-nitrilotriacetic acid (NTA) column. The processing profile in column volumes (CV) was as follows: 3 solubilization buffer, 12 wash buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 0.05% m/v DDM, and 30 mM imidazole), 5 elution buffer (50 mM Tris-HCl [pH 8.0], 300 mM NaCl, 0.05% m/v DDM, and 500 mM imidazole). The first elution fraction was dialyzed against 1 liter 4°C final buffer (10 mM HEPES [pH 7.5], 100 mM NaCl, 0.05% m/v) for 2 h and then analyzed by absorbance spectroscopy. Light and 50 µl of 23°C saturated hydroxylamine HCl was used to selectively remove retinal from 140 µl of sample to confirm the Schiff base. The resulting unbound retinal oxime absorbs at a wavelength maximum between those of retinal and retinol, where 247- and 257-nm peaks indicate 11-cis and all-trans oxime species, respectively.
Microelectrode pH trace acquisition.
The assay was similar to a published protocol and uses E. coli for expression of holo-ActR (14). All steps were carried out under red light generated by putting red cellophane over a Sylvania F20T12/2364 fluorescent bulb. E. coli cells (500 ODml) were harvested by centrifugation at 3,300 × g for 15 min at 4°C. Cells were suspended in 45 ml 23°C assay solution (10 mM NaCl, 10 mM MgSO4 · 7H2O, 100 μM CaCl2 · 2H2O) and centrifuged at 3,300 × g for 10 min. The latter wash step was repeated. The final suspension was in 20 ml 23°C assay solution to yield an OD of 25. Cells were incubated for 60 min at 23°C to stabilize in darkness before the onset of the assay. The assay was set up with the sample in a glass test tube surrounded by a 300-ml 23°C water bath in a 400-ml beaker. A Mettler-Toledo InLab microprobe (model number 51343160) was clamped in place above the sample tube and connected to a datalogger (model number 850060; Sper Scientific). The entire setup was surrounded on four sides by foil-lined cardboard. Two FE15T8 bulbs (15 W, 700 lumens each) in an 11 × 45 cm housing with 90° reflectors were placed 15 cm from the center of the sample tube, such that light illuminated the entire length of the closest tube side. pH was recorded every second after 3.5 min of equilibration time for 60 min as follows: three cycles of 10 min with fluorescent lights off and 10 min with fluorescent lights on. Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) (L06932, lot 10181844; Alfa Aesar) was added to a final concentration of 20 μM during dark stabilization after 45 min.
Data availability.
All genomic and metatranscriptomic sequences are available through the Integrated Microbial Genomes (IMG) and National Center for Biotechnology Information (NCBI) databases, respectively. Genome sequences can be accessed using IMG taxon ID numbers provided in Table S1. The raw RNA sequences can be found in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information under BioProject accession no. PRJNA362825.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institute of Food and Agriculture, U.S. Department of Agriculture (grants 2016-67012-24709 to J.J.H. and WIS01789 to K.D.M.), the U.S. National Science Foundation (NTL-LTER DEB-1440297 and DEB-1344254 to K.D.M. and MCB-1518160 to K.T.F.) and the National Oceanic and Atmospheric Administration (NA10OAR4170070 to K.D.M. and K.T.F. via the Wisconsin Sea Grant College Program Project #HCE-25).
We thank former and current McMahon Laboratory members for help obtaining transcript data, Diego Yanez for assisting with RNA methodology, and James Steele for supplying plasmid iGEM DNA.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01678-18.
REFERENCES
- 1.Ghai R, McMahon KD, Rodriguez-Valera F. 2012. Breaking a paradigm: cosmopolitan and abundant freshwater actinobacteria are low GC: freshwater actinobacteria are low GC. Environ Microbiol Rep 4:29–35. doi: 10.1111/j.1758-2229.2011.00274.x. [DOI] [PubMed] [Google Scholar]
- 2.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghylin TW, Garcia SL, Moya F, Oyserman BO, Schwientek P, Forest KT, Mutschler J, Dwulit-Smith J, Chan L-K, Martinez-Garcia M, Sczyrba A, Stepanauskas R, Grossart H-P, Woyke T, Warnecke F, Malmstrom R, Bertilsson S, McMahon KD. 2014. Comparative single-cell genomics reveals potential ecological niches for the freshwater acI Actinobacteria lineage. ISME J 8:2503. doi: 10.1038/ismej.2014.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kang I, Kim S, Islam MR, Cho J-C. 2017. The first complete genome sequences of the acI lineage, the most abundant freshwater Actinobacteria, obtained by whole-genome-amplification of dilution-to-extinction cultures. Sci Rep 7:42252. doi: 10.1038/srep42252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuenschwander SM, Ghai R, Pernthaler J, Salcher MM. 2018. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J 12:185–198. doi: 10.1038/ismej.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warnecke F, Amann R, Pernthaler J. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ Microbiol 6:242–253. doi: 10.1111/j.1462-2920.2004.00561.x. [DOI] [PubMed] [Google Scholar]
- 7.Warnecke F, Sommaruga R, Sekar R, Hofer JS, Pernthaler J. 2005. Abundances, identity, and growth state of actinobacteria in mountain lakes of different UV transparency. Appl Environ Microbiol 71:5551–5559. doi: 10.1128/AEM.71.9.5551-5559.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia SL, McMahon KD, Martinez-Garcia M, Srivastava A, Sczyrba A, Stepanauskas R, Grossart H-P, Woyke T, Warnecke F. 2013. Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J 7:137. doi: 10.1038/ismej.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 10.Šimek K, Nedoma J, Znachor P, Kasalický V, Jezbera J, Hornňák K, Sed’a J. 2014. A finely tuned symphony of factors modulates the microbial food web of a freshwater reservoir in spring. Limnol Oceanogr 59:1477–1492. doi: 10.4319/lo.2014.59.5.1477. [DOI] [Google Scholar]
- 11.Sharma AK, Sommerfeld K, Bullerjahn GS, Matteson AR, Wilhelm SW, Jezbera J, Brandt U, Doolittle WF, Hahn MW. 2009. Actinorhodopsin genes discovered in diverse freshwater habitats and among cultivated freshwater Actinobacteria. ISME J 3:726. doi: 10.1038/ismej.2009.13. [DOI] [PubMed] [Google Scholar]
- 12.Sharma AK, Zhaxybayeva O, Papke RT, Doolittle WF. 2008. Actinorhodopsins: proteorhodopsin-like gene sequences found predominantly in non-marine environments. Environ Microbiol 10:1039–1056. doi: 10.1111/j.1462-2920.2007.01525.x. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton JJ, Garcia SL, Brown BS, Oyserman BO, Moya-Flores F, Bertilsson S, Malmstrom RR, Forest KT, McMahon KD. 2017. Metabolic network analysis and metatranscriptomics reveal auxotrophies and nutrient sources of the cosmopolitan freshwater microbial lineage acI. mSystems 2:e00091-17. doi: 10.1128/mSystems.00091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keffer JL, Hahn MW, Maresca JA. 2015. Characterization of an unconventional rhodopsin from the freshwater actinobacterium Rhodoluna lacicola. J Bacteriol 197:2704–2712. doi: 10.1128/JB.00386-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura S, Kikukawa T, Tamogami J, Kamiya M, Aizawa T, Hahn MW, Ihara K, Kamo N, Demura M. 2016. Photochemical characterization of actinorhodopsin and its functional existence in the natural host. Biochim Biophys Acta 1857:1900–1908. doi: 10.1016/j.bbabio.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Sabehi G, Loy A, Jung K-H, Partha R, Spudich JL, Isaacson T, Hirschberg J, Wagner M, Béjà O. 2005. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol 3:e273. doi: 10.1371/journal.pbio.0030273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchman DL, Hanson TE. 2013. Bioenergetics of photoheterotrophic bacteria in the oceans. Environ Microbiol Rep 5:188–199. doi: 10.1111/j.1758-2229.2012.00367.x. [DOI] [PubMed] [Google Scholar]
- 18.Spudich JL, Yang C-S, Jung K-H, Spudich EN. 2000. Retinylidene proteins: structures and functions from archaea to humans. Annu Rev Cell Dev Biol 16:365–392. doi: 10.1146/annurev.cellbio.16.1.365. [DOI] [PubMed] [Google Scholar]
- 19.Man D, Wang W, Sabehi G, Aravind L, Post AF, Massana R, Spudich EN, Spudich JL, Béjà O. 2003. Diversification and spectral tuning in marine proteorhodopsins. EMBO J 22:1725–1731. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beja O, Lanyi JK. 2014. Nature’s toolkit for microbial rhodopsin ion pumps. Proc Natl Acad Sci U S A 111:6538–6539. doi: 10.1073/pnas.1405093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson JA, Krinsky NI. 1995. Introduction: the colorful, fascinating world of the carotenoids: important physiologic modulators. FASEB J 9:1547–1550. doi: 10.1096/fasebj.9.15.8529833. [DOI] [PubMed] [Google Scholar]
- 22.Gruszecki WI, Strzałka K. 2005. Carotenoids as modulators of lipid membrane physical properties. Biochim Biophys Acta BBA - Mol Basis Dis 1740:108–115. doi: 10.1016/j.bbadis.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Britton G, Liaaen-Jensen S, Pfander H (ed). 2004. Carotenoids. Springer Basel AG, Basel, Switzerland. [Google Scholar]
- 24.Graham JE, Bryant DA. 2009. The biosynthetic pathway for myxol-2′ fucoside (myxoxanthophyll) in the cyanobacterium Synechococcus sp. strain PCC 7002. J Bacteriol 191:3292–3300. doi: 10.1128/JB.00050-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luecke H, Schobert B, Stagno J, Imasheva ES, Wang JM, Balashov SP, Lanyi JK. 2008. Crystallographic structure of xanthorhodopsin, the light-driven proton pump with a dual chromophore. Proc Natl Acad Sci U S A 105:16561–16565. doi: 10.1073/pnas.0807162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balashov SP. 2005. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science 309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balashov SP, Imasheva ES, Choi AR, Jung K-H, Liaaen-Jensen S, Lanyi JK. 2010. Reconstitution of gloeobacter rhodopsin with echinenone: role of the 4-keto group. Biochemistry 49:9792–9799. doi: 10.1021/bi1014166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steindler L, Schwalbach MS, Smith DP, Chan F, Giovannoni SJ. 2011. Energy starved Candidatus Pelagibacter ubique substitutes light-mediated ATP production for endogenous carbon respiration. PLoS One 6:e19725. doi: 10.1371/journal.pone.0019725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casey JR, Ferrón S, Karl DM. 2017. Light-enhanced microbial organic carbon yield. Front Microbiol 8. doi: 10.3389/fmicb.2017.02157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia SL, McMahon KD, Grossart H-P, Warnecke F. 2014. Successful enrichment of the ubiquitous freshwater acI Actinobacteria. Environ Microbiol Rep 6:21–27. doi: 10.1111/1758-2229.12104. [DOI] [PubMed] [Google Scholar]
- 31.Kim S, Kang I, Seo J-H, Cho J-C. 2018. Culturing the ubiquitous freshwater actinobacterial acI lineage by supplying a biochemical “helper” catalase. bioRxiv 10.1101/343640. [DOI] [PMC free article] [PubMed]
- 32.Subramaniam S. 1999. The structure of bacteriorhodopsin: an emerging consensus. Curr Opin Struct Biol 9:462–468. doi: 10.1016/S0959-440X(99)80065-7. [DOI] [PubMed] [Google Scholar]
- 33.Ozaki Y, Kawashima T, Abe-Yoshizumi R, Kandori H. 2014. A color-determining amino acid residue of proteorhodopsin. Biochemistry 53:6032–6040. doi: 10.1021/bi500842w. [DOI] [PubMed] [Google Scholar]
- 34.Iniesta AA, Cervantes M, Murillo FJ. 2008. Conversion of the lycopene monocyclase of Myxococcus xanthus into a bicyclase. Appl Microbiol Biotechnol 79:793–802. doi: 10.1007/s00253-008-1481-7. [DOI] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes11Edited by F. Cohen. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 37.Arrach N, Fernández-Martín R, Cerdá-Olmedo E, Avalos J. 2001. A single gene for lycopene cyclase, phytoene synthase, and regulation of carotene biosynthesis in Phycomyces. Proc Natl Acad Sci U S A 98:1687–1692. doi: 10.1073/pnas.021555298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim Y-S, Kim N-H, Yeom S-J, Kim S-W, Oh D-K. 2009. In vitro characterization of a recombinant Blh protein from an uncultured marine bacterium as a β-carotene 15,15′-dioxygenase. J Biol Chem 284:15781–15793. doi: 10.1074/jbc.M109.002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee PC, Holtzapple E, Schmidt-Dannert C. 2010. Novel activity of Rhodobacter sphaeroides spheroidene monooxygenase CrtA expressed in Escherichia coli. Appl Environ Microbiol 76:7328–7331. doi: 10.1128/AEM.01606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rählert N, Fraser PD, Sandmann G. 2009. A crtA-related gene from Flavobacterium P99-3 encodes a novel carotenoid 2-hydroxylase involved in myxol biosynthesis. FEBS Lett 583:1605–1610. doi: 10.1016/j.febslet.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 41.Sun Z, Shen S, Wang C, Wang H, Hu Y, Jiao J, Ma T, Tian B, Hua Y. 2009. A novel carotenoid 1,2-hydratase (CruF) from two species of the non-photosynthetic bacterium Deinococcus. Microbiology 155:2775–2783. doi: 10.1099/mic.0.027623-0. [DOI] [PubMed] [Google Scholar]
- 42.Takaichi S, Maoka T, Takasaki K, Hanada S. 2010. Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2′-dirhamnoside. Microbiology 156:757–763. doi: 10.1099/mic.0.034249-0. [DOI] [PubMed] [Google Scholar]
- 43.Ahn J-W, Kim K-J. 2015. Crystal structure of 1′-OH-carotenoid 3,4-desaturase from Nonlabens dokdonensis DSW-6. Enzyme Microb Technol 77:29–37. doi: 10.1016/j.enzmictec.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Teramoto M, Rählert N, Misawa N, Sandmann G. 2004. 1-Hydroxy monocyclic carotenoid 3,4-dehydrogenase from a marine bacterium that produces myxol. FEBS Lett 570:184–188. doi: 10.1016/j.febslet.2004.05.085. [DOI] [PubMed] [Google Scholar]
- 45.Maresca JA, Bryant DA. 2006. Two genes encoding new carotenoid-modifying enzymes in the green sulfur bacterium Chlorobium tepidum. J Bacteriol 188:6217–6223. doi: 10.1128/JB.00766-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klassen JL. 2010. Phylogenetic and evolutionary patterns in microbial carotenoid biosynthesis are revealed by comparative genomics. PLoS ONE 5:e11257. doi: 10.1371/journal.pone.0011257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Breemen RBV, Huang CR. 1996. High-performance liquid chromatography-electrospray mass spectrometry of retinoids. FASEB J 10:1098–1101. doi: 10.1096/fasebj.10.9.8801173. [DOI] [PubMed] [Google Scholar]
- 48.Jang H-J, Ha B-K, Zhou J, Ahn J, Yoon S-H, Kim S-W. 2015. Selective retinol production by modulating the composition of retinoids from metabolically engineered E. coli. Biotechnol Bioeng 112:1604–1612. doi: 10.1002/bit.25577. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez GM, Atsumi S. 2014. Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab Eng 25:227–237. doi: 10.1016/j.ymben.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newton RJ, McMahon KD. 2011. Seasonal differences in bacterial community composition following nutrient additions in a eutrophic lake: Seasonal variation in nutrient-amended microcosms. Environ Microbiol 13:887–899. doi: 10.1111/j.1462-2920.2010.02387.x. [DOI] [PubMed] [Google Scholar]
- 51.Hall MW, Rohwer RR, Perrie J, McMahon KD, Beiko RG. 2017. Ananke: temporal clustering reveals ecological dynamics of microbial communities. PeerJ 5:e3812. doi: 10.7717/peerj.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berry MA, Davis TW, Cory RM, Duhaime MB, Johengen TH, Kling GW, Marino JA, Den Uyl PA, Gossiaux D, Dick GJ, Denef VJ. 2017. Cyanobacterial harmful algal blooms are a biological disturbance to Western Lake Erie bacterial communities. Environ Microbiol 19:1149–1162. doi: 10.1111/1462-2920.13640. [DOI] [PubMed] [Google Scholar]
- 53.Marasco EK, Vay K, Schmidt-Dannert C. 2006. Identification of carotenoid cleavage dioxygenases from Nostoc sp. PCC 7120 with different cleavage activities. J Biol Chem 281:31583–31593. doi: 10.1074/jbc.M606299200. [DOI] [PubMed] [Google Scholar]
- 54.Ahrazem O, Gómez-Gómez L, Rodrigo MJ, Avalos J, Limón MC. 2016. Carotenoid cleavage oxygenases from microbes and photosynthetic organisms: features and functions. Int J Mol Sci 17:E1781. doi: 10.3390/ijms17111781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu X, Jiang J, Hu J. 2013. Determination and occurrence of retinoids in a eutrophic lake (Taihu Lake, China): cyanobacteria blooms produce teratogenic retinal. Environ Sci Technol 47:807–814. doi: 10.1021/es303582u. [DOI] [PubMed] [Google Scholar]
- 56.Bertsova YV, Arutyunyan AM, Bogachev AV. 2016. Na+-translocating rhodopsin from Dokdonia sp. PRO95 does not contain carotenoid antenna. Biochemistry (Mosc) 81:414–419. doi: 10.1134/S000629791604012X. [DOI] [PubMed] [Google Scholar]
- 57.Imasheva ES, Balashov SP, Choi AR, Jung K-H, Lanyi JK. 2009. Reconstitution of Gloeobacter violaceus rhodopsin with a light-harvesting carotenoid antenna. Biochemistry 48:10948–10955. doi: 10.1021/bi901552x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian B, Hua Y. 2010. Carotenoid biosynthesis in extremophilic Deinococcus–Thermus bacteria. Trends Microbiol 18:512–520. doi: 10.1016/j.tim.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 59.Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joshi N, Fass J. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33). https://github.com/najoshi/sickle.
- 61.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kopylova E, Noé L, Touzet H. 2012. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 63.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, Finn RD. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 67.Clasquin MF, Melamud E, Rabinowitz JD. 2012. LC-MS data processing with MAVEN: a metabolomic analysis and visualization engine, p 14.11.1–14.11.23 In Baxevanis AD, Petsko GA, Stein LD, Stormo GD (ed), Current protocols in bioinformatics. John Wiley & Sons, Inc, Hoboken, NJ. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melamud E, Vastag L, Rabinowitz JD. 2010. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem 82:9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2014. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43:W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schrödinger, LLC. The PyMOL Molecular Graphics System, V2.0.0. Schrödinger, LLC, New York, NY. [Google Scholar]
- 76.Pei J, Kim B-H, Grishin NV. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 78.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 79.Huson DH, Scornavacca C. 2012. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic and metatranscriptomic sequences are available through the Integrated Microbial Genomes (IMG) and National Center for Biotechnology Information (NCBI) databases, respectively. Genome sequences can be accessed using IMG taxon ID numbers provided in Table S1. The raw RNA sequences can be found in the Sequence Read Archive (SRA) of the National Center for Biotechnology Information under BioProject accession no. PRJNA362825.