Abstract
Background
Ventilator-associated pneumonia (VAP) is one of the most commonly encountered intensive care unit (ICU) acquired infections worldwide. The objective of the study was to identify the immune alteration occurring in patients suffering from VAP at the transcriptomic level and explore its potential use for clinical diagnoses of this disease.
Methods
We performed a prospective observational study in five medical ICUs. Immunological gene expression profiles in the blood of VAP patients were compared with those of controls by using whole transcriptome microarrays and droplet digital polymerase chain reaction (ddPCR) in the first 24 hours following diagnosis.
Results
VAP patients showed significantly lower expression levels of HLA-DOA, HLA-DMA, HLA-DMB, ICOS, ICOSLG, IL2RA, CD1, CD3, CD28 and CD40LG. The molecules coded by these genes participate of the immunological synapse. CD1C, CD40LG and ICOS showed the highest values of area under the receiver operating characteristic curve (AUROC) with a good balance between sensibility and specificity.
Conclusions
Patients with VAP show a transcriptomic depression of genes participating of the immunological synapse. It takes a commonplace event, namely VAP, and highlights a quite significant underlying immune suppressive state. In effect this small study will change how we regard VAP, and proposes that we regard it as an infection in an immune compromised host, and that immunity has a central role for ICU acquired infections. This may in time change clinical practice, as it has profound implications for the role of protocolised care, or bundles, in the prevention of VAP. Quantifying the expression in blood of this genes using ddPCR could be a useful approach for the diagnosis of VAP.
Keywords: Ventilator-associated pneumonia (VAP), immunological synapse, microarrays, droplet digital polymerase chain reaction (ddPCR)
Introduction
Ventilator-associated pneumonia (VAP) is the most frequent intensive care unit (ICU)-acquired infection among patients on mechanical ventilation (MV). Experts estimate that VAP affects between 9% and 27% of intubated patients (1). This common complication had major consequences for intubated patient outcome, estimating the attributable mortality of VAP between 9% to 13% of all the deaths in mechanically ventilated patients (2). VAP, being a clinical diagnosis, is difficult to detect.
Increasing evidence shows that critically ill infected patients display alterations in their immune response that could impair infection control in these patients (3,4). Patients with severe infections, like sepsis, exhibit innate and adaptive immune response dysfunction which confers an increased risk of organ failure and mortality (4-6).
While there is an extensive literature describing the immune disorders occurring in sepsis (7,8), little has being done in this regard in the field of VAP. Transcriptomic analysis of circulating peripheral blood leucocytes has demonstrated to be a valuable approach for evaluating immunity in different infectious diseases (9-11). Moreover, this kind of analysis has been also used for identifying new molecular markers for diagnosis purposes (12).
The objective of the study was to identify immune alterations occurring in patients suffering from VAP at the transcriptomic level. For this, we conducted a prospective observational cohort study enrolling immune-competent adults with VAP, using a group of mechanically ventilated patients with no infection as controls. Two different molecular methods, whole transcriptome microarrays and droplet digital polymerase chain reaction (ddPCR) were employed for the transcriptomic analysis. Profiling immunological alterations in VAP could help to better understand the pathogenesis of this disease and contribute to improve its diagnosis, by providing an objective, gene expression-based test, which could distinguish infected from uninfected patients.
Methods
Study design and ethics statement
We performed a prospective observational study in five medical ICUs, in order to evaluate immunological alterations of patients with VAP. Patients under MV (for at least 48 hours) were recruited and monitored for the development of VAP. VAP was diagnosed when the patient fulfilled two or more of the following criteria: temperature >38 °C, white cell count >12,000 or <4,000/mm3, or presence of purulent respiratory secretions, plus a new or progressive pulmonary infiltrate on the chest X-ray (13). Exclusion criteria included the following: immunosuppression [patients with human immunodeficiency virus (HIV) infection and those undergoing radiotherapy or receiving immunosuppressive drugs, including chemotherapy or systemic steroids, in the last 3 months prior to admission to the ICU were considered to be immunosuppressed], patients presenting at the ICU with severe sepsis/septic shock without informed consent. A total of 30 patients with VAP were included in the study. A control group of 20 patients under MV who did not develop an infection during hospitalization at the ICU were enrolled in the study; 15 healthy controls recruited in the Blood Donation Center of Castilla y León (CHEMCYL) were also included for microarrays baseline normalization. Given the known difficulty with calculating sample size for microarray based studies, final “n” in our study groups was calculated based on our previous experience in similar studies (14). The study was approved by the Institutional Review Board of all participating hospitals, and informed consent was obtained from the patients’ relatives.
Clinical data
A specific standard survey was employed to collect the clinical data, including medical history, physical examination and hematological, biochemical, radiological and microbiological investigations. Treatment decisions were not standardized for all patients but were made by the treating physician.
Clinical data analysis
Data were analyzed by using the IBM SPSS 22.0 software (SPSS, Chicago, IL, USA). Differences in demographic and clinical characteristics between patient groups were assessed using the χ2 test for categorical variables and the Mann Whitney U test for continuous variables.
Sample collection
A sample of 2.5 mL of blood was collected in the first 24 hours following diagnosis of VAP using Paxgene venous blood vacuum collection tubes (Becton Dickinson, USA). Time of sample collection in the uninfected control group was pre-established at day 7 in the study protocol, to minimize the impact of ventilation duration on the potential differences in gene expression profiles between groups (VAP occurred at day 8 in median in our study).
Microarray processing
Gene expression levels of VAP patients and controls were examined using Agilent human oligo microarrays as previously described (9). Total RNA was extracted from blood samples using the PAXgene Blood RNA System (PreAnalytix, Hombrechtikon, Switzerland). RNA quality and concentration were analyzed by RNA Experion Bioanalyzer (BioRad, California, USA) assay and spectrometry (NanoDrop ND1000, NanoDrop Technologies, Wilminton, Delaware, USA). Samples from nine patients had insufficient RNA concentration for the microarray works [6 VAP and 3 mechanical ventilation controls (MVCs)], and in consequence they were removed from the study. Each RNA sample was concentrated with the RNeasy MinElute Cleanup kit (QIAGEN, Hilden, Germany). RNA was eluted with 10 microliters of RNase-free H2O; 100 ng of purified total RNA were used to produce Cyanine 3-CTP-labeled cRNA using the Quick Amp Labeling kit (Agilent p/n 5190-0442) according to the manufacturer’s instructions. cRNA amplification, hybridization, and scanning of hybridized arrays were performed according to “One-Color Microarray-Based Gene Expression Analysis” protocol Version 6.9. Data was extracted by using Agilent Feature Extraction Software 9.5.3 following the Agilent GE1-v5_95_Feb07 and the QC Metric Set GE1_QCMT_Jan08 protocols.
Microarray data accession number
Microarray expression data sets were uploaded at the Array Express microarray data repository and are available publicly under accession number E-MTAB-5638.
Microarray analysis
Raw data was collected with GeneSpring GX 12.0 software. The data sets were pre-processed as previously described (9). Briefly, local background was subtracted, log-transformed signal intensity values were globally normalized using the 75th percentile of each sample and baseline transformation was performed using the median of the healthy control samples. Before statistical analyses all microarrays were subjected to quality and filtering criteria. Quality of the microarray data was assessed on Principal Component analysis (PCA) plots. Mann-Whitney test (GeneSpring GX12.0) was used to identify genes differentially expressed between groups. The level of significance was fixed in P<0.05, with Benjamini-Hochberg multiple testing corrections and Fold change >1.5. Ingenuity pathway analysis (IPA) (Ingenuity Systems-Quiagen, Redwood City, CA, USA) was used to select the immunological pathways that were enriched among the differentially expressed genes. Only pathways related with “Cellular Immune Response”, “Cytokine Signaling” and “Humoral Immune Response” were included in IPA analysis.
ddPCR
Changes in microarray gene expression for the most representative genes of our analysis were verified by ddPCR (Bio-Rad, California, USA) using pre-designed TaqMan® Assay Primer/Probe Sets (FAM labelled MGB probes, Thermo Fisher/Scientific-Life Technologies, Waltham, MA, USA). Always exactly 1,000 ng RNA were transcribed to cDNA by using the iScript™ Advanced cDNA Synthesis Kit (Biorad, California, USA, cat: 1725038). The resulting product was further diluted (1/25) and 2.5 µL (5 ng of total mRNA) were employed for quantification of target gene expression according to the manufacturer instruction’s. ddPCR was performed using the Bio-Rad QX200™ Droplet Digital™ PCR system, ddPCR™ Supermix for Probes (No dUTP), and Bio-Rad standard reagents for droplet generation and reading. End-point PCR with 40 cycles was performed by using C1000 Touch™ Thermal Cycler (BioRad, California, USA) after splitting each sample into about 20,000 droplets. Next, the droplet reader used at least 10,000 droplets to determine the percentage of positive droplets and calculation of copy number of cDNA per ng of initial mRNA.
ddPCR statistical analysis
Differences in gene expression levels were assessed using Mann Whitney U test (IBM SPSS 22.0 software, Chicago, IL, USA). The correlation between gene expression levels and clinical variables was evaluated using the Spearman rank correlation test. Area under the receiver operating characteristic curve (AUROC) was used to assess accuracy of mRNA levels to differentiate VAP from uninfected controls (MVC). The optimal operating point (OOP) was the value for which the point on the curve had the minimum distance to the upper left corner (where sensitivity =1 and specificity =1), and was calculated as previously described (15).
Results
Clinical characteristics
Forty-one patients receiving MV were finally included in the study (24 patients with VAP and 17 non-infected MVC patients). Causes of ICU-admission and patient baseline characteristics are summarized in Table 1. Both groups were comparable in terms of age, gender and main comorbidities. The main reason for orotracheal intubation was low-consciousness level (72.5%), followed by cardiovascular failure. As expected, VAP group showed a lower PaO2/FiO2 ratio and higher Clinical Pulmonary Infection Score (CPIS) score compared with non-infected patients. Analytical data [including white cell count, procalcitonin (PCT) and C-reactive protein (CRP)], disease severity at ICU admission [assessed by Acute Physiology and Chronic Health Evaluation (APACHE) II and sepsis organ failure score (SOFA score)] and at diagnosis were also similar in both groups.
Table 1. Clinical characteristics of patients included in the study.
| Characteristics | MVC | VAP | All patients | P (VAP vs. MVC) |
|---|---|---|---|---|
| Number, n (%) | 17 (42.0) | 24 (58.0) | 41 | – |
| Age (years), median ± IQR | 68±26 | 54±23 | 55±25 | – |
| Sex (male), n (%) | 10 (58.8) | 17 (70.8) | 27 (65.9) | – |
| APACHE II score, median ± IQR | 22±11 | 21±23 | 22±12 | – |
| Comorbidities, n (%) | ||||
| Hypertension | 6 (35.3) | 1 (4.2) | 7 (17.1) | 0.01 |
| Diabetes mellitus | 5 (29.4) | 6 (25.0) | 11 (26.8) | – |
| Chronic cardiac failure | 2 (11.8) | 4 (16.7) | 6 (14.6) | – |
| Chronic renal failure | 1 (5.9) | 0 (0.0) | 1 (2.4) | – |
| Chronic lung disease/COPD | 2 (11.8) | 9 (37.5) | 11(26.8) | – |
| Chronic hepatic failure | 3 (17.6) | 1 (4.2) | 4 (9.8) | – |
| Reason for intubation, n (%) | ||||
| Respiratory failure | 0 (0.0) | 5 (21.7) | 5 (12.5) | 0.04 |
| Cardiovascular failure | 2 (11.8) | 4 (17.4) | 6 (15.0) | – |
| Coma (low-consciousness level) | 15 (88.2) | 14 (60.9) | 29 (72.5) | 0.05 |
| At diagnosis, median ± IQR | ||||
| SOFA score | 5±5 | 7±3 | 6±3.5 | – |
| CPIS score | 3±2 | 7±1 | 6±4 | <0,01 |
| PaO2/FiO2 | 235±62 | 160±90 | 214±102.5 | 0.03 |
| Temperature (°C) | 37±1 | 38±0 | 37.6±1 | <0.01 |
| CRP (mg/L) | 150±126 | 164±162 | 164±171 | – |
| PCT (ng/mL) | 0.4±1.55 | 0.1±0.85 | 0.19±1.07 | – |
| Creatinine (mg/dL) | 0.71±0.29 | 0.78±0.45 | 0.72±0.38 | – |
| Leukocytes (cells/mm3) | 10,590±2,000 | 11,630±5,875 | 11,110±4,365 | – |
| Lymphocytes (cells/mm3) | 1,160±683 | 1,208±623 | 1,180±721 | – |
| Neutrophils (cells/mm3) | 8,132±2,161 | 9,185±5,312 | 8,670±4,434 | – |
| Positive culture, n (%) | 0 (0.0) | 15 (62.5) | 15 (36.6) | N/A |
| ICU stay (days), median ± IQR | 17±13 | 25±30 | 20±16 | 0.01 |
| Hospital stay (days), median ± IQR | 28±49 | 32±32 | 31.5±40.25 | – |
| Days of MV, median ± IQR | 12±4 | 20±21.5 | 15±14.25 | <0,01 |
| 28-day mortality, n (%) | 3 (17.6) | 4 (16.7) | 7 (17.1) | – |
| Hospital mortality, n (%) | 4 (23.5) | 7 (29.2) | 11 (26.8) | – |
| ICU mortality, n (%) | 3 (17.6) | 7 (29.2) | 10 (24.4) | – |
MVC, mechanical ventilation control; VAP, ventilator-associated pneumonia; APACHE II, Acute Physiology and Chronic Health Evaluation II; COPD, chronic obstructive pulmonary disease; SOFA, sepsis organ failure score; CPIS, clinical pulmonary infection score; CRP, C-reactive protein; PCT, procalcitonin; PaO2/FiO2, ratio of partial pressure of arterial oxygen to fraction of inspired oxygen; MV, mechanical ventilation; ICU, intensive care unit; N/A; not applicable; IQR, interquartile range.
In the MVC group, microbiological cultures from the lower respiratory tract were negative during hospitalization. Positive cultures were observed in 62.5% of the patients in the VAP group. Most common agents of microbial diagnosis were bacterial. In the 25.0% of the cases, the infection was caused by Enterobacteriae, 16.7% by Staphylococcus aureus and 12.5% by non-fermenting Gram-negative bacteria.
VAP patients were mechanically ventilated for longer than controls. In addition, they stayed longer in the ICU and in the hospital. Finally, no differences in mortality were obvious between VAP and MVC groups.
Gene expression profiles in VAP
A microarray approach was employed to compare gene expression profiles in peripheral blood from VAP and MVC groups. A total of 1,231 genes showed significant differences in their expression levels between both groups: expression of 680 genes were increased in VAP patients while expression of 551 genes was decreased in this group.
IPA identified eight immunological pathways that were enriched among the differentially expressed genes (Figure 1). The vast majority of the genes participating in these pathways corresponded to genes that were less expressed in patients with VAP. Remarkably, the molecules coded by these genes participate of the immunological synapse, HLA-DOA, HLA-DMA, HLA-DMB, ICOS, ICOSLG, IL2RA, CD1, CD3, CD28 and CD40LG (Figure 2).
Figure 1.
Immunological signaling pathways identified by IPA. The upper x-axis represents the percent of genes of each pathway whose showed significant differences in their expression levels between VAP patients and MVC group. VAP, ventilator-associated pneumonia; MVC, mechanical ventilation control.
Figure 2.
T helper cell differentiation signaling pathway. “Ingenuity pathway analysis” identified this route as one of the most altered pathways of the analysis. Green: genes down-regulated in the VAP patients compared with MVC group. VAP, ventilator-associated pneumonia; MVC, mechanical ventilation control.
Expression quantification of the immunological synapse genes by ddPCR
Based on the IPA results, we selected some of the genes involved in the immunological synapse for validation of the microarray study. Accordingly, CD40LG, CD28, IL-2RA, ICOS, CD1, CD3 and CCR7 expression levels were quantified by using ddPCR. As occurred in the microarray study, in the ddPCR study VAP patients showed significantly lower expression levels of these genes as compared to the MVC group (Figure 3).
Figure 3.
Box plots showing gene expression levels of immunological synapse genes quantifying by ddPCR. *, represents significant differences between VAP patients and MVC group. VAP, ventilator-associated pneumonia; MVC, mechanical ventilation control; ddPCR, droplet digital polymerase chain reaction.
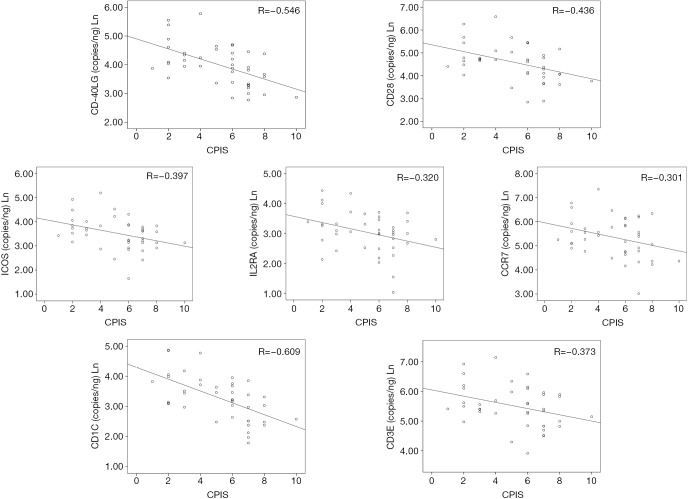
Correlation between immunological synapse gene expression levels and CPIS score
In order to evaluate the relationship between expression levels of the genes evaluated by ddPCR with the clinical presentation of the disease, a Spearman correlation test with the CPIS score was performed. The CPIS is a semi-objective assessment of several clinical factors predictive of pneumonia; temperature, white blood cell count, presence and character of respiratory secretions, PaO2/FiO2 (arterial oxygen partial pressure/inspired oxygen fraction) ratio, and chest radiograph findings (16). Expression levels of all the genes tested except those of CCR7 showed a significant inverse correlation with the CPIS score (Figure 4).
Figure 4.
Dot plots showing the correlations between gene expression levels and the CPIS score in the group of VAP patients. VAP, ventilator-associated pneumonia; R, correlation coefficient; CPIS, clinical pulmonary infection score.
Diagnostic accuracy of immunological synapse genes
Accuracy of the genes quantified by ddPCR for differentiating between VAP and MVC was evaluated by AUROC analysis. All the genes evaluated in this analysis yielded AUROCs >0.7, with CD1C, CD40LG and ICOS showing the highest AUROC values (Figure 5, Table 2). In addition, the OOP calculated for these genes showed a good balance between sensibility and specificity (Table 2).
Figure 5.
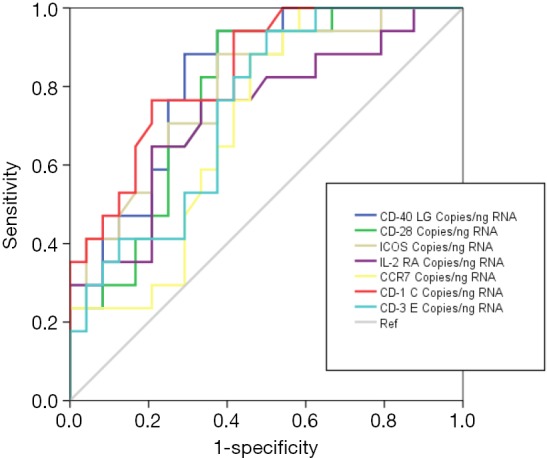
Graphical representation of the AUROCs for differential diagnosis between patients with VAP and MVCs (MVC is taken as the reference). VAP, ventilator-associated pneumonia; MVC, mechanical ventilation control.
Table 2. Diagnostic accuracy of immunological synapse genes.
| Gene | AUROC | |||||
|---|---|---|---|---|---|---|
| Area | 95% CI | P | OPP copies/ng total mRNA | Sensitivity | Specificity | |
| CD1C | 0.838 | 0.719–0.957 | <0.001 | 30.40 | 0.765 | 0.792 |
| CD40LG | 0.821 | 0.695–0.948 | 0.001 | 50.60 | 0.882 | 0.708 |
| ICOS | 0.793 | 0.654–0.931 | 0.002 | 38.00 | 0.706 | 0.750 |
| CD28 | 0.778 | 0.637–0.919 | 0.003 | 102.00 | 0.824 | 0.667 |
| CD3E | 0.743 | 0.593–0.892 | 0.009 | 22.60 | 0.765 | 0.625 |
| IL2RA | 0.733 | 0.573–0.892 | 0.012 | 20.80 | 0.765 | 0.667 |
| CCR7 | 0.707 | 0.549–0.866 | 0.025 | 158.60 | 0.882 | 0.541 |
AUROC for differential diagnosis between patients with VAP and MVCs are presented here. Results for gene expression levels are provided as cDNA copies/ng total mRNA. OPP, optimal operating point; AUROC, areas under receiving operating curves; VAP, ventilator-associated pneumonia; MVC, mechanical ventilation control.
Discussion
Our work shows for the first time the existence, in patients with VAP, of a transcriptomic signature denoting depression of the immunological synapse. Absolute quantification of expression levels in blood of genes participating of the immunological synapse by ddPCR is a potential useful tool for the diagnosis of VAP.
Increasing evidence supports the existence of immunological alterations in critically ill patients suffering from infection. In this regard, reports from our group and others have identified the existence of different features of immunological dysfunction conferring risk of organ failure and death in sepsis patients (4).
In the present study, we evaluated the immune alterations at the mRNA level of a group of patients who developed ventilator associated pneumonia by using two different approaches, microarrays (as a discovery tool) and ddPCR (as a validation tool, with more feasible clinical application). The microarray study identified eight immunological pathways whose genes showed depressed expression levels in VAP, compared with non-infected ventilated patients. The vast majority of these pathways were involved in the development of cellular immune responses, particularly in the immunological synapse between antigen presentation cells and lymphocytes. Immunological synapses are dynamically organized cell-cell interfaces formed between cells of the immune system (17). These cell connections involve a large number of receptors and co-receptors expressed on the surface of T and B cells and other antigen-presenting cells (APC). In our analysis, patients with VAP showed lower expression levels of genes coding for HLA-DOA, HLA-DMA, HLA-DMB, ICOS, ICOSLG, IL2RA, CD1, CD3, CD28, CD40LG and CCR7.
HLA-II molecules present peptides from outside the cell for inspection by CD4+ T cells (18). ICOS and ICOSLG have been shown to play roles in T-cell differentiation, cytokine secretion, and survival as well as providing signals required for humoral immune responses (19). The IL-2 receptor is involved in the regulation of immune tolerance by controlling regulatory T cells (TREGs) activity (20). CD1 mediates the presentation of primarily lipid and glycolipid antigens of self or microbial origin to T cells (21). The protein encoded by CD3 is the CD3-epsilon polypeptide, which forms part of the T-cell receptor-CD3 complex. It plays an essential role in T-cell development and defects in this gene cause immunodeficiency (22). CD40LG is a member of the tumor necrosis factor (TNF) family and is expressed on the surface of T cells. CD40LG together with CD28, is essential for T-cell proliferation and survival, cytokine production, and T-helper type-2 development (23). CCR7 controls the migration of memory T cells to inflamed tissues, as well as stimulating dendritic cell maturation (24). In summary, all these molecules play key roles in immune responses (T and B-cells activation, secretion of cytokines, generation of antigen specific antibodies) and are required for clearing the patient of the invading pathogen. Low expression levels of these genes could translate into a poor control of the infection in VAP patients.
Similar results have been described in the context of community-acquired pneumonia (CAP) and sepsis. In different microarray studies coming from our group and others, the depressed expression of HLA-DRA, CD40LG, CD3E, CD28, ICOS constituted a hallmark of sepsis (9,25). Davenport et al. identified down-regulation of different major histocompatibility complex class II genes and CD3, CD28, ICOS and CD40LG in patients with sepsis due to CAP (26). Our results, together with the aforementioned ones, reinforce the idea of the existence of immunosuppression in severe infections, with a particular impact on antigen presentation.
Profiling immunological alterations during VAP thus offers new opportunities to understand the pathological events that characterize this disease, and also to better diagnose its presence in intubated patients. Early diagnosis of VAP is challenging, and affects potential treatment initiation (27). In this sense, ddPCR is an accurate, fast and reproducible technology for achieving absolute quantification of gene expression levels in blood (28). This makes ddPCR attractive for clinical application. Our results in the AUROC analysis supports that quantification of the expression levels of those genes participating of the immunological synapse by ddPCR could constitute a good diagnostic test of VAP. The inverse association found between expression levels of these genes and the CPIS score reinforces the potential clinical utility of this approach.
The small sample size is an important limitation of this study, but our novel results suggest that quantifying the expression of immunological synapse genes could have a role in the diagnosis of VAP. Further studies with larger cohorts of patients should confirm the role of this approach for improving the detection of VAP. Our study was performed using peripheral blood. In consequence, the expression levels of immunological synapse genes at the respiratory level could not be assessed. Nonetheless, this limitation does not preclude the potential impact of our results in the diagnosis of this disease.
Conclusions
In conclusion, patients with VAP show a transcriptomic depression of the immunological synapse at the systemic level. It takes a commonplace event, namely VAP, and highlights a quite significant underlying immune suppressive state. In effect this small study will change how we regard VAP, and proposes that we regard it as an infection in an immune compromised host, and that immunity has a central role for ICU acquired infections. This may in time change clinical practice, as it has profound implications for the role of protocolised care, or bundles, in the prevention of VAP.
Acknowledgements
We thank the nurse teams of the participant hospitals for their help with sample collection for transcriptomic analysis through the years.
Funding: The study was supported by the ‘‘Instituto de Salud Carlos III’’ grant numbers (PI12/01815) and (PI16/01156).
Ethical Statement: The study was approved by the Institutional Review Board of all participating hospitals and written informed consent was obtained from all patients.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Schreiber MP, Shorr AF. Challenges and opportunities in the treatment of ventilator-associated pneumonia. Expert Rev Anti Infect Ther 2017;15:23-32. 10.1080/14787210.2017.1250625 [DOI] [PubMed] [Google Scholar]
- 2.Larsson J, Itenov TS, Bestle MH. Risk prediction models for mortality in patients with ventilator-associated pneumonia: A systematic review and meta-analysis. J Crit Care 2017;37:112-18. 10.1016/j.jcrc.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 3.Boomer JS, To K, Chang KC, et al. Immunosuppression in Patients Who Die of Sepsis and Multiple Organ Failure. JAMA 2011;306:2594-605. 10.1001/jama.2011.1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bermejo-Martin JF, Andaluz-Ojeda D, Almansa R, et al. Defining immunological dysfunction in sepsis: A requisite tool for precision medicine. J Infect 2016;72:525-36. 10.1016/j.jinf.2016.01.010 [DOI] [PubMed] [Google Scholar]
- 5.Patil NK, Bohannon JK, Sherwood ER. Immunotherapy: A promising approach to reverse sepsis-induced immunosuppression. Pharmacol Res 2016;111:688-702. 10.1016/j.phrs.2016.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delano MJ, Ward PA. Sepsis-induced immune dysfunction: can immune therapies reduce mortality? J Clin Invest 2016;126:23-31. 10.1172/JCI82224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shankar-Hari M, Fear D, Lavender P, et al. Activation-Associated Accelerated Apoptosis of Memory B Cells in Critically Ill Patients With Sepsis. Crit Care Med 2017;45:875-82. 10.1097/CCM.0000000000002380 [DOI] [PubMed] [Google Scholar]
- 8.Liao XL, Feng T, Zhang JQ, et al. Phenotypic Changes and Impaired Function of Peripheral γδ T Cells in Patients with Sepsis. Shock 2017;48:321-8. 10.1097/SHK.0000000000000857 [DOI] [PubMed] [Google Scholar]
- 9.Almansa R, Heredia-Rodríguez M, Gomez-Sanchez E, et al. Transcriptomic correlates of organ failure extent in sepsis. J Infect 2015;70:445-56. 10.1016/j.jinf.2014.12.010 [DOI] [PubMed] [Google Scholar]
- 10.Bermejo-Martin JF, Martin-Loeches I, Rello J, et al. Host adaptive immunity deficiency in severe pandemic influenza. Crit Care 2010;14:R167. 10.1186/cc9259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Q. Immunoinformatics and systems biology methods for personalized medicine. Methods Mol Biol 2010;662:203-20. 10.1007/978-1-60761-800-3_10 [DOI] [PubMed] [Google Scholar]
- 12.Sweeney TE, Shidham A, Wong HR, et al. A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med 2015;7:287ra71. 10.1126/scitranslmed.aaa5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallego M, Rello J. Diagnostic testing for ventilator-associated pneumonia. Clin Chest Med 1999;20:671-679, x. 10.1016/S0272-5231(05)70243-4 [DOI] [PubMed] [Google Scholar]
- 14.Almansa R, Socias L, Sanchez-Garcia M, et al. Critical COPD respiratory illness is linked to increased transcriptomic activity of neutrophil proteases genes. BMC Res Notes 2012;5:401. 10.1186/1756-0500-5-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almansa R, Ortega A, Ávila-Alonso A, et al. Quantification of Immune Dysregulation by Next-generation Polymerase Chain Reaction to Improve Sepsis Diagnosis in Surgical Patients. Ann Surg 2017. [Epub ahead of print]. 10.1097/SLA.0000000000002406 [DOI] [PubMed] [Google Scholar]
- 16.Pugin J, Auckenthaler R, Mili N, et al. Diagnosis of ventilator-associated pneumonia by bacteriologic analysis of bronchoscopic and nonbronchoscopic “blind” bronchoalveolar lavage fluid. Am Rev Respir Dis 1991;143:1121-9. 10.1164/ajrccm/143.5_Pt_1.1121 [DOI] [PubMed] [Google Scholar]
- 17.Agüera-Gonzalez S, Bouchet J, Alcover A. Immunological Synapse. eLS. Hoboken, New Jersey, USA: John Wiley & Sons, Ltd, 2001. [Google Scholar]
- 18.Miles JJ, McCluskey J, Rossjohn J, et al. Understanding the complexity and malleability of T-cell recognition. Immunol Cell Biol 2015;93:433-41. 10.1038/icb.2014.112 [DOI] [PubMed] [Google Scholar]
- 19.Yong PFK, Salzer U, Grimbacher B. The role of costimulation in antibody deficiencies: ICOS and common variable immunodeficiency. Immunol Rev 2009;229:101-13. 10.1111/j.1600-065X.2009.00764.x [DOI] [PubMed] [Google Scholar]
- 20.Chinen T, Kannan AK, Levine AG, et al. An essential role for IL-2 receptor in regulatory T cell function. Nat Immunol 2016;17:1322-33. 10.1038/ni.3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vartabedian VF, Savage PB, Teyton L. The processing and presentation of lipids and glycolipids to the immune system. Immunol Rev 2016;272:109-19. 10.1111/imr.12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soudais C, de Villartay JP, Le Deist F, et al. Independent mutations of the human CD3-epsilon gene resulting in a T cell receptor/CD3 complex immunodeficiency. Nat Genet 1993;3:77-81. 10.1038/ng0193-77 [DOI] [PubMed] [Google Scholar]
- 23.Blotta MH, Marshall JD, DeKruyff RH, et al. Cross-linking of the CD40 ligand on human CD4+ T lymphocytes generates a costimulatory signal that up-regulates IL-4 synthesis. J Immunol 1996;156:3133-40. [PubMed] [Google Scholar]
- 24.Pesce S, Moretta L, Moretta A, et al. Human NK Cell Subsets Redistribution in Pathological Conditions: A Role for CCR7 Receptor. Front Immunol 2016;7:414. 10.3389/fimmu.2016.00414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parnell GP, Tang BM, Nalos M, et al. Identifying key regulatory genes in the whole blood of septic patients to monitor underlying immune dysfunctions. Shock 2013;40:166-74. 10.1097/SHK.0b013e31829ee604 [DOI] [PubMed] [Google Scholar]
- 26.Davenport EE, Burnham KL, Radhakrishnan J, et al. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med 2016;4:259-71. 10.1016/S2213-2600(16)00046-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres A, Ramírez P. Risk factors for intensive care unit-acquired nosocomial pneumonia. Is there something new?. Med Clin (Barc) 2007;128:775-6. 10.1157/13106337 [DOI] [PubMed] [Google Scholar]
- 28.Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods 2013;10:1003-5. 10.1038/nmeth.2633 [DOI] [PMC free article] [PubMed] [Google Scholar]