Dairy products provide a substantial part of the food energy intake for many populations around the world. Fresh milk is an important source of many proteins (whey β-lactoglobulin and caseins), fats, vitamin D, calcium, and electrolytes—but it has only a single carbohydrate, lactose (1). We can all digest lactose as newborns but, after weaning, only some of us continue to express the lactase enzyme in our small intestine that digests lactose into glucose and galactose. Individuals with lactase persistence (LP) are thus lactose tolerant. Conversely, in lactase-nonpersistent individuals, the undigested lactose reaches the large intestine, where it undergoes bacterial fermentation, leading to a range of harmful symptoms, including diarrhea, flatulence, and constipation (2). The preparation of cheese and several milk beverages involves lactose fermentation, which is why lactose-intolerant persons can still benefit from the nutritional virtues of milk without suffering strong symptoms after consumption. With time, such food products have become part of the culinary identity of several countries, such as French cheese, Greek yogurt, and fermented mare’s milk known as kumis in Kazakhstan and airag in Mongolia.
Archaeological evidence shows that dairy consumption came soon after the domestication of ruminants. Cattle-specific β-lactoglobulin and casein peptides have been detected in ∼8,000-y-old vessel sherds from Çatalhöyük in Turkey, only two millennia after taurine cattle were first domesticated (3). In Europe, lipid signatures indicative of cheese making were found in ∼7,400- to 6,800-y-old strainer vessels (4) [the earliest cheese ever found was preserved in an Egyptian jar for ∼3,300 y (5)]. In the central Asian steppe, the isotopic content of fatty acids in pottery has suggested that dairy consumption accompanied the early stages of horse domestication, some ∼5,500 y ago (6, 7).
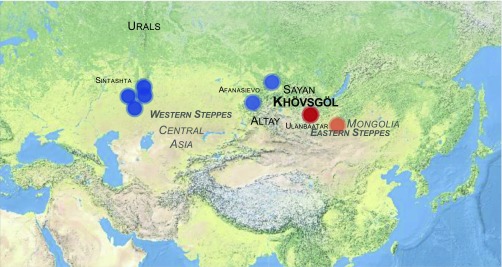
In contrast, the early origins of dairy consumption in the eastern Eurasian steppe, east of the Altai (or Altay) and Sayan mountains (ASMs; Fig. 1), are poorly understood. However, dairy pastoralism represents the main subsistence mode of traditional populations living in the Mongolian steppe. In PNAS, Jeong et al. (8) report that mobile pastoralists living in the Mongolian region of Khövsgöl aimag, located about 500 km northwest of the capital city of Ulaanbaatar (Fig. 1), obtain, on average, a third of their dietary energy from dairy. The authors, however, do not restrict their work to this ethnographic nutritional investigation; they instead embrace state-of-the-art techniques in biomolecular archaeology (see refs. 9 and 10 for reviews) to uncover the early origins of dairy pastoralism in Mongolia.
Fig. 1.
Location of the study sites and cultures. The figure was prepared using the R Leaflet package. GPS coordinates of the Sintashta (deep blue) and Afanasievo (light blue) sites are those reported in ref. 19. Ulaanbaatar and the Khövsgöl region are shown in orange and red, respectively.
The 22 study sites were from the Khövsgöl region and were all dated to ∼2,900 to 3,300 y ago, in the Late Bronze Age (LBA) period. At that time, the funerary traditions west of the ASMs often consisted of important mounds called kurgans, where people were buried together with a range of grave goods, which help archaeologists follow the spread of pastoralism and dairy animals in the region. East of the ASM range, however, the archaeological record is far scarcer, which makes the origins of pastoralism difficult to precisely track. The Khövsgöl LBA sites selected in the study consist of central stone mounds, often surrounded by a ring of stones called khirigsuurs. In most, a single individual was buried under the central stone mound. Ritually deposited ruminant bones found associated with the burials are suggestive of pastoralism, but whether dairy products were already consumed in the region is unknown. However, using mass spectrometry, Jeong et al. (8) could detect in the dental calculus of nine Khövsgöl LBA individuals peptides showing mass spectra typical of bovine, ovine, and caprine β-lactoglobulin and α-S1-casein, two milk-specific proteins. The peptides also showed evidence of postmortem degradation, with some amino acids deamidated and others oxidized; thus, they were not modern contaminants but authentic traces of dairy products surviving over ∼3,300 y in the dental plaque of deceased people. The findings leave no doubt that ruminant dairy pastoralism had penetrated the Eastern steppe by that time. The next logical step was to determine whether this developed after the arrival of new peoples, or simply through the adoption of a new lifestyle by local populations. Answering this question, though, required a different approach, this time leveraging not the proteins entrapped in dental calculus but the information encoded in the genomes of the prehistoric individuals.
Since 2010 when the first ancient genomes from the human family were sequenced (11–13), genome-wide sequence data from more than 1,000 ancient individuals have been collected (see refs. 14 and 15 for recent reviews). Such breakthroughs were made possible thanks to increasingly cheaper, faster, and deeper sequencing instruments (16), as well as through the development of wet and dry laboratory techniques tailored to the ultradamaged nature of ancient DNA (see ref. 9 for a review). The discovery that specific osseous remains, such as the petrosal bone from the inner ear (17), could preserve considerably better DNA than others was also instrumental. Jeong et al. (8) accessed only teeth and femur bones from the Khövsgöl LBA individuals, but they nonetheless managed to sequence the genome of two individuals and recover sequence data at ∼1.2 million sites across the genome for an additional 18 individuals. Similar to what was observed with proteins, the DNA molecules showed molecular evidence of postmortem damage. They also showed hardly any mitochondrial or nuclear DNA molecules originating from present-day humans. The DNA data were thus also authentic. None of the 12 males and 8 females for which sufficient data could be obtained were found to be close relatives. The alleles they carried at ∼600,000 SNP loci, also characterized in other ancient and modern human populations across the world, unveiled their genomic ancestry.
It appears that the Khövsgöl LBA individuals mainly descended from Early Bronze Age populations living in the Eastern steppe. In all but one individual, the genetic contribution of human groups originating from the western Eurasian steppe was no greater than 4 to 7%. This small contribution was best modeled as being derived from the Sintashta people, who developed the first spoke-wheeled chariots in the Ural region, at the border between Europe and Asia, some ∼4,100 to 3,800 y ago (18). Earlier migrations, however, reached the ASM range as the Afanasievo people, who lived in the ASM range northwest of Mongolia some ∼5,300 to 4,500 y ago (19) and who showed strong genetic affinities to Copper and Bronze Age people from the Western steppe. The presence of a minor component of Sintashta-like, but not Afanasievo-like, ancestry at Khövsgöl suggests that although western Eurasian steppe herders had reached the ASM border of Mongolia at least 1,000 y earlier, these early groups either did not penetrate Mongolia or left no descendants in the subsequent populations living there.
The findings of this study imply that the people who practiced dairy pastoralism in Mongolia ∼3,300 y ago were mainly local in origin and were not of western Eurasian steppe pastoralist descent. Moreover, this seems to be an example of mainly hunter-gatherers adopting the lifestyle of their neighbors without a corresponding massive migration wave. Whether the same process also took place among other or earlier Bronze Age Mongolian cultures remains untested. However, for now, the emerging picture is that of a migration of pastoralists from the western Eurasian steppe to the ASMs, followed by the cultural transmission of dairy pastoralism to the eastern Eurasian steppe of Mongolia. This contrasts with the European Bronze Age, where previous ancient DNA work has provided compelling evidence for large-scale population movements ∼5,000 y ago in which departing steppe populations migrated into Europe and strongly reshaped the local European population genetic makeup at that time (19–21).
The opposition between migrationism and diffusionism is a well-known debate in archaeology (22). The former posits that the migration of people with new technologies is the main driver of archaeological cultural change, while the latter invokes the adoption, in situ, of foreign ideas by local people. Ironically, the spread of steppe pastoralism in the Bronze Age shows that both processes contributed to the same archaeological phenomenon, with primarily a migration of domesticated animals and a diffusion of ideas into Mongolia, but a migration of people into Europe. It emphasizes that no single, strong, predefined theoretical view can be transposed to the past as a unique, unequivocal, and universal explanatory model.
There is one last important observation made by Jeong et al. (8) which challenges current models on the evolutionary history of LP in human populations in Eurasia. The most common view posits that people carrying the -13910*T allele, which is located almost 14 kb upstream of the lactase gene and underlies LP across Eurasia, provided a strong selective advantage to carriers as dairy consumption emerged and developed (2, 23). Previous ancient DNA work, however, did not detect the -13910*T allele in pre-5,000-y-old Europeans, including among early Neolithic farmers (24), despite the availability of dairy products. It only rose in frequency after the Bronze Age period (2, 19). Thus, dairy consumption in Western Europe does not seem to have necessarily kick-started a strong selective advantage for carriers of the -13910*T allele. Hence, selective advantage(s) other than just dairy consumption might explain the more-recent-than-expected rise of LP in Europe and the strong selection signatures measured in modern Western Europeans. It is also known that this allele only segregates at very low frequencies in modern Mongolians. Now, Jeong et al. (8) reveal that it was absent in the Khövsgöl LBA dairy pastoralists. Therefore, it appears that the eastern Eurasian steppe challenges the common view of LP coming hand-in-hand with dairy consumption.
How can Mongolian people thus consume substantial quantities of dairy products without developing harmful symptoms? The preparation of fermented cheese and milk beverages almost certainly provides at least part of the explanation. Perhaps the unusual gut microbiome structure of Mongolians, which is especially rich in beneficial microbes, also helps. Indeed, two of the elevated genera, Prevotella and Lactobacillus, are involved in carbohydrate fermentation, and their abundance varies in rural populations with seasonal consumption of milk (25, 26).
Future microbial studies of Mongolian foods and microbiota, combined with ancient DNA analyses of the oral microbiome of Khövsgöl LBA people as they incorporated dairy products in their diet, may help address these questions. This likely represents the next logical step for Jeong and colleagues, as recent studies have shown that in addition to proteins, microbiota DNA signatures can be preserved in ancient dental plaque (27–29). This will nicely complement the already visionary experimental design of their present study (8). Applied to complete archaeological time series, this approach holds the potential to reveal not only how dairy pastoralism developed in the region through space and time but also how this major dietary transition circled back to the taxonomic and functional composition of the human microbiome.
Acknowledgments
This work was supported by the European Research Council under the European Commission Horizon 2020 Research and Innovation Program Grant 681605.
Footnotes
The author declares no conflict of interest.
See companion article on page E11248.
References
- 1.Warinner C, et al. Direct evidence of milk consumption from ancient human dental calculus. Sci Rep. 2014;4:7104. doi: 10.1038/srep07104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ségurel L, Bon C. On the evolution of lactase persistence in humans. Annu Rev Genomics Hum Genet. 2017;18:297–319. doi: 10.1146/annurev-genom-091416-035340. [DOI] [PubMed] [Google Scholar]
- 3.Hendy J, et al. Ancient proteins from ceramic vessels at Çatalhöyük West reveal the hidden cuisine of early farmers. Nat Commun. 2018;9:4064. doi: 10.1038/s41467-018-06335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salque M, et al. Earliest evidence for cheese making in the sixth millennium BC in northern Europe. Nature. 2013;493:522–525. doi: 10.1038/nature11698. [DOI] [PubMed] [Google Scholar]
- 5.Greco E, et al. Proteomic analyses on an ancient Egyptian cheese and biomolecular evidence of Brucellosis. Anal Chem. 2018;90:9673–9676. doi: 10.1021/acs.analchem.8b02535. [DOI] [PubMed] [Google Scholar]
- 6.Outram AK, et al. The earliest horse harnessing and milking. Science. 2009;323:1332–1335. doi: 10.1126/science.1168594. [DOI] [PubMed] [Google Scholar]
- 7.Gaunitz C, et al. Ancient genomes revisit the ancestry of domestic and Przewalski’s horses. Science. 2018;360:111–114. doi: 10.1126/science.aao3297. [DOI] [PubMed] [Google Scholar]
- 8.Jeong C, et al. Bronze Age population dynamics and the rise of dairy pastoralism on the eastern Eurasian steppe. Proc Natl Acad Sci USA. 2018;115:E11248–E11255. doi: 10.1073/pnas.1813608115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orlando L, Gilbert MTP, Willerslev E. Reconstructing ancient genomes and epigenomes. Nat Rev Genet. 2015;16:395–408. doi: 10.1038/nrg3935. [DOI] [PubMed] [Google Scholar]
- 10.Hendy J, et al. Proteomic evidence of dietary sources in ancient dental calculus. Proc Biol Sci. 2018;285:20180977. doi: 10.1098/rspb.2018.0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reich D, et al. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasmussen M, et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llamas B, Willerslev E, Orlando L. Human evolution: A tale from ancient genomes. Philos Trans R Soc Lond B Biol Sci. 2017;372:20150484. doi: 10.1098/rstb.2015.0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marciniak S, Perry GH. Harnessing ancient genomes to study the history of human adaptation. Nat Rev Genet. 2017;18:659–674. doi: 10.1038/nrg.2017.65. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin S, McPherson JD, McCombie WR. Coming of age: Ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamba C, et al. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 2014;5:5257. doi: 10.1038/ncomms6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuznetsov PF. The emergence of Bronze Age chariots in eastern Europe. Antiquity. 2006;90:638–645. [Google Scholar]
- 19.Allentoft ME, et al. Population genomics of Bronze Age Eurasia. Nature. 2015;522:167–172. doi: 10.1038/nature14507. [DOI] [PubMed] [Google Scholar]
- 20.Haak W, et al. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature. 2015;522:207–211. doi: 10.1038/nature14317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damgaard PB, et al. 137 ancient human genomes from across the Eurasian steppes. Nature. 2018;557:369–374. doi: 10.1038/s41586-018-0094-2. [DOI] [PubMed] [Google Scholar]
- 22.Adams WY, Gerven DPV, Levy RS. The retreat from migrationism. Annu Rev Anthropol. 1978;7:483–532. [Google Scholar]
- 23.Gerbault P, et al. Evolution of lactase persistence: An example of human niche construction. Philos Trans R Soc Lond B Biol Sci. 2011;366:863–877. doi: 10.1098/rstb.2010.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci USA. 2007;104:3736–3741. doi: 10.1073/pnas.0607187104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, et al. Mongolians core gut microbiota and its correlation with seasonal dietary changes. Sci Rep. 2014;4:5001. doi: 10.1038/srep05001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, et al. Unique features of ethnic Mongolian gut microbiome revealed by metagenomic analysis. Sci Rep. 2016;6:34826. doi: 10.1038/srep34826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adler CJ, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet. 2013;45:450–455, e1. doi: 10.1038/ng.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warinner C, et al. Pathogens and host immunity in the ancient human oral cavity. Nat Genet. 2014;46:336–344. doi: 10.1038/ng.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weyrich LS, et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature. 2017;544:357–361. doi: 10.1038/nature21674. [DOI] [PubMed] [Google Scholar]