Significance
TRPA1 channels are expressed primarily in sensory neurons with human genetic linkage to pain disorders. Despite the strong confidence for TRPA1 as a therapeutic target, few modulators have advanced to the clinic being hindered by poor physiochemical properties and narrow structure-activity relationships related to their site of interaction. We report the discovery of a potent and selective thiadiazole small molecule class of TRPA1 inhibitor that interacts with the N-terminal ankyrin repeat (ankyrinR) domain. The discovery of this interaction site provides opportunities to broaden TRPA1 small molecule drug discovery for the treatment of pain and adds insight into TRPA1 channel gating mechanisms.
Keywords: TRPA1, ankyrin repeat, pain, thiadiazole, ion channel
Abstract
TRPA1, a member of the transient receptor potential channel (TRP) family, is genetically linked to pain in humans, and small molecule inhibitors are efficacious in preclinical animal models of inflammatory pain. These findings have driven significant interest in development of selective TRPA1 inhibitors as potential analgesics. The majority of TRPA1 inhibitors characterized to date have been reported to interact with the S5 transmembrane helices forming part of the pore region of the channel. However, the development of many of these inhibitors as clinical drug candidates has been prevented by high lipophilicity, low solubility, and poor pharmacokinetic profiles. Identification of alternate compound interacting sites on TRPA1 provides the opportunity to develop structurally distinct modulators with novel structure-activity relationships and more desirable physiochemical properties. In this paper, we have identified a previously undescribed potent and selective small molecule thiadiazole structural class of TRPA1 inhibitor. Using species ortholog chimeric and mutagenesis strategies, we narrowed down the site of interaction to ankyrinR #6 within the distal N-terminal region of TRPA1. To identify the individual amino acid residues involved, we generated a computational model of the ankyrinR domain. This model was used predictively to identify three critical amino acids in human TRPA1, G238, N249, and K270, which were confirmed by mutagenesis to account for compound activity. These findings establish a small molecule interaction region on TRPA1, expanding potential avenues for developing TRPA1 inhibitor analgesics and for probing the mechanism of channel gating.
The transient receptor potential (TRP) channel family is a group of weakly selective cation channels activated by extracellular or intracellular ligands as well as by changes in temperature or membrane voltage (1). The TRP family comprises some 28 distinct channels, many of which have been implicated as drug targets for a wide variety of disease states including pain, inflammatory bowel disease, and asthma (2, 3). Among the TRP family members, TRPA1 is the most divergent being expressed primarily in sensory neurons where they are activated by inflammatory mediators and noxious environmental substances. These substances covalently modify cysteine residues within the ankryin repeat rich N terminus to activate the channel (4, 5). Nonreactive agonists, such as menthol, can also activate TRPA1 by interacting with the pore lining helices (6). When activated in neurons, TRPA1 channels preferentially conduct Na+ and Ca2+ producing a generator potential to depolarize the membrane potential toward the action potential threshold (7).
A TRPA1 gain-of-function mutation (N855S) in patients with familial episodic pain syndrome directly links TRPA1 activity to pain in humans (8). TRPA1 small molecule antagonists have been shown to reverse pain in rodent models of inflammatory pain and to inhibit excitability of nociceptors (9–11). These and related data have driven significant interest in developing potent and selective TRPA1 inhibitors as a new class of analgesics. However, it has proven challenging to transition these compound classes into clinical candidates often due to undesirable physicochemical properties (12–14). The most advanced clinical compound to date, GRC-17536 developed by Glenmark, was efficacious in a Phase IIa clinical trial for a subset of patients with diabetic nerve pain when dosed as a suspension but has not progressed further, in part, due to the limitations imposed by its pharmaceutical properties (12, 15).
The majority of small molecule TRPA1 inhibitors characterized to date has been reported to interact with the pore region of the channel (6, 16–22). Generally, these compounds possess high lipophilicity, narrow structure-activity relationships (SARs), and can functionally flip from channel inhibition to activation with only subtle structural changes. Identification of alternate TRPA1 small molecule interaction sites has the potential to support the discovery of new chemical series with different SARs and more desirable physicochemical properties than existing modulators. Herein, we report the discovery of a thiadiazole class of TRPA1 inhibitor that interacts with amino acid residues within ankyrin repeat (ankyrinR) #6 of the distal N terminus of the channel. This class of inhibitor provides the opportunity to probe the mechanism by which the distal N-terminal ankyrinRs are involved in TRPA1 gating and potentially provide a binding site that is amenable to the development of a new class of therapeutic agent for the treatment of pain.
Results
Discovery of a Novel and Selective TRPA1 Small Molecule Inhibitor Chemotype.
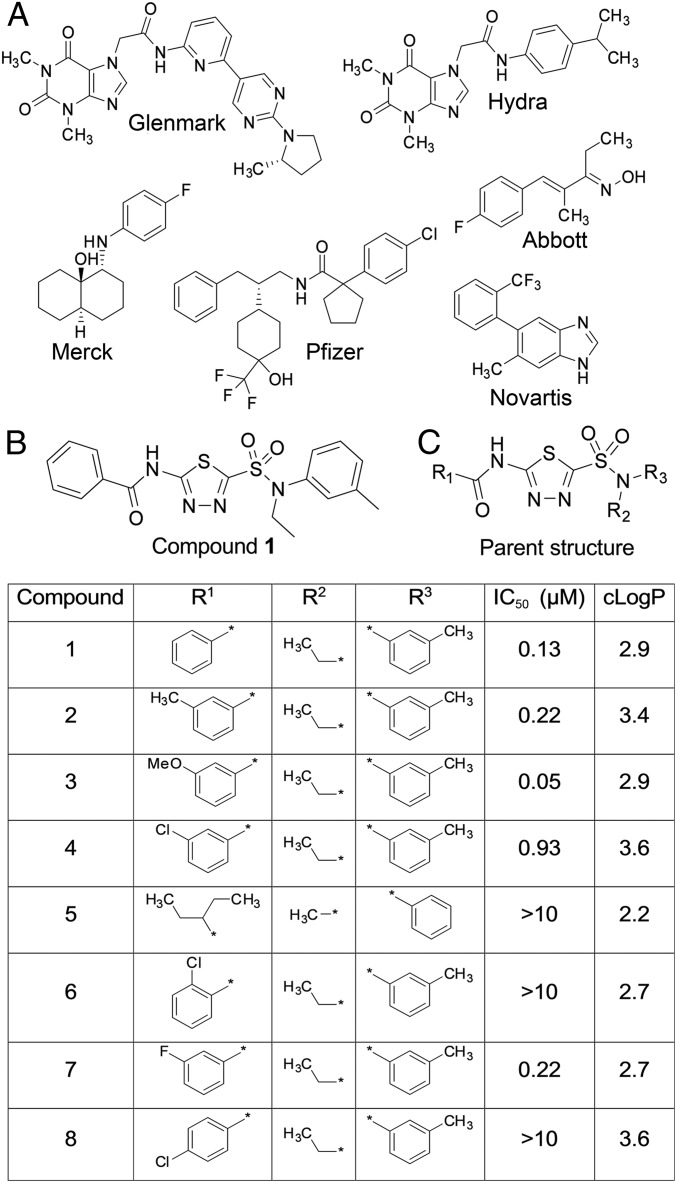
To identify a new starting point for novel TRPA1 inhibitors, we screened a portion of the Pfizer compound collection using a high throughput fluorescent FLUO-4 FLIPR based Ca entry assay in HEK-293 cells stably expressing human TRPA1 (hTRPA1). One class of compound that emerged from this screen was structurally unlike those previously reported (see Fig. 1A for examples) being based on a thiadiazole core (Compound 1, Fig. 1B). Compound 1 was commercially available and had not been previously reported as a TRPA1 inhibitor. In FLIPR Ca entry studies in HEK-hTRPA1 cells, Compound 1 inhibited AITC stimulated Ca2+ fluorescence with an IC50 of 0.13 µM. Given the low molecular weight, relative polarity, and structural features distinct from previously reported TRPA1 chemotypes, we further explored the structure activity relationship, selectivity over other TRP channel family members, and the mechanism of action.
Fig. 1.
A high throughput FLIPR screen identifies a thiadiazole series of small molecule hTRPA1 inhibitors. (A) Examples of literature TRPA1 antagonists. (B) Compound 1 structure. (C) SAR of the thiadiazole series showing substitution patterns, mean FLIPR IC50 (n = 3–12 per compound), and cLogP.
A number of close analogs of Compound 1 (see Fig. 1C, full structures are shown in SI Appendix, Fig. S1) were commercially available, and when we screened a small set of thiadiazole analogs we identified an interesting SAR pattern. The amide aryl group was sensitive to substitution changesin that addition of polar substituents at the meta position, such as a methoxy group (3) were more potent than the unsubstituted 1, whereas lipophilic or halogen substituents, such as a methyl (2), a chloro group (4), or fluoro groups (7) were slightly less potent. The ortho chloro derivative 6 and the para analog 8 were inactive in the FLIPR assay as was the nonaryl containing pentyl amide 5. There was some concern that the amide group may be subject to amidolysis and therefore potentially chemically unstable, but when Compound 1 was incubated with glutathione, no depletion of the parent compound was observed, confirming its stability (SI Appendix, Fig. S2). The lack of capture by glutathione and lack of TRPA1 channel activation were good indicators that the compound was not functioning as a cysteine trap and thus unlikely to be a covalent modifier of the channel.
TRPA1 channels are known to be modulated by a variety of covalent modulators, but they can also be activated by nonelectrophilic agonists, such as PF-4840154 (23). To test whether the thiadiazole chemotype specifically inhibited covalently activated channels, we extended the FLIPR experiments to evaluate the most potent compounds (1, 2, 3, and 7) against PF-4840154 activated channels. These compounds inhibited AITC and PF-4840154 modulated activity with similar potency. Representative FLIPR traces are shown in SI Appendix, Figs. S3 and S4 for the AITC and PF-4840154 experiments, respectively.
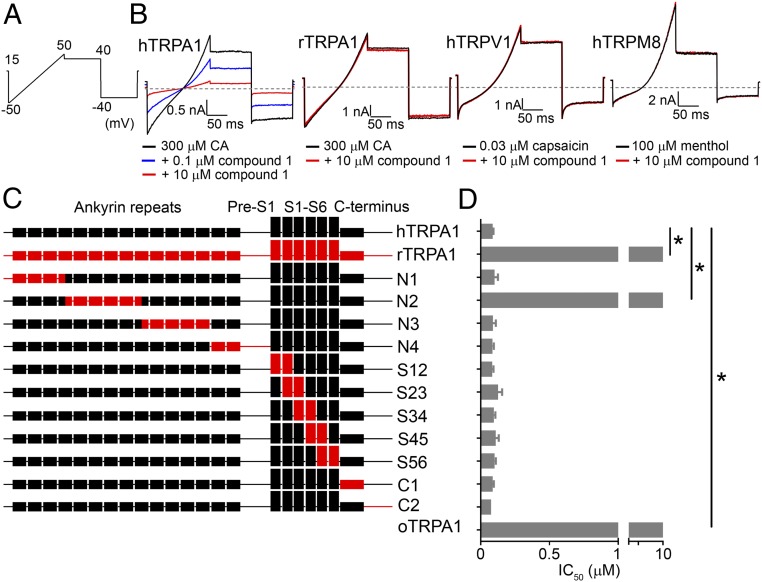
The selectivity of the most potent analogs 1, 2, 3, and 7 against members of the TRP gene family was evaluated to further understand the thiadiazole series profile. TRPV1 and TRPM8 were chosen as representative TRP channels since they are also involved in pain signaling. Taking this approach allowed for simultaneous sampling of gene family selectivity and assessment of the potential utility of compounds as probes in animal pain models. Inhibitory potency for hTRPA1 in conventional manual patch-clamp electrophysiology was similar to that determined in fluorescence based Ca entry assays. None of the tested thiadiazole compounds significantly inhibited human TRPV1 and TRPM8 at 10 µM, indicating a TRPA1 selectivity ratio of >30. Furthermore, thiadiazole compounds exhibited selectivity for human TRPA1 over the rat TRPA1 ortholog (rTRPA1) with IC50s ranging from 90 to 170 nM for hTRPA1 to >10 µM for rTRPA1. Mean IC50 ± standard error of the mean (SEM) for hTRPA1 and percent inhibition at 10 µM for rTRPA1, hTRPV1, and hTRPM8 are summarized in SI Appendix, Table S1. Representative current traces for human and rat TRPA1, hTRPV1, and hTRPM8 channels in the presence and absence of Compound 1 are shown in Fig. 2B. Traces for compounds 2, 3, and 7 are shown in SI Appendix, Fig. S5.
Fig. 2.
hTRPA1 N-terminal region within ankyrinRs #4–#9 confers species specificity of thiadiazole interaction. (A) Voltage-clamp protocol used in all TRPA1 and TRPV1 manual patch-clamp electrophysiology experiments. Note that, for TRPM8, a similar protocol was used with the voltage ramp ranging from −80 to +80 mV with steps to +50 and −50 mV. (B) Representative current traces showing Compound 1 inhibition of hTRPA1, selectivity over hTRPV1 and hTRPM8, and species selectivity over rTRPA1. Compound 1 was tested in the continued presence of agonist: TRPA1 = 300 μM cinnamaldehyde (CA), TRPV1 = 0.03 μM capsaicin, and TRPM8 = 100 μM menthol. The dashed gray line denotes zero current level. (C) Illustration of 11 human TRPA1 chimeras with domains replaced by rat TRPA1 sequence. (D) Mean ± SEM FLIPR AITC activated calcium entry IC50 of Compound 1 against human, rat, opossum TRPA1, and 11 hTRPA1-rTRPA1 chimeras. Chimera N2 was the only chimeric construct with significant loss in potency being comparable to rTRPA1 and oTRPA1: Chimera N2 IC50 > 10 μM, wild type (WT) hTRPA1 IC50 = 0.09 ± 0.01 µM [*P < 0.001, analysis of variance (ANOVA) with post hoc Dunnett’s t test, n = 3–6 per construct]. The Chimera N2 construct contained amino acid swaps of rTRPA1 with hTRPA1 residues 180–358 of ankyrinRs #4–#9.
The next stage of the characterization of the thiadiazole inhibitor class was to establish if inhibition was mediated via an interaction with the S5 pore lining helix, such as the majority of published TRPA1 modulators. Human-species ortholog chimeras of TRPA1 have proven useful to identify regions of the channel involved in species-dependent compound interactions. Previously, we demonstrated the opossum TRPA1 (oTRPA1) variant which shares 67% amino acid homology with hTRPA1 to be insensitive to carboxamide (Pfizer) and indazole (Novartis) TRPA1 inhibitors (24, 25). Likewise, Compound 1 was found to be inactive vs. oTRPA1 (Fig. 2D) up to 10 µM. However, in contrast to the carboxamide, indazole, and A-967079 (Abbott) TRPA1 inhibitors, the latter of which has been shown in a ligand bound cryoelectron microscopy (cryo-EM) structure to interact with the pore region, Compound 1 behaved differently on human-opossum TRPA1 chimeras (21). Using chimeras in which the human TRPA1 S5 helix was replaced by the opossum S5, Compound 1 retained its inhibitory potency, whereas A-967079 lost activity (SI Appendix, Fig. S6). Evaluating the reverse chimera in which opossum S5 was replaced by the human S5 helix, Compound 1 was found to be inactive up to 10 µM, whereas A-967079 retained inhibitory potency (SI Appendix, Fig. S6). These findings suggested that the thiadiazole class of TRPA1 inhibitors interact with the channel in a manner different from previously published chemical structures.
Thiadiazole Chemotype Interacts with Residues in the Distal N-Terminal AnkyrinR Domain.
Most previously characterized TRPA1 inhibitors exhibit similar potencies for inhibition of human and rat TRPA1 which contrasts with the clear human vs. rat selective inhibition observed with the current thiadiazole class of inhibitors. rTRPA1 shares an 80% sequence homology with hTRPA1 at the protein level. Using a similar approach to the studies described above evaluating human vs. opossum TRPA1, a series of 11 human-rat chimeras encompassing the entire TRPA1 channel were generated to aid the identification of the compound interaction region accounting for rat vs. human selectivity (Fig. 2C). All chimeric constructs yielded functional TRPA1 channels as measured using fluorescence based calcium entry assay with the AITC activated responses being similar in magnitude to WT hTRPA1. Of the 11 chimeras tested, only chimera N2 exhibited a reduced sensitivity to Compound 1 with a potency similar to rTRPA1 (>10 µM). Chimera N2 replaced residues 180–358 from human TRPA1 with the equivalent residues from rat TRPA1. The observed loss of Compound 1 inhibitory activity suggested the thiadiazole interaction site to be located between ankyrinRs #4 and #9 in the N terminus. The remaining 10 chimeras all exhibited Compound 1 IC50 values that were not statistically different from hTRPA1 (Fig. 2D) suggesting no involvement of the other ankyrinRs in inhibition by this compound class. Representative FLIPR traces from these experiments are shown in SI Appendix, Fig. S7.
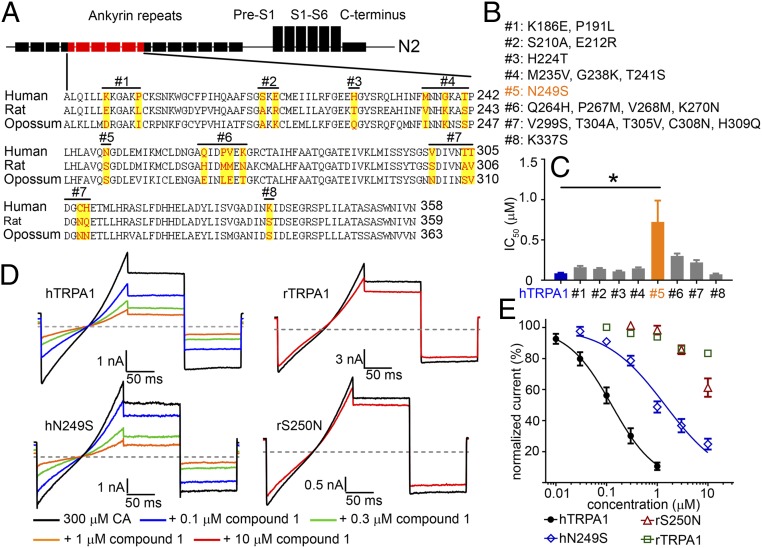
The evidence that a subregion of the N-terminal ankyrinRs contained the potential interaction site for Compound 1 resulted in a closer analysis and comparison of the amino acid sequences of human, rat, and opossum TRPA1 to look for residues that were different in Compound 1 sensitive (human) vs. nonsensitive (rat and opossum) TRPA1 species orthologs (Fig. 3A). The differences in sequences were divided into eight groups with the number of amino acid residues in each group ranging from one to five. We then performed site-directed mutagenesis in each group to generate eight human TRPA1 constructs with residues mutated to the corresponding rat ortholog residues (Fig. 3B). Of the eight constructs tested, only construct #5 showed a statistically significant reduction in Compound 1 potency compared with the hTRPA1 value (Construct #5 IC50 = 0.72 ± 0.26 µM, WT hTRPA1 IC50 = 0.09 ± 0.01 µM, P < 0.001 ANOVA, post hoc Dunnett’s t test, n = 6). Summary data are shown in Fig. 3C. Interestingly, construct #5 carried a single N249S residue swap within ankyrinR #6 of the human TRPA1 channel.
Fig. 3.
Residue N249 in hTRPA1 is critical but not sufficient for thiadiazole activity. (A) Sequence alignment among human, rat, and opossum TRPA1 within the N2 chimeric region. Residues targeted for site-directed mutagenesis are highlighted. hTRPA1 residues were mutated to rTRPA1 to produce eight mutant constructs as depicted above the sequence. (B) List of point mutation(s) for all eight constructs. (C) FLIPR IC50 of the mutant hTRPA1 constructs compared with WT hTRPA1 (n = 6 each group). Compound 1 was significantly less potent on Construct #5, IC50 = 0.72 ± 0.26 µM compared with hTRPA1, IC50 = 0.09 ± 0.01 µM (*P < 0.001 ANOVA with post hoc Dunnett’s t test). (D) Representative steady-state current traces showing Compound 1 inhibition of hN249S and rS250N constructs compared with WT hTRPA1 and rTRPA1 channels. Channels were activated with 300 µM cinnamaldehyde (CA) and Compound 1 was applied in the continued presence of agonist. The gray line denotes zero current level, and the voltage protocol is shown in Fig. 2A. (E) Concentration response curves of Compound 1 from manual patch-clamp electrophysiology. Steady-state current amplitudes were taken from the mean of the −40 mV step and normalized to 300 μM CA control. Each data point represents the normalized mean ± SEM current for the tested concentration. Potencies with 95% confidence intervals (n = 3 to 4 per construct): hTRPA1 IC50 = 0.13 µM (95% CI = 0.10–0.16 µM), hN249S IC50 = 1.43 µM (95% CI = 1.11–1.86 µM), rS250N IC50 > 10 µM, and rTRPA1 IC50 > 10 µM.
The reduced potency of Compound 1 on the human N249S TRPA1 mutation was confirmed using conventional manual patch-clamp electrophysiology employing a ramp step protocol evoked every 5 s in the continued presence of 300 µM CA to activate the channels (Fig. 3D). Similar to the FLIPR Ca entry assay results, Compound 1 was less potent on hN249S compared with WT hTRPA1 [hN249S IC50 = 1.43 µM, 95% confidence interval (CI) = 1.11–1.86 µM, n = 4, WT hTRPA1 IC50 = 0.13 µM, 95% CI = 0.10–0.16 µM, n = 4]. As a complementary experiment, the corresponding rat S250N humanized mutation was generated to test for gain-of-function activity. Against rat S250N TRPA1, Compound 1 inhibited 38 ± 4% at 10 µM (n = 7) similar to WT rTRPA1 which was inhibited by 17 ± 1% at 10 µM (n = 3, Fig. 3 D and E). These results demonstrate human TRPA1 residue N249 to be necessary for Compound 1 inhibition. However, because only partial loss of function was observed with the human N249S mutation and rat S250N failed to demonstrate significant gain of function, it was hypothesized that additional residues were involved in forming the complete Compound 1 interaction site. Instead of embarking on a random mutagenesis exercise to identify the additional amino acid residues, a computational model based on a previously solved ankyrinR domain was generated. In the absence of a ligand bound TRPA1 structure, we believed that a docking model that could guide and predict mutagenesis results would provide stronger evidence in support of the actual binding mode.
Computational Modeling of AnkyrinR Region Reveals Additional Interactions.
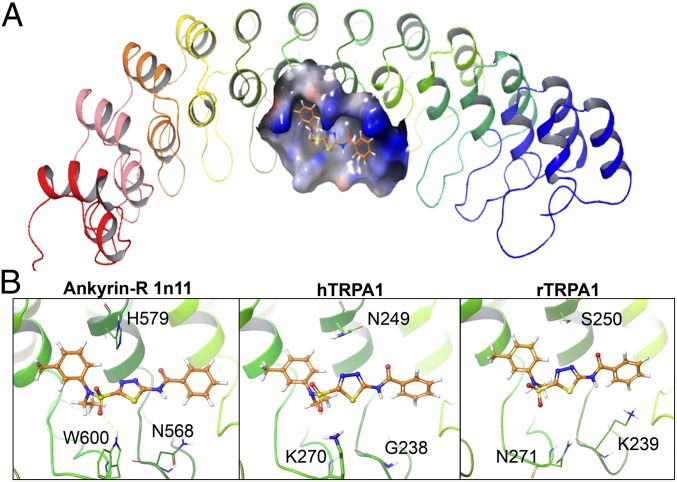
To identify the full complement of residues important for the interaction of the thiadiazole chemotype within TRPA1 N-terminal ankyrinR domains, we focused our computational modeling studies around the N2 region of human ankyrinR identified by the human-rat TRPA1 chimera experimental results. We used the crystal structure of the D34 region of human ankyrinR (PDB ID: 1N11) as a surrogate crystal structure for the modeling studies as the N-terminal region of TRPA1 was not completely resolved in the published cryo-EM structure (26). Compound docking was performed using Glide v5.0 software (Schrödinger, LLC) to provide some preliminary models. We observed that Compound 1 makes good van der Waals contacts in the putative binding site and complements the shape of the pocket very well (Fig. 4A). Aligning various sequences to the model (Fig. 4B), we noted that a key H-bonding acceptor pharmacophore was based on the sulfonamide functional group of Compound 1. The sulfonamide oxygen makes a key H-bonding interaction with K270 in humans but does not with the corresponding N271 in rats. We also propose a water mediated interaction between other sulfonamide oxygen and N249 in humans that is not possible with S250 in rats. A second key residue that we envisioned is based on the H-bonding donor pharmacophore, the amide SAR, and our screening results. A smaller glycine residue (G238 in humans) provides space for the amide to position the phenyl in a pocket, but K239 in rats could have deleterious effects due to a donor–donor negative interaction. Based on these observations, we highlighted residues K270 and G238 as additional key residues in addition to N249 which had already been identified from mutagenesis experiments.
Fig. 4.
Model of Compound 1 in the human ankyrinR crystal structure. (A) Overview of human ankyrinR D34 region crystal structure (PDP: 1n11) in ribbon representation; putative binding site shown as a solid surface with Compound 1 (ball and stick). (B) Close-in view of the binding conformation of Compound 1 in AnkyrinR 1n11 (Left), hTRPA1 (Middle), and rTRPA1 models (Right). Compound 1 (orange) key residues are shown as green sticks and labeled.
Human TRPA1 Residues G238, N249, and K270 of AnkyrinR #6 Fully Account for Thiadiazole Potency.
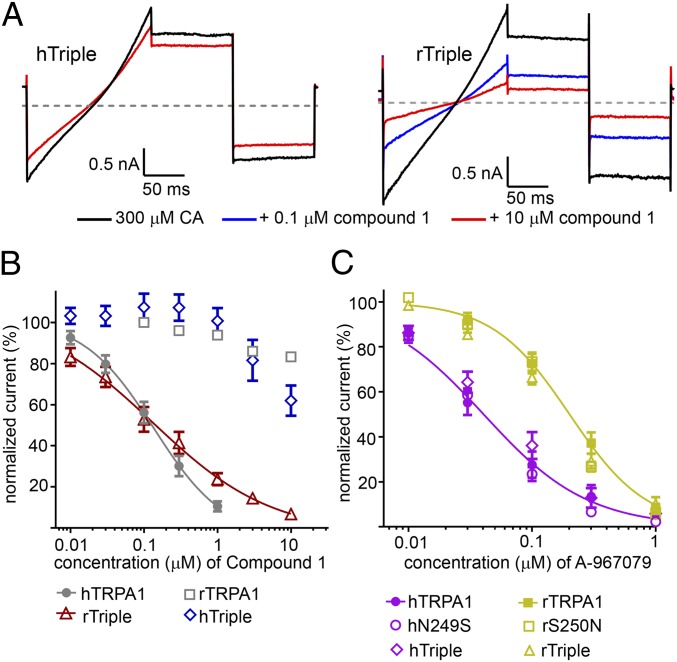
Based on the computational modeling results, two additional mutant forms of TRPA1 were generated. In the first, the mutations G238K, N249S, and K270N (hTriple) were introduced, replacing human residues with the equivalent rat residues. In the second construct, the equivalent residues in rat TRPA1 were replaced with the corresponding human residues K239G, S250N, and N271K (rTriple). Using conventional manual patch-clamp electrophysiology, it was found that the hTriple mutated form of hTRPA1 was only weakly sensitive to Compound 1 which inhibited 38 ± 5% at 10 µM (n = 8) similar to WT rTRPA1 and S250N rTRPA1 (Fig. 5 A and B). Compounds 2, 3, and 7 were also insensitive at 10 µM concentration with percent inhibitions of 2 ± 3%, 0 ± 2%, and 1 ± 2%, respectively (n = 4, SI Appendix, Fig. S8). In the gain-of-function study, Compound 1 was tested on the rTriple mutated form of rTRPA1 and gained sensitivity (Fig. 5 A and B) with an IC50 value of 0.15 µM (95% CI = 0.11–0.19 µM, n = 4) being comparable to the WT hTRPA1 IC50 of 0.13 µM (95% CI = 0.10–0.16 µM, n = 4). As a specificity control, the effect of A-967079 on the triple mutation constructs was evaluated (18, 20, 21). In these experiments, A-967079 was fivefold more potent on human vs. rat TRPA1 with IC50 values of 0.04 (95% CI = 0.03–0.05 µM) and 0.20 µM (95% CI = 0.17–0.24 µM). The potency of the single and triple mutation constructs overlapped with these values and were not shifted (Fig. 5C). These results demonstrate that the thiadiazole TRPA1 structural class of inhibitor interacts specifically with residues G238, N249, and K270 and that these mutations were unlikely to be introducing allosteric effects or structural changes that indirectly influence channel pharmacology.
Fig. 5.
hTRPA1 Residues G238, N249, and K270 in AnkyrinR #6 mediate thiadiazole Interaction. (A) Representative steady-state traces of Compound 1 effects on hTRPA1 G238K/N249S/K270N (hTriple) and rTRPA1 K239G/S250N/N271K (rTriple) channels. Channels were activated with 300 µM CA and Compound 1 was applied in the continued presence of agonist. The gray line denotes zero current level, and the voltage protocol is shown in Fig. 2A. (B) Concentration response curves of Compound 1 on hTriple and rTriple channels with WT hTRPA1 and rTRPA1 shown for reference. Potencies with 95% confidence intervals (n = 4–8 per construct): rTriple IC50 = 0.15 μM (95% CI = 0.11–0.19 μM), hTRPA1 = 0.13 µM (95% CI = 0.10–0.16 µM), hTriple IC50 > 10 µM, rTRPA1 IC50 > 10 µM. (C) A-967069 which interacts with the S5 helix of the channel pore is unaffected by the mutations that affect Compound 1 potency. Only the WT TRPA1 fits are shown for clarity (n = 5 per construct): hTRPA1 IC50 = 0.04 µM (95% CI = 0.03–0.05 µM) and rTRPA1 IC50 = 0.20 µM (95% CI = 0.17 = 0.24 µM).
In conclusion, we have discovered a potent and selective TRPA1 inhibitor structural class with a previously uncharacterized site of action. Using a combination of mutagenesis and functional readouts, in part, guided by computational prediction, we have identified amino acid residues G238, N249, and K270 within N-terminal ankyrinR #6 to account for the potency of Compound 1 of the thiadiazole chemotype. This small molecule interaction site serves as a possible alternative to the pore region for the development of TRPA1 inhibitors, and these compounds may serve as valuable tools to probe the mechanism by which the distal N terminus modulates TRPA1 channel gating.
Discussion
The thiadiazole chemotype we report was identified in a high throughput screen. Compounds from this series are low molecular weight, relatively polar, and structurally orthogonal to any previous TRPA1 inhibitors and therefore an attractive starting point to explore further. Given that the thiadiazoles appeared to be interacting differently than previously reported TRPA1 channel modulators, a human-rat chimeric strategy was employed to further define the key regions. The functional studies using these chimeras unequivocally pointed to a distal N-terminal ankyrinR as the interaction site. The majority of the TRPA1 N terminus is a composed of ankyrinRs ranging from 14 to 18 in number as determined from a primary amino acid sequence. Although not fully resolved, the general structure of the distal N terminus can be seen in the cryo-EM as four crescentlike structures extending perpendicular to the axis in a propellerlike fashion with the concave sides, or ankyrin grooves, facing the lipid bilayer. By combining sequence alignments with known ankyrin structures and TRPA1 cryo-EM, ankyrinR #6 is predicted to sit at the apex of the crescent (21).
Using a mutagenesis strategy guided by the chimera results and species variant sequence homology, the amino acid residue N249 was identified as critical for the Compound 1 inhibitor interaction. Whereas there was a partial loss of potency vs. the N249S mutant construct, there was no gain of function in the complementary S250N construct suggesting a role for additional residues. To aid the identification of these residues, a computational homology model based on the 2.7 Å X-ray crystal structure of the D34 domain of the human ankyrinR protein (PDP ID: 1n11) (27) was developed. This domain was selected as it consists of 12 ankyrinRs, similar in length to the ankyrinRs that form the crescent structure in TRPA1 and has been previously used to model the N terminus of TRPA1 (28). The docking results for Compound 1 predicted residues K270 and G238 along with N249 to form the putative binding pocket. It is important to note that the computational modeling was used predictively to guide the mutagenesis strategy and not retrospectively. The ability of this model to fully predict the full gain of function mutagenesis results in a single iteration gives strength to the argument that the K270, G238, and N249 residues are primary contributors to the Compound 1 binding pocket.
Inhibition of TRPA1 gating via an ankyrinR #6 interaction was initially surprising being significantly upstream from the cysteines that are modulated by covalent activators. However, there are interesting parallels for a role of this region in channel gating. For example, caffeine modulates TRPA1 channels weakly with an EC50 of ∼5 mM with species-dependent pharmacology in activating mouse TRPA1 but inhibiting hTRPA1 channels with similar potencies. The key amino acid for this interaction in the human channel is P267 which resides within ankyrinR #6. When P267 is swapped for the corresponding mouse M286, caffeine switches from an inhibitor to an activator (29). In rattlesnakes, TRPA1 activity is modulated by heat rather than covalent activation as in humans (30). Swapping ankyrinR domains (including ARD 6), switches the modality of channel activation (31). Interestingly the homologous residue to N249 in mouse TRPA1 (S250) has been reported to be important in modulating the temperature sensitivity of voltage-dependent activation and channel open probability (32). The overlapping role of residue N249 in both temperature-dependent gating and pharmacological inhibition by thiadiazoles suggests that the residue may undergo conformational change or may interact with a regulatory factor that modulates channel gating.
The precise mechanism by which the thiadiazoles inhibit TRPA1 channel gating requires further study. AnkyrinR domains are known to mediate protein–protein interactions. However, they can also bind sugars and lipids (33, 34). Whereas, protein–protein interactions are largely governed by the secondary structure involving interactions across clusters of residues, smaller regulatory molecules can bind to more defined binding pockets (35). It is unlikely although not impossible that the thiadiazoles block by interfering protein–protein interaction. A more likely scenario with precedent is that the compounds may compete for regulatory molecules, such as phosphate containing molecules, that are important for lowering the free energy barrier for channel gating. For example, phosphoinositides regulate TRPV4 channel activity, and PI(4,5)P2 has been shown by X-ray crystallography to interact specifically with AR domain 4 of a six repeat domain (26). Perhaps a similar interaction of a regulatory molecule important for activation occurs in TRPA1 and for which the thiadiazoles compete.
The potential to leverage the ankyrin repeat #6 site for drug discovery remains to be seen. The thiadiazole compounds possess acceptable physicochemical property starting points, but further analoging is required to know if the site can support robust SAR while retaining selectivity. If TRPA1 species selectivity is a feature of ankyrinR #6, a humanized mouse of the rTriple construct would enable the establishment of pharmacokinetic-pharmacodynamic efficacy models. A potential advantage for this site is that multiple cytosolic ankyrinR structures have been solved by X-ray crystallography to ∼2 Å resolution. Thus, a high resolution ligand bound structure of the TRPA1 distal N terminus along with molecular dynamic simulation could enable an in silico screen toward identification of new chemical matter and SAR development for modulators of this important pain target.
Methods
Whole-Cell Patch-Clamp Electrophysiology.
WT and mutant TRPA1 channels were recombinantly expressed in T-Rex-293 cells (Thermo Fisher). Protein accession IDs for the WTs TRPA1 channels used are CAA71610 (human), NP_997491.1 (rat), and XP_001378427 (opossum). Whole-cell manual patch-clamp recordings were recorded with an internal solution, consisting of (in millimoles) 90 CsCl, 32 CsF, 10 Hepes, 10 glycol bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), 10 Cs4 BAPTA, 1 MgCl2, 5 Mg ATP, and 0.1 Na GTP, pH 7.3 with CsOH. The external solution consisted of (in millimoles) 132 NaCl, 5.4 KCl, 1.8 CaCl2, 0.8 MgCl2, 10 Hepes, and 5 glucose, pH 7.4. Voltage protocols were evoked every 5 min and mean steady-state current analyzed from the −40 mV step (−50 mV for TRPM8).
Computational Modeling.
A computational model of human ankyrinR with Compound 1 was built based on the crystal structure of the D34 region of human ankyrinR, (1N11.pdb) (27). The protein was prepared and minimized for modeling using the protein preparation workflow (Schrödinger) in Maestro. The receptor docking grid was centered around the N2, and the size of the inner and outer grid boxes was set to 10 Å in each direction. The structure of Compound 1 for docking was generated with Maestro (Schrödinger). The molecules were further prepared using LigPrep with the OPLS2005 force field. Docking was carried out using XP precision with Glide v. 5 (Schrödinger, LLC, 2018).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808142115/-/DCSupplemental.
References
- 1.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 2.Bautista DM, Pellegrino M, Tsunozaki M. TRPA1: A gatekeeper for inflammation. Annu Rev Physiol. 2013;75:181–200. doi: 10.1146/annurev-physiol-030212-183811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran MM, McAlexander MA, Bíró T, Szallasi A. Transient receptor potential channels as therapeutic targets. Nat Rev Drug Discov. 2011;10:601–620. doi: 10.1038/nrd3456. [DOI] [PubMed] [Google Scholar]
- 4.Macpherson LJ, et al. Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature. 2007;445:541–545. doi: 10.1038/nature05544. [DOI] [PubMed] [Google Scholar]
- 5.Hinman A, Chuang HH, Bautista DM, Julius D. TRP channel activation by reversible covalent modification. Proc Natl Acad Sci USA. 2006;103:19564–19568. doi: 10.1073/pnas.0609598103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao B, et al. Identification of transmembrane domain 5 as a critical molecular determinant of menthol sensitivity in mammalian TRPA1 channels. J Neurosci. 2008;28:9640–9651. doi: 10.1523/JNEUROSCI.2772-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nilius B, Szallasi A. Transient receptor potential channels as drug targets: From the science of basic research to the art of medicine. Pharmacol Rev. 2014;66:676–814. doi: 10.1124/pr.113.008268. [DOI] [PubMed] [Google Scholar]
- 8.Kremeyer B, et al. A gain-of-function mutation in TRPA1 causes familial episodic pain syndrome. Neuron. 2010;66:671–680. doi: 10.1016/j.neuron.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, et al. Selective blockade of TRPA1 channel attenuates pathological pain without altering noxious cold sensation or body temperature regulation. Pain. 2011;152:1165–1172. doi: 10.1016/j.pain.2011.01.049. [DOI] [PubMed] [Google Scholar]
- 10.da Costa DS, et al. The involvement of the transient receptor potential A1 (TRPA1) in the maintenance of mechanical and cold hyperalgesia in persistent inflammation. Pain. 2010;148:431–437. doi: 10.1016/j.pain.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 11.McGaraughty S, et al. TRPA1 modulation of spontaneous and mechanically evoked firing of spinal neurons in uninjured, osteoarthritic, and inflamed rats. Mol Pain. 2010;6:14. doi: 10.1186/1744-8069-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moran MM. TRP channels as potential drug targets. Annu Rev Pharmacol Toxicol. 2018;58:309–330. doi: 10.1146/annurev-pharmtox-010617-052832. [DOI] [PubMed] [Google Scholar]
- 13.Skerratt S. Recent progress in the discovery and development of TRPA1 modulators. Prog Med Chem. 2017;56:81–115. doi: 10.1016/bs.pmch.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Hackos DH. TRPA1 as a drug target–Promise and challenges. Naunyn Schmiedebergs Arch Pharmacol. 2015;388:451–463. doi: 10.1007/s00210-015-1088-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhopadhyay I, et al. Transient receptor potential ankyrin 1 receptor activation in vitro and in vivo by pro-tussive agents: GRC 17536 as a promising anti-tussive therapeutic. PLoS One. 2014;9:e97005. doi: 10.1371/journal.pone.0097005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moldenhauer H, Latorre R, Grandl J. The pore-domain of TRPA1 mediates the inhibitory effect of the antagonist 6-methyl-5-(2-(trifluoromethyl)phenyl)-1H-indazole. PLoS One. 2014;9:e106776. doi: 10.1371/journal.pone.0106776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takaishi M, Uchida K, Fujita F, Tominaga M. Inhibitory effects of monoterpenes on human TRPA1 and the structural basis of their activity. J Physiol Sci. 2014;64:47–57. doi: 10.1007/s12576-013-0289-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banzawa N, et al. Molecular basis determining inhibition/activation of nociceptive receptor TRPA1 protein: A single amino acid dictates species-specific actions of the most potent mammalian TRPA1 antagonist. J Biol Chem. 2014;289:31927–31939. doi: 10.1074/jbc.M114.586891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta R, et al. Structural basis of TRPA1 inhibition by HC-030031 utilizing species-specific differences. Sci Rep. 2016;6:37460. doi: 10.1038/srep37460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatsuka K, et al. Identification of molecular determinants for a potent mammalian TRPA1 antagonist by utilizing species differences. J Mol Neurosci. 2013;51:754–762. doi: 10.1007/s12031-013-0060-2. [DOI] [PubMed] [Google Scholar]
- 21.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ton HT, Phan TX, Abramyan AM, Shi L, Ahern GP. Identification of a putative binding site critical for general anesthetic activation of TRPA1. Proc Natl Acad Sci USA. 2017;114:3762–3767. doi: 10.1073/pnas.1618144114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryckmans T, et al. Design and pharmacological evaluation of PF-4840154, a non-electrophilic reference agonist of the TrpA1 channel. Bioorg Med Chem Lett. 2011;21:4857–4859. doi: 10.1016/j.bmcl.2011.06.035. [DOI] [PubMed] [Google Scholar]
- 24.Pryde DC, et al. The discovery of a potent series of carboxamide TRPA1 antagonists. MedChemComm. 2016;7:2145–2158. [Google Scholar]
- 25.Pryde DC, et al. Discovery of a series of indazole TRPA1 antagonists. ACS Med Chem Lett. 2017;8:666–671. doi: 10.1021/acsmedchemlett.7b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi N, et al. TRPV4 channel activity is modulated by direct interaction of the ankyrin domain to PI(4,5)P2. Nat Commun. 2014;5:4994. doi: 10.1038/ncomms5994. [DOI] [PubMed] [Google Scholar]
- 27.Michaely P, Tomchick DR, Machius M, Anderson RG. Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J. 2002;21:6387–6396. doi: 10.1093/emboj/cdf651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zayats V, et al. Regulation of the transient receptor potential channel TRPA1 by its N-terminal ankyrin repeat domain. J Mol Model. 2013;19:4689–4700. doi: 10.1007/s00894-012-1505-1. [DOI] [PubMed] [Google Scholar]
- 29.Nagatomo K, Ishii H, Yamamoto T, Nakajo K, Kubo Y. The Met268Pro mutation of mouse TRPA1 changes the effect of caffeine from activation to suppression. Biophys J. 2010;99:3609–3618. doi: 10.1016/j.bpj.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gracheva EO, et al. Molecular basis of infrared detection by snakes. Nature. 2010;464:1006–1011. doi: 10.1038/nature08943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cordero-Morales JF, Gracheva EO, Julius D. Cytoplasmic ankyrin repeats of transient receptor potential A1 (TRPA1) dictate sensitivity to thermal and chemical stimuli. Proc Natl Acad Sci USA. 2011;108:E1184–E1191. doi: 10.1073/pnas.1114124108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jabba S, et al. Directionality of temperature activation in mouse TRPA1 ion channel can be inverted by single-point mutations in ankyrin repeat six. Neuron. 2014;82:1017–1031. doi: 10.1016/j.neuron.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim DH, et al. An ankyrin repeat domain of AKR2 drives chloroplast targeting through coincident binding of two chloroplast lipids. Dev Cell. 2014;30:598–609. doi: 10.1016/j.devcel.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodford CR, Thoden JB, Holden HM. New role for the ankyrin repeat revealed by a study of the N-formyltransferase from Providencia alcalifaciens. Biochemistry. 2015;54:631–638. doi: 10.1021/bi501539a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Islam Z, Nagampalli RSK, Fatima MT, Ashraf GM. New paradigm in ankyrin repeats: Beyond protein-protein interaction module. Int J Biol Macromol. 2018;109:1164–1173. doi: 10.1016/j.ijbiomac.2017.11.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.