Significance
We used a hybrid design combining epidermis samples from both deep phenotyping of 20 individuals and snapshot data from more than 200 other human subjects to find biomarkers of the human circadian phase. Using CYCLOPS to reconstruct the temporal order of all samples, we identified hundreds of genes cycling at population level and found that phase relationships of human skin epidermis output genes are conserved with the mouse skin. Finally, we showed that human epidermis has a stronger clock than blood and developed a panel of biomarkers that can phase individuals to within 3 hours from a single sample.
Keywords: human skin, hybrid design, circadian medicine, population-level rhythms, biomarkers
Abstract
Skin is the largest organ in the body and serves important barrier, regulatory, and sensory functions. The epidermal layer shows rhythmic physiological responses to daily environmental variation (e.g., DNA repair). We investigated the role of the circadian clock in the transcriptional regulation of epidermis using a hybrid experimental design, in which a limited set of human subjects (n = 20) were sampled throughout the 24-h cycle and a larger population (n = 219) were sampled once. We found a robust circadian oscillator in human epidermis at the population level using pairwise correlations of clock and clock-associated genes in 298 epidermis samples. We then used CYCLOPS to reconstruct the temporal order of all samples, and identified hundreds of rhythmically expressed genes at the population level in human epidermis. We compared these results with published time-series skin data from mice and found a strong concordance in circadian phase across species for both transcripts and pathways. Furthermore, like blood, epidermis is readily accessible and a potential source of biomarkers. Using ZeitZeiger, we identified a biomarker set for human epidermis that is capable of reporting circadian phase to within 3 hours from a single sample. In summary, we show rhythms in human epidermis that persist at the population scale and describe a path to develop robust single-sample circadian biomarkers.
Skin is the largest organ in the body, functioning in the biosynthesis of vitamins and protection from the environment (1, 2). Anticipated daily variation in stressors (e.g., temperature, humidity, UV light) is managed by corresponding changes in skin physiology, especially in the epidermal layer. For example, genetic programs regulating responses to transepidermal water loss, cell proliferation, and DNA repair all undergo daily change (3–5). These changes depend on an intact clock in mouse epidermis (5–7). Human epidermis is no exception, as a pilot study identified hundreds of transcripts that vary in the daytime (8).
In addition to knowledge gained in exploring the biology of human epidermis, it is also a potential source of circadian biomarkers. For circadian medicine to influence health, such as when to take a drug (9) or undergo a procedure (10), a practical measure of circadian phase is needed. The current gold standard tool for assessing human circadian phase is the dim-light melatonin-onset (DLMO) assay. This assay requires a subject to sit in a dim room for repeated saliva sample collection, a difficult practice to standardize and perform at scale and unsuitable for clinical practice. Furthermore, DLMO is a biomarker for the circadian phase of locomotor activity. This may or may not be aligned with clocks in the periphery regulating drug metabolism or action. Which is the best source of circadian biomarkers? What aspects of circadian biology should these biomarkers measure? These remain important and open questions.
Over the last decade, many groups have sought to develop more practical and informative circadian biomarkers. Originally, this research focused on whole blood transcriptomics and metabolomics; however, there is little evidence this work has influenced clinical care (11, 12). Moreover, whether whole blood is the best source for circadian biomarkers has been called into question. The composition of blood varies greatly over time and between subjects due to internal (e.g., endocrine, body temperature, individual variation) and external (e.g., diet, infection) factors. Recently, Wittenbrink et al. (13) explored a blood cell type, CD14+ monocytes, as a source of circadian biomarkers in a two-stage process. The discovery study was longitudinal and included 12 healthy young people in a hospital environment. The validation study focused on 28 individuals, following gene expression measured in their home environment. These excellent studies showed accurate assignment of DLMO phase from a single sample to within 3 h; however, these studies remain relatively small scale (40 total individuals), the study population was generally young (18–41 y), and only part of the circadian cycle was predicted (morning and afternoon phases). Will these markers extend to larger populations? To different age groups? Ethnicities? Can they accurately report the entire circadian cycle?
Here we explore epidermis as an alternative source of robust biomarkers of circadian phase. We describe a hybrid experimental design in which a limited set of subjects (n = 20; age 21–49 y) was sampled throughout the 24-h cycle and a larger population (n = 219; age 20–74 y) was sampled once. Using a recently described algorithm, cyclic ordering by periodic structure (CYCLOPS) (14), we reconstructed the circadian transcriptome from human epidermis and found hundreds of clock-regulated genes with rhythms persisting at the population level. We compared these results with time-series data collected from mouse skin and found strong concordance in circadian phase across species at the transcript and pathway levels. Finally, we applied a second algorithm, ZeitZeiger, to these reconstructed time course data. We describe a biomarker set for human epidermis capable of reporting circadian phase to within 3 h from a single sample. In summary, we present a survey of circadian rhythms in human epidermis and point to skin as a source of robust single-sample circadian biomarkers suitable for population-scale research.
Results
Clock-Regulated Genes Identified from Time-Series Analysis of Human Epidermis.
To explore the potential of human skin as a source of circadian phase biomarkers, we performed a time-series analysis of human epidermis every 6 h across a 24-h sampling period from 19 individuals (subject 115 was excluded; SI Appendix, Table S1). In each subject, an epidermal biopsy specimen was collected every 6 h starting at 12 PM, for a total of four specimens. We used MetaCycle’s meta3d function to analyze these data. We identified 110 genes (P < 0.05; SI Appendix, Fig. S1 A and B) that varied with a rhythmic pattern over a 24-h period. These genes show a bimodal distribution, with peak phases clustered at 8–9 AM and 8–9 PM (SI Appendix, Fig. S1C). We reasoned that genes with a circadian pattern showing evolutionary conservation between mice and humans would represent a robust source for circadian biomarkers. If it was conserved across 80 My of evolution, it should be conserved across different populations of humans. To identify these genes using a common statistical framework, we applied MetaCycle’s meta2d function to a recently published time-series dataset of mouse telogen skin (containing small and resting hair follicles) and anagen skin (containing large and growing hair follicles) (7). Our reanalysis found 1,280 and 294 circadian genes in telogen and anagen skin, respectively (P < 0.05). Similar to our observations in human epidermis, we noticed a pronounced bimodal phase distribution from mouse telogen, as has been reported in analyses of many other mouse tissues (15) (SI Appendix, Fig. S1D).
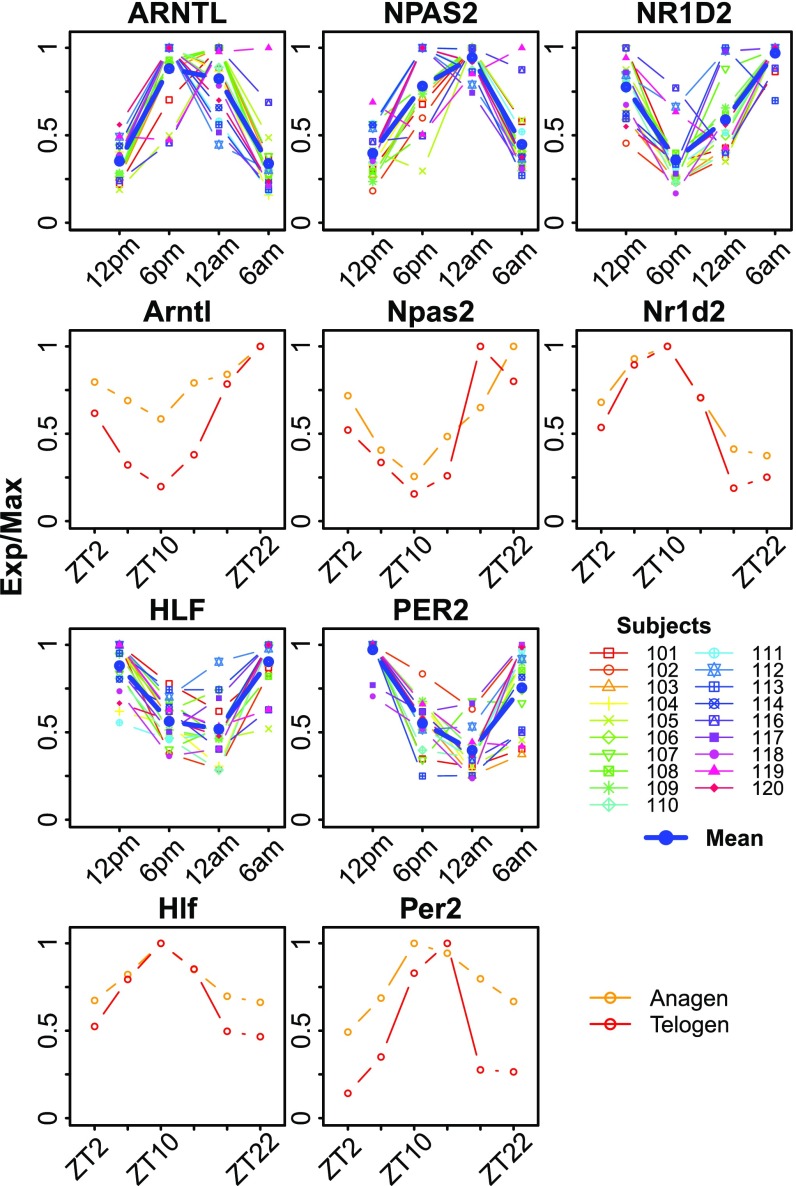
Expression patterns for five genes—ARNTL, NPAS2, NR1D2, HLF, and PER2—were conserved across all three datasets (Fig. 1 and SI Appendix, Fig. S1E). We compared the expression profiles of each of these five genes across all human subjects. The patterns of expression for these five genes generally followed the same phase relationship (Fig. 1); however, there were clear interindividual differences. For example, subjects 116 and 119 show delayed clock phases of genes, salivary melatonin, and cortisol (SI Appendix, Fig. S2). These five clock genes were also robustly rhythmic in mouse telogen and anagen, albeit with a nocturnal pattern of expression (Fig. 1). As noted previously, the phase of clock gene expression reflects locomotor activity; that is, ARNTL peaks at 8–9 PM in humans and ZT23 in mouse, predicting the sleep phase in both species. The phase of clock gene expression matches the established phase relationships: ARNTL precedes NR1D2, which precedes PER2 (16).
Fig. 1.
Expression profiles of conserved circadian genes in human and mouse skin. Epidermis from forearms of human subjects (n = 19) was sampled every 6 h for one circadian cycle starting at 12 PM. Expression profiles from all 19 human subjects along with the mean (blue) are shown (rows 1 and 3). Expression profiles for mouse anagen (orange) and telogen (red) skin samples are shown (rows 2 and 4). Mouse data are from Geyfman et al. (7) (SI Appendix, Table S1). Exp/Max indicates the expression value at each time point normalized to the maximum expression across the time series.
Reproducing Human Epidermis Phase Without Wall Time.
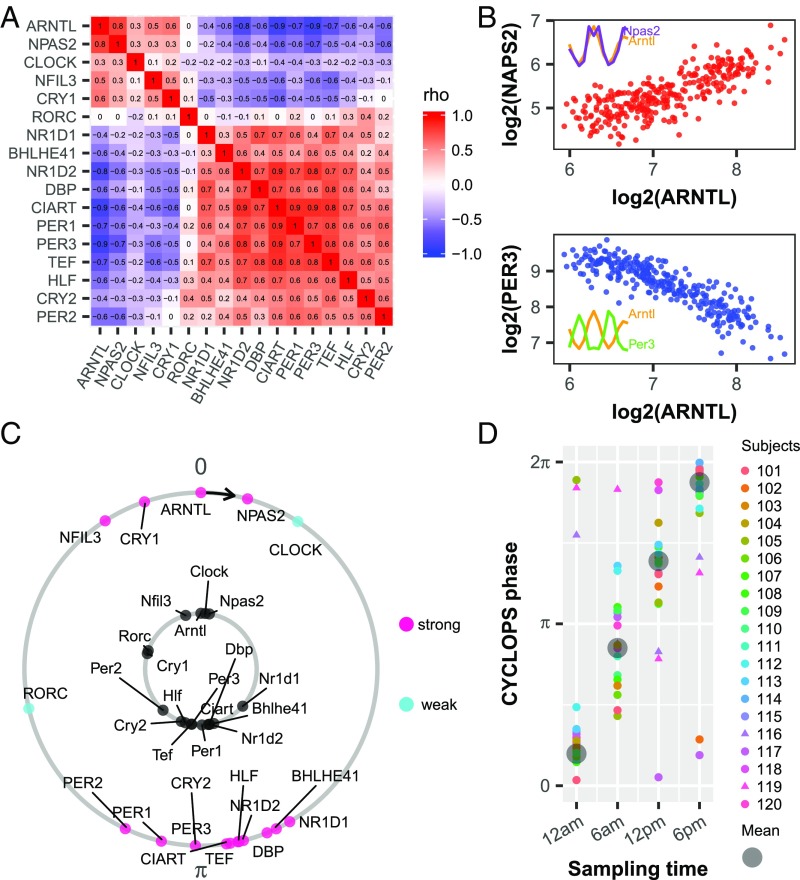
For skin epidermis to be a good source for determining human circadian phase, clock gene phase relationships should be conserved at a population scale. In other words, the circadian variation in clock gene expression must exceed interindividual variation. We analyzed a gene expression dataset from human epidermis from 20 subjects sampled around a 24-h clock and 219 subjects each sampled once irrespective of wall time (SI Appendix, Table S1). This hybrid design (SI Appendix, Fig. S3) captures advantages of both longitudinal and population-based studies while mitigating disadvantages. Using a set of clock and clock-associated genes (Materials and Methods) (17–20), we applied Spearman’s rho to evaluate the correlation of each gene against all others (21) (Fig. 2A). If the clock is intact, then ARNTL should correlate positively with its partner transcriptional activator, NPAS2, and negatively with its target, NR1D1, which represses ARNTL. As expected, we saw strong positive correlations among ROR/REV-ERB-response element (RORE) targets ARNTL, NPAS2, and CLOCK and strong negative correlations between these genes and their E-box targets DBP, NR1D1, and PER3 (Fig. 2 A and B).
Fig. 2.
Evaluation of circadian function in human epidermis. (A) Heat map of Spearman’s ρ for clock and clock-associated genes from ordered data (n = 20; SI Appendix, Table S1) and unordered data (n = 219; SI Appendix, Table S1) showing a conserved correlation structure. (B) Examples of clock genes with positively correlated (ARNTL and NPAS2, red) and negatively correlated (ARNTL and PER3, blue) expression. Each point represents one human sample. (Inset) Expression profiles of corresponding clock genes from mouse telogen. (C) CYCLOPS was used to order all 298 human samples. In an intact clock, ROR- phased genes (e.g., ARNTL, NPAS2, CLOCK) always peak before E-box–phased genes (e.g., NR1D1, DBP, PER1). Conserved phase relationships are shown for clock and clock-associated genes (internal circle, mouse; external circle, human). For human genes, pink indicate strong cycling (P < 0.01, rAMP >0.1, rsq >0.1) and cyan depicts weaker cycling. The phase of Arntl (mouse) or ARNTL (human) is set as 0 to facilitate comparison. (D) CYCLOPS accurately recalls circadian phase from 20 subjects. Different colors indicate different subjects, and the circular average phases for all samples is shown in gray. Samples from subjects 116 and 119 are indicated by triangle points.
To reconstruct the daily rhythm of genome-wide gene expression in human epidermis, we applied CYCLOPS (Materials and Methods). We first evaluated the accuracy of the reconstructed sample order by comparing clock gene phase order between humans and mice (Fig. 2C). By both visual inspection and statistical analysis (Fisher’s circular correlation, 0.715), the reconstruction of the human circadian cycle (Fig. 2C, outer circle) matched that of the mouse (Fig. 2C, inner circle). In addition, we compared CYCLOPS-predicted sample phase for the 20 subjects for which sampling time was available. The strong linear relationship between the mean CYCLOPS-predicted phase and sampling time suggests accurate sample ordering (Fisher’s circular correlation, 0.955; Fig. 2D). As expected, the interindividual circadian phase variability was also reflected in the CYCLOPS-predicted phase (e.g., samples from subject 116 and 119 were separated from the mean phase at each time point; Fig. 2D).
Human Transcriptional Rhythms Are Evolutionarily Conserved.
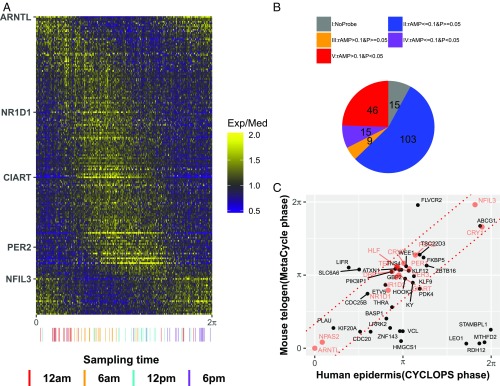
We expanded our analysis of rhythms in human epidermis to the broader transcriptome from 298 samples collected from 238 donors. Using the CYCLOPS modified cosinor regression algorithm, we selected 188 circadian genes [P < 0.01, relative amplitude (rAMP) >0.1, goodness-of-fit (rsq) >0.1, fitmean >16] (Fig. 3A, SI Appendix, Fig. S4, and Dataset S1). While RORE and E-box phases had dominant signatures, other phases were represented as well (Fig. 3A). Our central hypothesis is that high-amplitude oscillatory genes with conserved phase relationships across species will be the most robust biomarkers. Comparing periodic genes from mouse telogen, we defined five categories of genes, including those with (i) no mouse homologs, (ii) low amplitude and no significant cycling in mouse, (iii) high amplitude and no significant cycling, (iv) low amplitude and cycling, and (v) high amplitude and cycling (Fig. 3B). For those genes that were robustly cycling in mouse telogen, we compared their phases and found a strong linear correlation for most (Fig. 3C). These evolutionary conserved clock and clock output genes suggest the improved power of a hybrid design compared with a longitudinal design.
Fig. 3.
Population analysis of human epidermis data revealing conserved circadian transcriptional output. (A) Heatmap of median centered gene expression values for all human samples (n = 298) ordered by CYCLOPS phase and all significant circadian genes (n = 188). Clock genes that cover a range of circadian phases (e.g., ARNTL, NR1D1) are labeled on the y-axis. Samples with known sample acquisition times (n = 79) are shown along the x-axis. (B) All circadian genes in human epidermis. Groups are no mouse homolog (gray), low amplitude (rAMP ≤0.1) and not cycling (P ≥0.05) (blue), high amplitude (rAMP >0.1) and not cycling (orange), low amplitude and cycling (P <0.05) (purple), and high amplitude and cycling (red). (C) A comparison of high-amplitude cycling genes (n = 46) revealing strong phase conservation between mice and humans. Clock and clock-associated genes are shown in red. The predicted gene phases from CYCLOPS and MetaCycle were adjusted to ARNTL/Arntl phase (set to 0).
We investigated the underlying biology using phase set enrichment analysis (22) and identified temporal regulation of eight significantly enriched pathways at the population level (SI Appendix, Fig. S5A). Three of these pathways are also enriched in mouse telogen skin (SI Appendix, Fig. S5 B and C), including pathways related to the cell cycle, adaptive immune system, and matrisome. Interestingly, the phase order of the pathways is also conserved (SI Appendix, Fig. S5C), demonstrating the orderly progression of the skin’s circadian gene expression program.
Population-Level Biomarkers of Circadian Phase in Human Epidermis.
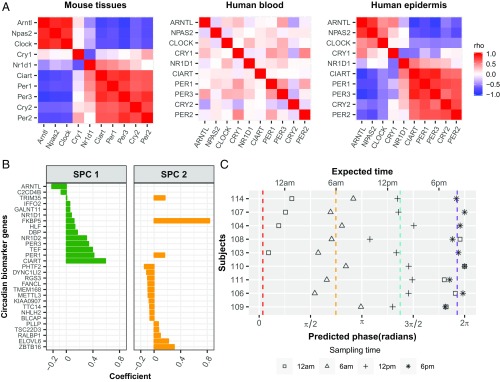
In addition to analyzing clock-regulated biology, these datasets provide an opportunity to discover robust biomarkers of circadian phase that persist at the population level. To date, studies performed on human whole blood suggest high variability and low-amplitude rhythms from mixed cell types (23). Furthermore, when Brown et al. (24) analyzed single cell types from blood, they found a significantly weaker autonomous oscillator in blood monocytes than in fibroblasts. To clarify which human tissue is a better source of circadian biomarkers, we compared the expression correlation matrices of 10 core clock genes (18–20, 25) from time-series samples of human whole blood (n = 201) (26), skin epidermis (n = 79), and multiple mouse tissues (n = 144) (15). The coexpression pattern of these 10 core clock genes across 12 mouse tissues is similar (Mantel test, P = 3e-06) to human epidermis, but not blood (Mantel test, P = 0.897) (Fig. 4A). At population level, the coexpression pattern of clock genes is also stronger in epidermis (Fig. 2A) than in isolated monocytes or T cells collected from hundreds of individuals (SI Appendix, Fig. S6 A and B). This suggests that skin epidermis is a strong alternative tissue for identifying robust circadian biomarkers at the population level.
Fig. 4.
Population-level biomarkers of circadian phase from a single human epidermal sample. (A) Evaluation of circadian clock function from 12 mouse tissues (Left), human blood (Middle), and human epidermis (Right). The mouse data are from Zhang et al. (15) and are included as a benchmark. Red and blue indicate positive and negative Spearman’s ρ, respectively. (B) ZeitZeiger (27) was used to select a set of 29 circadian marker gene candidates. (C) Validation of predicted markers. Using the circadian marker set and ZeitZeiger, we analyzed samples collected every 6 h over a circadian day for nine subjects who were excluded from the training set. Average predicted phases of samples collected at 12 AM, 6 AM, 12 PM, and 6 PM are indicated with red, orange, cyan, and purple dashed lines, respectively.
We used ZeitZeiger (27) to identify biomarkers that can accurately report circadian phase. ZeitZeiger was trained with CYCLOPS-assigned phase from all 219 samples without sampling time and 43 samples with sampling time (Materials and Methods). This resulted in a set of 29 candidate biomarkers (Fig. 4B), with approximately one-half expressed in two different phases, represented by two sparse principal components, SPC1 and SPC2 (Fig. 4B). To validate these biomarkers, we analyzed a set of nine human subjects each measured at four separate times of day. The average predicted phase was within 3 h at all four time points compared with the expected time (Fig. 4C). For six of the subjects, it was difficult to distinguish between the 6 PM and 12 AM samples. When we evaluated the sample relationships for different times of day, the mean absolute error was 2.95 h for the nine subjects in this study (SI Appendix, Fig. S7); nevertheless, we were able to accurately assign 30 of 36 samples to their correct circadian phase.
Discussion
Here we report a population-based analysis of circadian rhythms in human epidermis. We used a hybrid experimental design to examine circadian gene expression, in which 20 subjects were sampled longitudinally and 219 subjects were sampled once without regard to circadian time. We found that clock gene correlations are strong and evident in unordered data from human epidermis. We used CYCLOPS (14) to order these samples and explore population-level transcriptional rhythms. As predicted, the phases of core clock machinery and most clock outputs are shared between mice and humans. For pathways that are clock- regulated in both species, the phase order is preserved, with the cell cycle peaking early in the inactive phase, followed by the immune system and then the matrisome. Finally, prompted by the observation that human epidermis appears to be highly rhythmic, we used ZeitZeiger to search for a set of biomarkers that accurately reports circadian phase from a single sample. These results have broad implications for skin physiology and circadian medicine.
The gold standard sampling resolution for genome-wide circadian transcriptomics requires sampling every 2 h for 2 d (28). This is not practical for human studies, however. An alternative strategy is a hybrid design that incorporates a small subset of longitudinally sampled subjects with hundreds of single time point samples. This design overcomes key limitations of traditional time-series experiments in humans, including (i) low statistical power, (ii) large interindividual variation, and (iii) cost. We believe that the hybrid design could be advantageous for circadian studies in many different human tissues.
Among the evolutionarily conserved clock output genes found in skin, several are components of the cell cycle machinery. These include CDC20, CDC25B, KIF20A, and WEE1, all of which are associated with mitosis and may contribute to circadian phase-dependent proliferation of epidermal cells (7, 29). Interestingly, liver cells regenerate in a Wee1-dependent fashion at a single phase in the circadian cycle (30). The cellular clock in human skin may also use WEE1 to gate epidermal stem cell division. This process is key to epidermal homeostasis, as new cells replace those lost during turnover or injury (31).
Biomarkers capable of reporting circadian phase are essential for circadian medicine. Our data showing strong population-level rhythms in human epidermis suggest the epidermis as a source for single-sample circadian biomarkers suitable for population scale research. Recent studies have shown that the timing of medical interventions can impact patient response (10; reviewed in ref. 9). The current standard in the field, DLMO, is impractical and burdensome. The identification of a biomarker set that can accurately predict circadian phase to within 3 h from a single sample is a major step toward translating circadian medicine.
Numerous groups have sought circadian biomarkers from whole blood, including analysis of metabolites and gene expression (11, 12, 32). However, whole blood is a heterogeneous mixture that changes composition in response to diet, infection, exercise, and many other factors, likely explaining the weak rhythmic expression of clock genes in time-series whole blood samples (23). Analysis of publicly available datasets including hundreds of subjects showed that the oscillator is far stronger in human epidermis than in isolated T cells or monocytes (SI Appendix, Fig. S6 A–C); however, an excellent recent study identified promising circadian biomarkers from isolated monocytes from young subjects (13). This raises the possibility that certain blood cell types may be a useful source of biomarkers. There are several important differences between our present study and that previous study, however, including varying gene expression platforms and experimental designs. We analyzed a larger population (239 subjects vs. 40 subjects) with a wider age range (20–74 y vs. 18–41 y), and validated the biomarker set against time points covering the entire circadian cycle (morning/noon/afternoon/midnight vs. morning/afternoon). Head to head comparisons are needed to define the best source of biomarkers for each application of circadian medicine.
Clinical use of circadian biomarkers will require fast, cost-effective, and noninvasive sampling techniques (e.g., tape strip-based, hair follicle- or oral mucosa- based) (33) and standardization across platforms (e.g., qPCR or NanoString). Larger studies would also benefit from a comparison with other standardized techniques (e.g., DLMO, temperature, actigraphy). Finally, the consistency of results should be confirmed across a range of disease states and pathologies. Solving these problems will help unlock the potential of circadian medicine.
Materials and Methods
Human Clinical Experimental Design and Sample Collection.
Our clinical studies adhered to the guidelines of the International Council on Harmonization of Good Clinical Practices and the principles expressed in the Declaration of Helsinki. Associated protocols were approved by Aspire’s Institutional Review Board, and each subject provided informed consent. For collecting the ordered samples, 20 healthy Caucasian male subjects, aged 21–49 y, were recruited and housed in a facility that specializes in sleep studies (Community Research). Subject inclusion criteria and rules of Community Research are described in detail in SI Appendix, SI Methods. Saliva samples were collected every 3 h over the same 24-h period to confirm normal cycling of cortisol and melatonin (SI Appendix, Figs. S8 and S9). Full-thickness (2 mm) punch biopsy specimens were collected from sites on the forearms of each subject at 6-h intervals over a 24-h period (6 AM, 12 PM, 6 PM, and 12 AM). The biopsy specimens were processed for separation into epidermal and dermal compartments by laser capture microdissection (SI Appendix, SI Methods). Collection of the unordered samples from 152 Caucasian and 67 African-American females has been described previously (34). The mRNA target labeling and processing steps are described in detail in SI Appendix, SI Methods.
Time-Series Analysis with MetaCycle.
The RMA algorithm was used to extract expression profiles from the raw CEL files. MetaCycle (35) was used to detect circadian genes (P < 0.05 and rAMP >0.1) from time-series expression profiles of human epidermis samples and mouse anagen and telogen samples (7), as detailed in SI Appendix, SI Methods.
Ordering Human Epidermal Samples with CYCLOPS.
Before CYCLOPS ordering, we verified the presence of an intact circadian clock in human epidermis. To do this, we constructed pairwise gene correlation matrices for 17 clock and clock-associated genes, using a method similar to that described by Shilts et al. (21). Our criteria for selecting clock and clock-associated genes were high-amplitude rhythms in most (8 of 12) mouse tissues from Zhang et al. (15) and coverage over multiple phases of expression (driven by E-box, D-box, and RORE elements) (17–20). Next, all skin samples (298 in total) were ordered with CYCLOPS (Julia version 0.3.12) (14). To select eigengenes for CYCLOPS, we incorporated features of a second algorithm, Oscope (36), to filter periodic out-of-phase eigengenes. The implementation of CYCLOPS and Oscope are described in detail in SI Appendix, Methods. Using the best sample ordering, modified cosinor regression was applied to each expressed gene. Genes with P < 0.01, fitmean >16, rAMP >0.1, and rsq >0.1 were designated as circadian.
Using ZeitZeiger to Identify Epidermal Biomarkers of Circadian Phase.
ZeitZeiger (27) was used to identify circadian biomarkers from CYCLOPS-ordered human epidermis samples. All samples were divided into two datasets: testing and training. The testing set included 36 time-stamped samples from nine subjects (each subject with four samples). Clock genes in these nine subjects followed the mean expression profile of all subjects (Fig. 1). These nine subjects were selected for testing because they had CYCLOPS-predicted phases surrounding the mean phase at each time point (Fig. 2D), enhancing the ability to evaluate prediction accuracy. The training set included all 219 samples without sampling time and the remaining 43 time-stamped samples collected from 11 subjects. Circadian biomarkers were selected from the training dataset and used for predicting sample phases in the test set, as detailed in SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank Dr. Joseph Kaczvinsky and Kathleen Werchowski for clinical protocol development and Jim Li for statistical analysis for the time-series clinical study. We also thank Drs. Bhavani Kasibhatla, Charles Bascom, and Rosemarie Osborne for helpful discussions and Tiago de Andrade and Garrett Rogers for thoughtful discussions. Procter and Gamble paid for 100% of the costs of clinical work reported in this paper. This work is supported by National Institute of Neurological Disorders and Stroke Grants 2R01NS054794 and 1R21NS101983 (to J.B.H.), National Human Genome Research Institute Grant 2R01HG005220 (to J.B.H.), Defense Advanced Research Projects Agency Grant D17AP00003 (to R.C.A.), and National Institute of General Medical Sciences Grant R35GM124685 (to J.J.H.).
Footnotes
Conflict of interest statement: K.J.M., J.E.O., J.D.S., and R.T. are employees of the Procter & Gamble Company, which markets skin care products.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE112660).
See Commentary on page 12095.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809442115/-/DCSupplemental.
References
- 1.Solanas G, Benitah SA. Regenerating the skin: A task for the heterogeneous stem cell pool and surrounding niche. Nat Rev Mol Cell Biol. 2013;14:737–748. doi: 10.1038/nrm3675. [DOI] [PubMed] [Google Scholar]
- 2.Engelsen O, Brustad M, Aksnes L, Lund E. Daily duration of vitamin D synthesis in human skin with relation to latitude, total ozone, altitude, ground cover, aerosols and cloud thickness. Photochem Photobiol. 2005;81:1287–1290. doi: 10.1562/2004-11-19-RN-375. [DOI] [PubMed] [Google Scholar]
- 3.Yosipovitch G, et al. Time-dependent variations of the skin barrier function in humans: Transepidermal water loss, stratum corneum hydration, skin surface pH, and skin temperature. J Invest Dermatol. 1998;110:20–23. doi: 10.1046/j.1523-1747.1998.00069.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown WR. A review and mathematical analysis of circadian rhythms in cell proliferation in mouse, rat, and human epidermis. J Invest Dermatol. 1991;97:273–280. doi: 10.1111/1523-1747.ep12480379. [DOI] [PubMed] [Google Scholar]
- 5.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci USA. 2011;108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plikus MV, et al. The circadian clock in skin: Implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J Biol Rhythms. 2015;30:163–182. doi: 10.1177/0748730414563537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geyfman M, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci USA. 2012;109:11758–11763. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spörl F, et al. Krüppel-like factor 9 is a circadian transcription factor in human epidermis that controls proliferation of keratinocytes. Proc Natl Acad Sci USA. 2012;109:10903–10908. doi: 10.1073/pnas.1118641109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dallmann R, Okyar A, Lévi F. Dosing-time makes the poison: Circadian regulation and pharmacotherapy. Trends Mol Med. 2016;22:430–445. doi: 10.1016/j.molmed.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Montaigne D, et al. Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbα antagonism: A single-centre propensity-matched cohort study and a randomised study. Lancet. 2017;391:59–69. doi: 10.1016/S0140-6736(17)32132-3. [DOI] [PubMed] [Google Scholar]
- 11.Laing EE, et al. Blood transcriptome-based biomarkers for human circadian phase. eLife. 2017;6:e20214. doi: 10.7554/eLife.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hughey JJ. Machine learning identifies a compact gene set for monitoring the circadian clock in human blood. Genome Med. 2017;9:19. doi: 10.1186/s13073-017-0406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wittenbrink N, et al. High-accuracy determination of internal circadian time from a single blood sample. J Clin Invest. 2018;128:3826–3839. doi: 10.1172/JCI120874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anafi RC, Francey LJ, Hogenesch JB, Kim J. CYCLOPS reveals human transcriptional rhythms in health and disease. Proc Natl Acad Sci USA. 2017;114:5312–5317. doi: 10.1073/pnas.1619320114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci USA. 2014;111:16219–16224. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korenčič A, et al. Timing of circadian genes in mammalian tissues. Sci Rep. 2014;4:5782. doi: 10.1038/srep05782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ukai H, Ueda HR. Systems biology of mammalian circadian clocks. Annu Rev Physiol. 2010;72:579–603. doi: 10.1146/annurev-physiol-073109-130051. [DOI] [PubMed] [Google Scholar]
- 18.Anafi RC, et al. Machine learning helps identify CHRONO as a circadian clock component. PLoS Biol. 2014;12:e1001840. doi: 10.1371/journal.pbio.1001840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goriki A, et al. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 2014;12:e1001839. doi: 10.1371/journal.pbio.1001839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Annayev Y, et al. Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression. J Biol Chem. 2014;289:5013–5024. doi: 10.1074/jbc.M113.534651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shilts J, Chen G, Hughey JJ. Evidence for widespread dysregulation of circadian clock progression in human cancer. PeerJ. 2018;6:e4327. doi: 10.7717/peerj.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang R, Podtelezhnikov AA, Hogenesch JB, Anafi RC. Discovering biology in periodic data through phase set enrichment analysis (PSEA) J Biol Rhythms. 2016;31:244–257. doi: 10.1177/0748730416631895. [DOI] [PubMed] [Google Scholar]
- 23.Arnardottir ES, et al. Blood-gene expression reveals reduced circadian rhythmicity in individuals resistant to sleep deprivation. Sleep (Basel) 2014;37:1589–1600. doi: 10.5665/sleep.4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown SA, et al. The period length of fibroblast circadian gene expression varies widely among human individuals. PLoS Biol. 2005;3:e338. doi: 10.1371/journal.pbio.0030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ripperger JA, Jud C, Albrecht U. The daily rhythm of mice. FEBS Lett. 2011;585:1384–1392. doi: 10.1016/j.febslet.2011.02.027. [DOI] [PubMed] [Google Scholar]
- 26.Möller-Levet CS, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110:E1132–E1141. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughey JJ, Hastie T, Butte AJ. ZeitZeiger: Supervised learning for high-dimensional data from an oscillatory system. Nucleic Acids Res. 2016;44:e80. doi: 10.1093/nar/gkw030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes ME, et al. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janich P, et al. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 30.Matsuo T, et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- 31.Blanpain C, Fuchs E. Epidermal homeostasis: A balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akashi M, et al. Noninvasive method for assessing the human circadian clock using hair follicle cells. Proc Natl Acad Sci USA. 2010;107:15643–15648. doi: 10.1073/pnas.1003878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimball AB, et al. Age-induced and photoinduced changes in gene expression profiles in facial skin of Caucasian females across 6 decades of age. J Am Acad Dermatol. 2018;78:29–39.e7. doi: 10.1016/j.jaad.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 35.Wu G, Anafi RC, Hughes ME, Kornacker K, Hogenesch JB. MetaCycle: An integrated R package to evaluate periodicity in large- scale data. Bioinformatics. 2016;32:3351–3353. doi: 10.1093/bioinformatics/btw405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leng N, et al. Oscope identifies oscillatory genes in unsynchronized single-cell RNA-seq experiments. Nat Methods. 2015;12:947–950. doi: 10.1038/nmeth.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.