Loss of fragile X mental retardation protein (FMRP) in fragile X syndrome is the most common cause of inherited intellectual deficiency and is associated with additional neurodevelopmental issues, including increased risks of autism and epilepsy and characteristic physical changes (1). FMRP is an mRNA binding protein (RBP) implicated in many distinct modes of mRNA regulation (2). Recently, the increased proliferation and dysregulated differentiation of adult neural stem cells has been proposed to be an underlying cause of behavioral changes in a mouse model of fragile X syndrome (3). In PNAS, Liu et al. (4) use a comprehensive combination of transcriptome analysis and ribosome profiling in a purified population of neural stem cells to define changes caused by the loss of FMRP in this cell type. This work shows how the loss of FMRP has effects on many pathways through both direct and indirect mechanisms, providing insight into both the phenotype of FMRP loss and the more general issue of how to use a systems approach to study the loss of a single RBP.
There are over 750 RBPs in the genome, ∼3.8% of the protein-coding regions (5). Not only are there many different RBPs, but each individual RBP can play distinct roles in the life of the mRNA, controlling splicing, nuclear export, stability, translatability, and transport. Moreover, over 200 RBPs are associated with a heritable disease (6). Thus, it is important to find ways of establishing how to study the impact of the loss or mutation of an individual RBP.
In their paper, Liu et al. (4) combine RNA sequencing (RNA-seq; to examine changes in the transcriptome) and ribosome profiling (to examine changes in the loading of ribosomes on mRNAs) to define changes caused by the loss of FMRP. The results of the loss of an RBP will be cell specific due to many factors: which mRNAs are expressed in that cell, competition between RBPs for similar binding sites, and cofactors that affect RBP actions. Thus, examination of the role of an RBP in a heterogeneous cell population will be diluted by distinct changes caused by the loss in different cells. Similarly, work in cell lines may not recapitulate results from primary cells. Unlike RNA-seq, which can now be done from single cells, albeit with low coverage of the whole transcriptome, as of now, ribosome profiling still requires a fairly large amount of material (>100,000 cells) because sedimentation of polysomes and subsequent purification of the resultant monosomes before sequencing are required. The ability to grow neuronal precursor cells isolated from the brain in neurospheres is therefore a major advantage for the present approach. While neurospheres are not completely homogeneous, they are largely made up of neural stem cells with only a small proportion of more-differentiated cells (7).
A major result of the paper by Liu et al. (4) is that by combining the effects on mRNA levels and ribosome loading (“translation”), six distinct types of regulation by FMRP were defined and the processes affected by these six modes were distinct, suggesting that these classifications define subgroups of FMRP-regulated mRNAs. FMRP is a known translational repressor, so it is not surprising that loss of FMRP leads to an increase in ribosome loading (“translation up” group) for many of the mRNAs known to directly bind to FMRP (8–10). In some cases, these changes are associated with decreases in the levels of mRNAs that act to normalize the expected translational output of the mRNA (“buffering up” group). Somewhat surprisingly, direct targets of FMRP binding were also enriched for mRNAs that were down-regulated without a change in ribosome loading (4), suggesting a role for FMRP in mRNA stabilization.
Although increased abundance of ribosomes on a message is normally interpreted as increased translational efficiency, slowing the elongation rate should increase the abundance of ribosomes on mRNAs while actually decreasing translation rates. FMRP has been implicated in slowing elongation in neurons (10, 11); however, the relatively small numbers of mRNAs found to have a significant decrease in the ribosomal density (“translation down”) after loss of FMRP were not the FMRP targets shown to be present in stalled polysomes in neurons (10). Stalled polysomes are postulated to be a constituent of the RNA granules important for mRNA transport in neurons (12), but since neural precursors and stem cells lack this requirement for mRNA transport to dendrites, it is not surprising that they appear to lack this form of regulation by FMRP. Interestingly, the “buffering down” group (increased mRNA, decreased ribosome loading) was enriched in mitochondrial ribosomal proteins and may be linked to changes in mitochondrial function after loss of FMRP seen in the neuronal precursor cells. Whether this group is primarily defined by changes in transcription of these mRNAs, followed by homeostatic decreases in translation (or vice versa) will require further study.
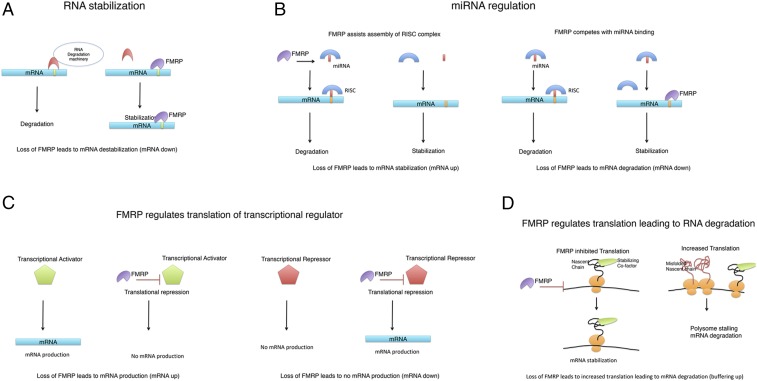
Why is there such a large effect on levels of mRNAs through loss of an RBP such as FMRP? There are four major possibilities (Fig. 1): (i) Loss of FMRP affects mRNA stability; (ii) loss of FMRP affects miRNA regulation of mRNAs, leading to changes in mRNA levels; (iii) loss of FMRP regulates the translation of transcriptional regulators that, in turn, regulate transcription of these mRNAs; and (iv) loss of FMRP leads to increased translation of mRNAs, which then stimulates a feedback decrease in the levels of mRNA, a form of buffering to conserve the amount of protein produced. Below, we expand on these possibilities.
Fig. 1.
Schematic for how loss of FMRP leads to changes in mRNA levels. (A) Sites in the mRNA (yellow bar) that bind to RBP link to the RNA degradation machinery, leading to RNA degradation. FMRP stabilizes mRNAs by competing for these sites. (B) FMRP assists in miRNA loading, leading to RNA degradation of the mRNAs targeted by that miRNA. Alternatively, FMRP binding to mRNA can prevent miRNA binding and stabilize mRNAs. (C) FMRP normally translationally represses both transcriptional activators and transcriptional repressors. Loss of FMRP increases levels of these proteins, leading to transcriptional changes. (D) FMRP increases ribosome loading on a subset of mRNAs. If these mRNAs are part of a complex, the altered stoichiometry of the complex may lead to unfolded nascent chains, ribosome stalling, and degradation of the mRNAs. RISC, RNA-induced silencing complex.
RNA Stability
RBPs decrease RNA stability by linking mRNAs to the RNA degradation machinery, whereas they increase RNA stability by binding to these same sites but not coupling to RNA degradation, competing with the destabilizing RBPs. Recently, FMRP has been shown to bind to mRNAs modified with an N6-methyladenosine (m6A) and has been proposed to stabilize those mRNAs by competing with m6A readers that lead to down-regulation of m6A-modified mRNA (13). It will be interesting to see whether the subset of mRNAs down-regulated without concomitant changes in translation in this study are enriched in m6A modifications.
miRNA Regulation
There is considerable evidence that FMRP regulates miRNA processing, and, in some cases, it regulates mRNA levels by competing for miRNA binding (14). Thus, changes in mRNA levels could be due to altered miRNA regulation. It would be interesting to compare the mRNAs whose levels were increased by loss of FMRP for a shared regulation with a set of miRNAs known to interact with FMRP (15), or to determine whether the mRNAs that are down-regulated after loss of FMRP contain FMRP binding sites that compete with miRNA binding sites.
Regulation of Translation of Proteins Involved in Transcription
The indirect effect of FMRP regulating the translation of transcriptional regulators is a likely way to explain many of the changes in mRNA levels seen in this study. FMRP regulation of translation of transcriptional regulators is a major mediator of the phenotypic expression of FMRP in other studies (16). Moreover, down-regulation of necdin, a protein that regulates transcription in neural stem cells (17) and whose mRNA levels are increased by the loss of FMRP, rescued the perturbed neuronal precursor differentiation caused by loss of FMRP in this study (4). This highlights how important effects of loss of an RBP can be quite indirect, as necdin transcriptional regulation is probably not a direct effect of FMRP; thus, the transcriptional changes downstream of necdin are presumably at least two steps away from FMRP (regulating translation of a protein that regulates necdin levels, which then regulates transcription of neuronal differentiation factors).
Buffering Changes in Translation
A considerable number of the mRNA decreases were accompanied by increased ribosome loading, and many of these buffering up mRNAs directly bind to FMRP. Thus, one must consider that down-regulation of the mRNAs is a direct consequence of the increase in translation caused by the loss of FMRP. One possibility is that for proteins that are part of a complex, there is a cotranslational requirement for assembly (18). Increased translation of only one part of a complex leads to stalling of translation due to misfolding of the nascent chain in the absence of the stabilizing cocomplex members. Stalling then recruits machinery to degrade the mRNA and protein (19). In this scenario, increased translation leads to increased degradation of the mRNA.
There has been considerable interest on the relative roles of transcription, translation, and degradation on controlling the levels of a protein. The concept of buffering was introduced to explain how fairly large changes in RNA expression, particularly between species, led to only small changes in protein levels (20). Although buffering due to translation was initially thought to be the major mechanism (20), more recent studies have suggested that for most buffering, posttranslational effects are more important (21). Thus, an important missing aspect of the present study is confirmation that the changes observed in the present study at the transcriptional and translational levels are actually present at the protein level. It is possible that some of the changes in this study could be buffered posttranscriptionally.
In summary, loss of a single RBP can have complex and cell-specific changes. Combining transcriptome analysis with ribosome profiling allowed classification of these changes in a specific cell type, demonstrating how distinct classes of mRNAs are regulated by distinct mechanisms. This systems-level approach to the study of RBPs, perhaps combined with proteomic analysis in the future, should lead to fundamental improvements in our understanding of the phenotypic effects of the loss of an RBP, with important implications for our understanding of many genetic diseases.
Footnotes
The author declares no conflict of interest.
See companion article on page E11397.
References
- 1.Garber KB, Visootsak J, Warren ST. Fragile X syndrome. Eur J Hum Genet. 2008;16:666–672. doi: 10.1038/ejhg.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee A, Ifrim MF, Valdez AN, Raj N, Bassell GJ. Aberrant RNA translation in fragile X syndrome: From FMRP mechanisms to emerging therapeutic strategies. Brain Res. 2018;1693:24–36. doi: 10.1016/j.brainres.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luo Y, et al. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu B, et al. Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc Natl Acad Sci USA. 2018;115:E11397–E11405. doi: 10.1073/pnas.1809588115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013;29:318–327. doi: 10.1016/j.tig.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Jensen JB, Parmar M. Strengths and limitations of the neurosphere culture system. Mol Neurobiol. 2006;34:153–161. doi: 10.1385/MN:34:3:153. [DOI] [PubMed] [Google Scholar]
- 8.Anderson BR, Chopra P, Suhl JA, Warren ST, Bassell GJ. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016;44:6649–6659. doi: 10.1093/nar/gkw593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Udagawa T, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graber TE, et al. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci USA. 2013;110:16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F, et al. Fragile X mental retardation protein modulates the stability of its m6A-marked messenger RNA targets. Hum Mol Genet. August 9, 2018 doi: 10.1093/hmg/ddy292. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muddashetty RS, et al. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin SL. microRNAs and fragile X syndrome. Adv Exp Med Biol. 2015;888:107–121. doi: 10.1007/978-3-319-22671-2_7. [DOI] [PubMed] [Google Scholar]
- 16.Korb E, et al. Excess translation of epigenetic regulators contributes to fragile X syndrome and is alleviated by Brd4 inhibition. Cell. 2017;170:1209–1223.e20. doi: 10.1016/j.cell.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshikawa K. Cell cycle regulators in neural stem cells and postmitotic neurons. Neurosci Res. 2000;37:1–14. doi: 10.1016/s0168-0102(00)00101-2. [DOI] [PubMed] [Google Scholar]
- 18.Shiber A, et al. Cotranslational assembly of protein complexes in eukaryotes revealed by ribosome profiling. Nature. 2018;561:268–272. doi: 10.1038/s41586-018-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuller AP, Green R. Roadblocks and resolutions in eukaryotic translation. Nat Rev Mol Cell Biol. 2018;19:526–541. doi: 10.1038/s41580-018-0011-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McManus CJ, May GE, Spealman P, Shteyman A. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 2014;24:422–430. doi: 10.1101/gr.164996.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]