Significance
All proteins are synthesized by the ribosome and therefore have the opportunity to fold for the first time while they are still attached to it. Although protein folding in vitro has been extensively characterized, we are just beginning to understand how proteins fold on the ribosome, which is essential to understanding protein folding in vivo. Here, we characterize and compare the folding pathway of a single protein domain on the ribosome and in bulk solution. By probing the response of the protein to mechanical force, chemical denaturant, and point mutations, and comparing experimental results to simulations, we show that this single protein domain folds through the same pathway on and off the ribosome.
Keywords: protein folding, ribosome, cotranslational folding, single-molecule force spectroscopy, optical tweezers
Abstract
In vivo, proteins fold and function in a complex environment subject to many stresses that can modulate a protein’s energy landscape. One aspect of the environment pertinent to protein folding is the ribosome, since proteins have the opportunity to fold while still bound to the ribosome during translation. We use a combination of force and chemical denaturant (chemomechanical unfolding), as well as point mutations, to characterize the folding mechanism of the src SH3 domain both as a stalled ribosome nascent chain and free in solution. Our results indicate that src SH3 folds through the same pathway on and off the ribosome. Molecular simulations also indicate that the ribosome does not affect the folding pathway for this small protein. Taken together, we conclude that the ribosome does not alter the folding mechanism of this small protein. These results, if general, suggest the ribosome may exert a bigger influence on the folding of multidomain proteins or protein domains that can partially fold before the entire domain sequence is outside the ribosome exit tunnel.
Proteins function in a complex cellular context where they are exposed to countless perturbing conditions that influence the folding process (1–3). Environmental conditions such as temperature, solutes, and mechanical stress are well known to modulate protein energy landscapes (4–10). To fully understand protein folding in vivo, it is essential to consider all conditions that a protein may experience in the cell.
A potential influence on the folding of every protein is the ribosome, which synthesizes proteins via translation. Because folding often occurs on a faster timescale than translation (11), proteins have the opportunity to fold as they are being translated. However, we are just beginning to learn how the ribosome affects the folding process (11–18). There are many aspects of folding that may differ between a protein tethered to the ribosome during translation and free in solution. For instance, the emerging peptide chain can explore conformational space before the entire protein is synthesized. Indeed, small protein domains have been shown to fold in the mouth of the ribosome or even deeper in the exit tunnel, creating a strong spatial and steric constraint on the folding process (18–20). Another possibility is that, because the ribosome is a highly charged macromolecule, short-range chemical interactions, such as hydrophobic interactions and hydrogen bonding, and longer-range coulombic interactions of the emerging protein with the ribosome could also affect the folding process (21).
Recent advances in experimental methodologies have begun to shed light on how the ribosome alters protein folding. Structural studies on ribosome nascent chain complexes (RNCs) using NMR and cryo-EM demonstrate that, while secondary structure and simple tertiary motifs can form inside the ribosome exit tunnel, most proteins cannot find their native structure until they are outside the tunnel (12, 18, 20). Recent experiments harnessing the ability of the force generated by folding to release stalled nascent chains provide insight into the point during translation when different types of proteins fold (17–19, 22). FRET and force spectroscopy experiments reveal the partially folded conformations some domains access before their entire sequence has left the exit tunnel (16, 23). Pulse proteolysis and optical trapping experiments have been used to explore the energy landscape of RNCs outside the exit tunnel, showing that the ribosome significantly alters the stability and dynamics of proteins (13, 14, 23). Moreover, translation rates, which can be modulated by codon usage and mRNA secondary structure, affect the folding efficiency of a protein (24–28).
How does the ribosome affect the protein-folding pathway? For multidomain proteins, the vectorial nature of cotranslational folding can affect the pathway by altering which domain folds first (15, 29, 30). Other studies have suggested the ribosome and/or translation may alter the stable intermediates that single domains sample during folding (16, 23). Here, we focus on how attachment to the ribosome changes the folding barrier (i.e., the transition state) of a single protein domain that folds without stable intermediates. Simulations of an Ig domain on the ribosome suggest that the ribosome does not affect its folding mechanism (19). Using a unique approach combining multiple probes, we are able to experimentally probe the folding transition state for a protein while still a ribosome-bound nascent chain in a way that has not been possible with other techniques.
Previously, we combined denaturant and force in a technique termed chemomechanical unfolding to probe the folding pathway of the src SH3 domain (5, 6). src SH3 can access at least three different folding routes and small changes in denaturant concentration, force, and sequence can alter the flux between them. We now use this same approach to explore another environmental condition: the ribosome. The src SH3 domain occurs in nature as part of multidomain proteins. It folds and unfolds rapidly and therefore will have the opportunity to sample these different states while still attached to the ribosome as other domains are being synthesized. Employing the methodology developed by Kaiser et al. (13, 31), we use optical tweezers to apply force on stalled RNCs containing src SH3. To distinguish between the different folding pathways, we measure four parameters that characterize the transition state: urea m‡-values, force x‡-values, mutational ϕ-values, and the extrapolated unfolding rate in the absence of force or denaturant. We measure those same parameters on and off the ribosome (RNCs and free protein) and use them to determine which pathway src SH3 folds through when tethered to the ribosome. Our results indicate that this version of src SH3 folds through the same pathway on and off the ribosome. Folding simulations both on and off the ribosome support these findings. Thus, small single-domain proteins can access the same folding pathway both as ribosome nascent chains and free in solution. The ribosome may more easily modulate the folding pathway of larger multidomain proteins.
Results and Discussion
Constructs Used for Optical Trap Studies On and Off the Ribosome.
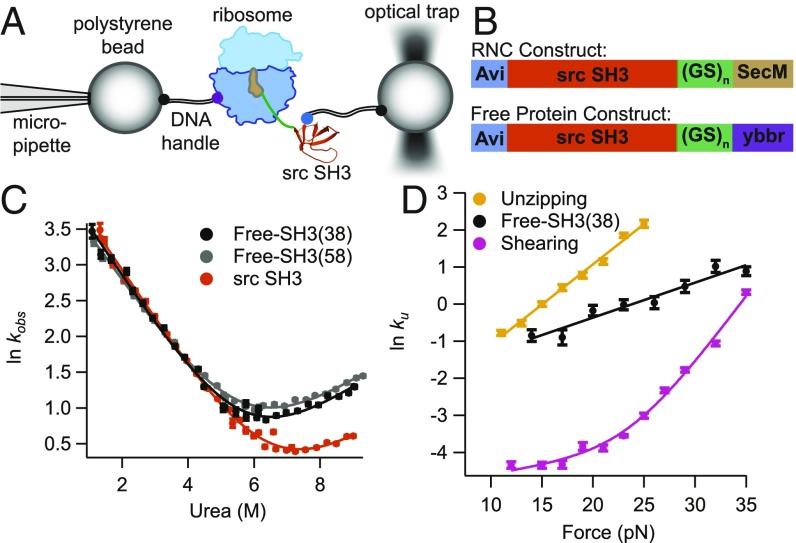
Protein constructs used in the optical tweezers are tethered between two beads via DNA handles attached to genetically encoded Avi and ybbr peptide tags (32, 33); one bead is held on a micropipette and the other is held in an optical trap to apply force (Fig. 1A). To monitor free src SH3, these tags are encoded at the N and C termini of the protein. To monitor RNCs, the Avi tag is encoded in the N terminus of src SH3 and the ybbr tag is encoded on protein L17 of the ribosome (the C terminus of the RNC construct does not contain a peptide tag, ensuring that our optical trapping experiments will not capture any free protein released by the ribosome) (13). The RNC constructs (Fig. 1B) contain a strong variant of the SecM stalling sequence (34) at the C terminus to tether the protein to the ribosome and a glycine–serine (GS) linker to ensure that src SH3 is fully outside the ribosome exit tunnel. To alter the distance between src SH3 and the ribosome, we use constructs with either 10 or 20 GS repeats, which, together with the 18-aa SecM sequence, create linkers of 38 and 58 aa, respectively, between src SH3 and the peptidyl transferase center [these constructs are referred to as SH3(38) and SH3(58), respectively]. The 38-aa linker is known to be enough to ensure that the protein sequence is entirely outside of the ribosome exit tunnel (14). These GS linkers are also added to the free protein construct (Fig. 1B) to make the free protein and RNC constructs as similar as possible.
Fig. 1.
Overview of experimental setup and src SH3 constructs studied. (A) Schematic diagram of the setup used to apply force to stalled RNCs using the optical tweezers. DNA handles are attached via an N-terminal Avi tag and a ybbr tag on protein L17 of the ribosome. (B) Constructs used in experiments for RNCs and free protein. SH3(38) and SH3(58) constructs contain linkers with 10 and 20 GS repeats, respectively. (C) Kinetic chevron plots of ln kobs against [urea] for src SH3 without the tags and linkers used in this work, free-SH3(38) and free-SH3(58). Data were collected in 25 mM Hepes, 150 mM KCl, 5 mM Mg-acetate, pH 7.4 (HKM) buffer. (D) Plots of ln kU against force for free-SH3(38) compared with previously published data with force applied in unzipping and shearing geometries. Unzipping and shearing data were collected in 100 mM Tris, 250 mM NaCl, pH 7, buffer, and free-SH3(38) were collected in HKM buffer.
Ensemble-Based Folding Studies with Linkers and Tags.
To determine whether the added linkers and tags affect folding of src SH3, we measured folding/unfolding kinetics for free-SH3(38), free-SH3(58), and src SH3 without the tags and linkers. Kinetic data were collected using stopped-flow fluorescence and were well fit by a single exponential for all of the constructs. The resulting chevron plots (ln kobs vs. [urea]) (Fig. 1C) were fit assuming a two-state model:
| [1] |
where kobs is the observed relaxation rate constant, and are the unfolding/folding rate constants in the absence of urea, and and are the unfolding and folding m‡-values (the slope of the lnku and lnkf against [urea]). While the tags and linkers increase the unfolding rate of SH3, they do not affect the folding rate, or (Table 1, Ensemble data). Kinetic m‡-values are related to the change in accessible surface area (ASA) for folding and unfolding to the transition state (35, 36). Since they are the same for all of these constructs, the structure of the transition state is likely the same. Therefore, the tags and linkers appear to increase the unfolding rate of src SH3 mostly by destabilizing the folded state. The SH3(58) rate is within error of the SH3(38) rate, suggesting that this change does not depend on linker length.
Table 1.
Results from kinetic analysis of ensemble and single-molecule force experiments
| Construct | Linker length, aa | Urea , M−1 | , nm | |
| Ensemble data | ||||
| Src SH3* | 0 | 0.29 ± 0.04 | NA | −2.03 ± 0.38 |
| Free-SH3† | 38 | 0.30 ± 0.04 | NA | −1.47 ± 0.31 |
| Free-SH3† | 58 | 0.29 ± 0.02 | NA | −1.23 ± 0.15 |
| Free-SH3 L24A† | 38 | 0.28 ± 0.02 | NA | 1.08 ± 0.13 |
| Single-molecule force data | ||||
| Free-SH3† | 38 | 0.31 ± 0.23 | 0.37 ± 0.02§ | −2.12 ± 0.16 |
| RNC-SH3¶ | 38 | 0.33 ± 0.23 | 0.37 ± 0.02§ | −2.63 ± 0.16 |
| RNC-SH3¶ | 58 | NA | 0.37 ± 0.02§ | −2.76 ± 0.19 |
| RNC-SH3 L24A¶ | 38 | NA | 0.37 ± 0.02§ | −1.85 ± 0.18 |
Contains no tags of linkers.
Contains N-terminal Avi tag and C-terminal ybbr tag.
These values were linked in global analysis.
Contains N-terminal Avi tag and C-terminal SecM sequence.
Effect of Force on Unfolding Rates off the Ribosome.
The DNA handles required for optical trap studies are attached via genetically encoded peptide tags, so force is applied to the N and C termini of the protein. This differs from our previous work with src SH3, where we attached handles to cysteines within the protein and pull along different geometries [shearing (A7C/N59C) and unzipping (R19C/N59C)] (5). To compare this N-to-C geometry to the previous studies, we pulled on the free-SH3(38) construct and determined the unfolding rate as a function of force using force-jump experiments (7). The ln kU values are plotted against force and fit using the Bell equation (37):
| [2] |
where gives the effect of force on the unfolding rate, is kU in the absence of force, kB is the Boltzmann constant, and T is the absolute temperature. The -value is related to the distance between the native state and transition state (by microscopic reversibility, the transition state for unfolding and folding are the same) along the mechanical reaction coordinate (38). Fig. 1D compares this free-SH3(38) data to the previously published unzipping and shearing data. Unlike the upward curvature in the force dependence of ln kU with a shearing force, which suggests parallel unfolding pathways (5), the free SH3(38) data, like the unzipping data, can be approximated by a linear relationship between ln kU and force, indicating that when pulled from the termini, src SH3 unfolds through a single pathway over the range of force probed. The and ln values differ between unzipping and free-SH3(38) data; however, these datasets use different pulling geometries and buffers (in this work, we use a buffer compatible with RNCs), so we cannot determine whether these differences are due to the different experimental conditions or different transition states. We showed previously that the unzipping pathway is likely the same as the pathway seen in ensemble experiments. Therefore, a comparison of the denaturant dependence of free-SH3(38) force data to ensemble kinetic data will help us to determine whether this is the same as the unzipping pathway (see Using Chemomechanical Unfolding to Compare the Folding Pathway On and Off the Ribosome).
Determining the Effect of the Ribosome on the Force Dependence of Unfolding.
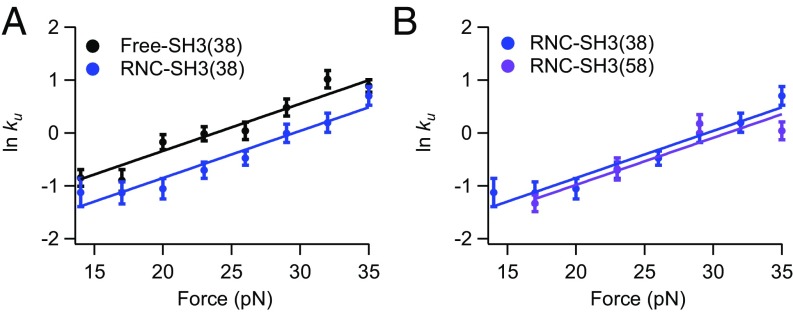
To explore the effect of the ribosome, we apply force to RNC-SH3(38) and again use force-jump experiments analyzed as above (Fig. 2A). The force-dependent RNC-SH3(38) data run parallel to that for free-SH3(38), indicating that is the same on and off the ribosome. Therefore, the ribosome does not alter the distance to the transition state, suggesting that src SH3 folds through the same pathway on and off the ribosome. The RNC rates are slightly lower than the rates for free protein, indicating that the ribosome may affect the stability of the folded state or the transition state. This difference, however, is very small and may well arise from small differences between the experimental handles (DNA vs. DNA and the ribosome), which may affect the kinetics in the trap (SI Appendix). In principle, weak interactions of the protein may increase or decrease unfolding rates depending on their magnitude (SI Appendix, Supplemental Text and Fig. S1).
Fig. 2.
Effect of force on free-SH3 and RNC-SH3 constructs. Plots of ln kU against force for (A) free protein and RNCs with a 38-aa linker and (B) RNCs with a 38- and 58-aa linker.
To explore the effect of increasing the distance from the ribosome, we performed the same force-jump experiments with RNC-SH3(58). The RNC-SH3(38) and RNC-SH3(58) data (Fig. 2B) look quite similar, suggesting that increasing the distance from the ribosome does not significantly affect the unfolding rate or pathway. Simulations (see Simulations of the Effect of the Ribosome on the Folding of SH3 at Different Linker Lengths) suggest that our linker length of 38 aa is as close as we can get to the ribosome and still observe folding. Consequently, all subsequent experimental analysis will use only SH3(38) constructs.
Using Chemomechanical Unfolding to Compare the Folding Pathway On and Off the Ribosome.
The above ensemble and force-dependent results suggest that src SH3 may unfold through the same pathway on and off the ribosome and at different distances from the ribosome. To test this hypothesis directly, we turn to chemomechanical unfolding to determine an additional parameter, the m‡-value, that is also related to the structure of the transition state and can distinguish between pathways (5, 35, 36). Moreover, because m‡-values can be measured in standard ensemble experiments (Fig. 1), they provide a means to compare the pathways seen in single-molecule force data to those seen in ensemble data.
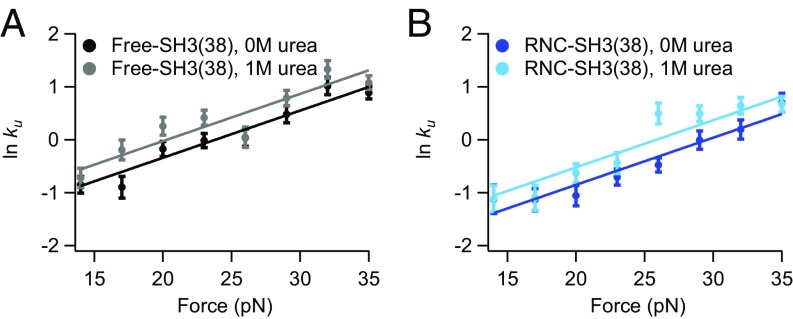
Using chemomechanical unfolding, we simultaneously determine the effect of force and urea on unfolding of free-SH3(38) and RNC-SH3(38). Fig. 3 shows a comparison of the data in 1 M urea to that in 0 M urea. Urea affects the free protein and RNCs similarly; for both, the 1 M urea data show a linear relationship with the same slope as the 0 M urea data but a slightly higher rate. In fact, when all force data collected in this work are fit separately to Eq. 2, the resulting -values are very close (SI Appendix, Table S1). Therefore, to minimize differences due to experimental error, these data were all fit together in a global analysis to determine a single -value. Figs. 2, 3, and 4B show the results of this global analysis (SI Appendix, Fig. S2 compares individual and global fits). Table 1 reports and ln kU in the absence of urea and force for each construct. The -value from the global fit is within error of all -values from individual fits, validating our hypothesis that is the same for all constructs studied here.
Fig. 3.
Chemomechanical unfolding analysis of free-SH3 and RNC-SH3. Plots of ln kU against force for (A) free protein and (B) RNCs with a 38-aa linker in 0 and 1 M urea.
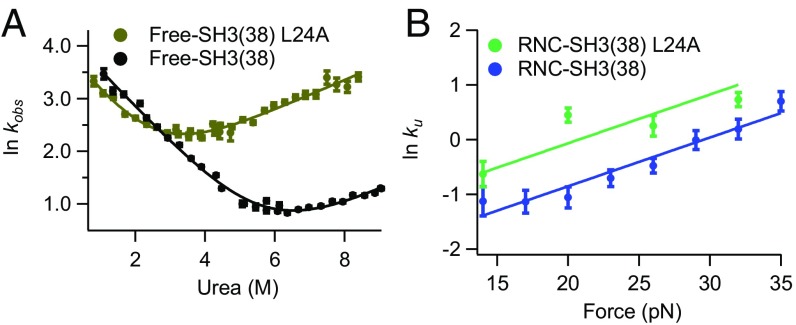
Fig. 4.
Effect of L24A mutation on free-SH3 and RNC-SH3. (A) Kinetic chevron plots of ln kobs against [urea] for WT and L24A SH3 free protein construct. (B) Plots of ln kU against force for WT and L24A RNC-SH3(38).
The effect of urea on unfolding rate is quantified by -values via Eq. 3 as follows:
| [3] |
where [urea] is the molar concentration of urea and is kU in the absence of urea. The -value is related to the solvent ASA exposed in unfolding to the transition state, providing another parameter to compare transition states (35, 36). The resulting -values for free-SH3(38) and RNC-SH3(38), determined by the difference between ln for 1 M urea and 0 M urea data, reported in Table 1, are the same within error, indicating that the solvent ASA exposed in unfolding to the transition state is the same for free-SH3 and RNC-SH3. This again indicates that the ribosome does not affect the folding transition state.
We have also determined -values for free-SH3 constructs in the absence of force from kinetic chevron experiments (Fig. 1). These -values can be used to compare the unfolding pathway in force experiments and standard ensemble experiments. The -values for free-SH3(38) and RNC-SH3(38) force experiments are the same within error as the -values from chevron plots for all free-SH3 constructs used in this work, suggesting that the folding pathway in force experiments is the same folding pathway seen in standard ensemble experiments: SH3 folds through the same transition state on the ribosome that it folds through in bulk solution.
In our previous work (5), the different high- and low-force pathways obtained when pulling in a shearing direction could be distinguished via their urea -values. On the other hand, pulling in an unzipping direction resulted in a similar -value to that obtained in the absence of force, suggesting a similar pathway. Thus, the similar -values obtained here from N-to-C pulling and bulk chevron experiments are consistent with src SH3 unfolding through the pathway seen in the absence of force (the bulk pathway) in this pulling direction and when tethered to the ribosome.
Simulations of the Effect of the Ribosome on the Folding of SH3 at Different Linker Lengths.
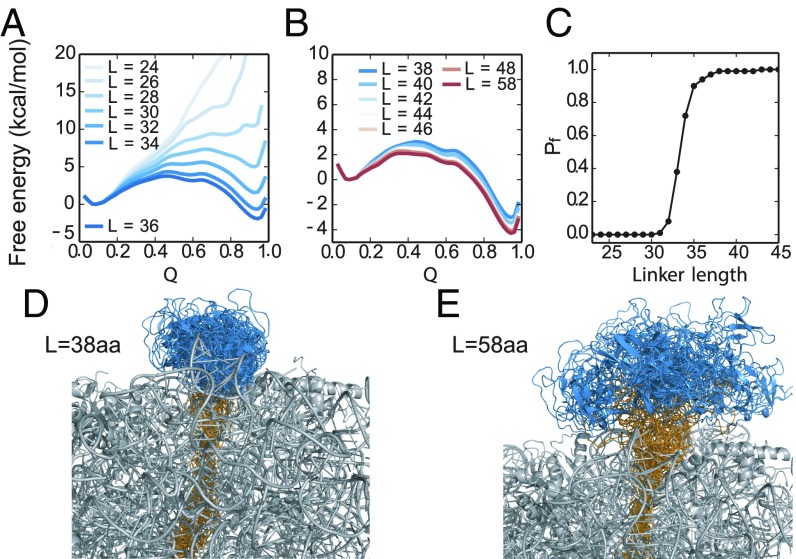
To gain further insight into the folding of src SH3 at different distances from the ribosome, we determined the native state stability of the protein by molecular-dynamics (MD) simulations of RNC-SH3 at different linker lengths (L). All free energies are calculated under conditions where the stability of RNC-SH3 with L = 58 aa is equal to the stability of the isolated src SH3 domain (3.6 kcal/mol). Free-energy profiles projected onto the fraction of native contacts (Fig. 5 A and B), Q (39), show that src SH3 becomes more stable with longer linker lengths (Fig. 5 A–C). The fraction of folded protein approaches ∼100% (Fig. 5C) after a sharp transition centered at L ∼ 34 aa. In these simulations, at the linker length of L = 38 aa used in the experiments, the src SH3 domain has not completely emerged yet from the mouth of the tunnel. Several C-terminal residues remain in the tunnel, and so the domain is less stable (∆∆G ≅ 1.1 kcal/mol) than the WT. A representative set of folded states of src SH3 from unbiased MD folding simulations are shown in Fig. 5 D and E, from which we observe that src SH3 is folding at the mouth of ribosome tunnel at L = 38 aa and completely outside of the tunnel at L = 58 aa. The simulations also show that the application of force only slightly reduces the number of contacts between the protein and ribosome (SI Appendix, Fig. S3). Thus, simulations suggest that the 38-aa linker used in experiments is a good model to study the effect of folding close to the ribosome.
Fig. 5.
MD simulations of RNC-SH3 with different linker lengths. (A and B) Free energy profiles for src SH3 folding on the ribosome with linker lengths from 24 to 58 aa. Q is the fraction of native contacts. (C) Fraction of folded src SH3 population (Pf) as a function of linker length. Ensemble of folded states of src SH3 with linker L = 38 aa (D) and L = 58 aa (E). Each ensemble contains conformations obtained from 2-µs MD simulations (saved every 40 ns) where the protein remained folded. The SH3 and linker residues are shown in blue and yellow respectively.
Characterizing Folding Pathways Using Point Mutation.
Like urea and force, the effect of mutation is related to the folding pathway (40). Therefore, a point mutation can be used to test our hypothesis that src SH3 unfolds through the same pathway on the ribosome and in bulk solution. In previous work, we determined the effect of various mutations on the different folding pathways of src SH3; the L24A mutation significantly increases the unfolding rate for the bulk pathway but has a much smaller effect on the other observable unfolding pathways (5). Here, we make the same L24A mutation in free-SH3(38) and RNC-SH3(38) to see whether it also increases the unfolding rate in these experiments. In ensemble studies, we determine folding and unfolding rates for free-SH3(38) from a kinetic chevron plot fit to Eq. 1 (Fig. 4A) and find that free-SH3(38) L24A behaves very similarly to src SH3 without the tags and linkers (5)—the mutation significantly destabilizes src SH3, primarily by increasing the unfolding rate.
For RNC-SH3(38) L24A, we determine the unfolding rate as a function of force and find a linear relationship with a similar slope to the other constructs (Fig. 4B). These data were included in the global analysis, linking all of the -values. Comparison with WT RNC-SH3(38) data shows that, as expected for the bulk pathway, the L24A mutation increases the unfolding rate for src SH3 on the ribosome. Mutations are often used in a Φ-value analysis to characterize a transition state. Unfortunately, we are unable to determine the stability of src SH3 on the ribosome and so cannot calculate an experimental Φ-value (40). We can only measure unfolding rates for RNC-SH3 constructs and not folding rates because src SH3 folds at very low forces where the optical tweezers have poor resolution. Instead, we use simulations to determine Φ-values for src SH3 on and off the ribosome.
Simulations of Φ-Values for Src SH3 On and Off the Ribosome.
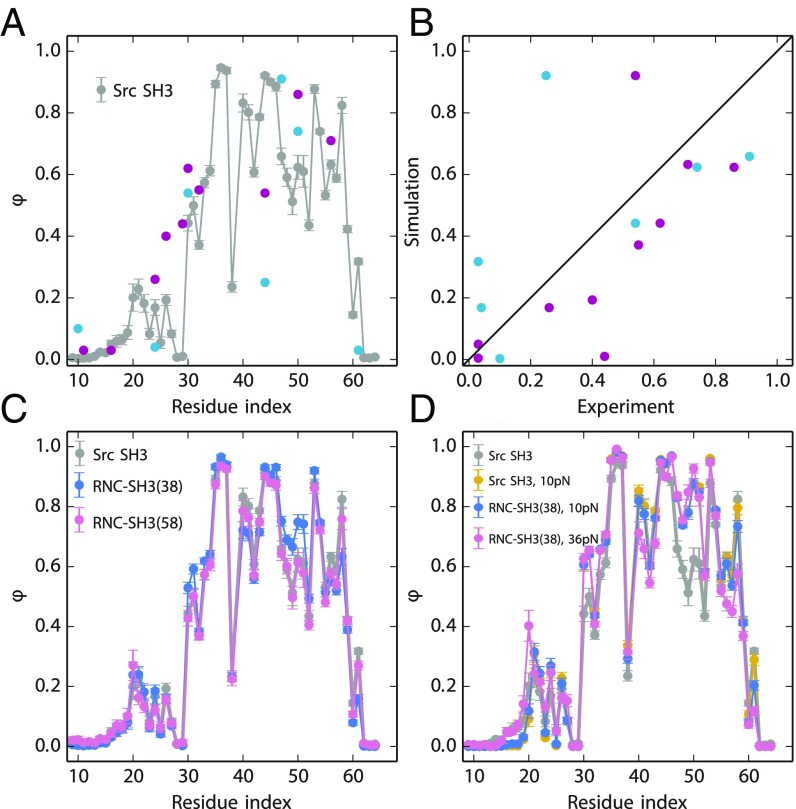
Previous studies have shown that MD simulations with a coarse-grained model can reproduce the transition state ensemble of the isolated src SH3 domain (41–44). In our study, unbiased MD folding simulations at constant temperature are carried out for the isolated src SH3 domain, RNC-SH3(38) and RNC-SH3(58). We approximate Φ-values for each residue from folding transition paths, here defined as the segments of a trajectory between Q ∼ 0.3 and Q ∼ 0.6 (45). The simulated Φ-values are averaged from 50 transition paths for each construct. The simulated Φ-value of residue 24 on isolated src SH3 is 0.12 (Fig. 6A), close to the experimental value (0.04) (5). Furthermore, the simulations reproduce well the overall Φ-value profile of src SH3 from previous experiments (Fig. 6A) (5, 46). The largest discrepancy between simulation and experiments is at residue 44, where the measurements obtained by two independent experimental groups also differ the most. The Spearman rank correlation coefficient between simulation and experimental data are 0.75 (Fig. 6B), using Φ-values for mutations whose ∆∆G is larger than 7 kJ/mol (47). We also estimated the Φ-values for RNC-SH3(38) and RNC-SH3(58) (Fig. 6C), obtaining Spearman correlations between these two cases and the experimental Φ-values for isolated src SH3 of 0.78 and 0.75, respectively. We can compute Φ-values for each individual folding event as well as an average over all folding events. The folding pathways are all very similar, indicating that the fractional Φ-values come largely from the partial formation of contacts rather than heterogeneous folding mechanisms (SI Appendix, Fig. S4).
Fig. 6.
Φ-values from MD simulations and experiments. (A) Φ-values of the isolated src SH3 domain from two experimental studies are shown in cyan (5) and purple (46). Simulated Φ-values are calculated from the transition path ensemble of isolated src SH3 with no tags or linkers (gray). (B) The correlation between simulated and experimental Φ-values. (C) Simulated Φ-values of RNC-SH3(38) (blue) and RNC-SH3(58) (red) compared with isolated src SH3 (gray). (D) Φ-values for isolated src SH3 subjected to 10 pN of force (yellow) and for RNC-SH3(38) subjected to 10 pN (blue) and 36 pN (red) of force.
Urea -values can be used to compare transition states in simulations and experiments. The -values obtained from MD simulations of free SH3 and RNC-SH3(38) using an empirical relation between ASA and m-value (36) are 0.42 and 0.44, respectively. These values are in reasonable agreement with the experimental -value of 0.29 ± 0.04 (Table 1) here, considering that an earlier study obtained an -value of 0.46 ± 0.04 in different buffer conditions (5).
Taken together, these results suggest that the transition-state ensemble of src SH3 is not affected much by the ribosome or the linker length. We also carried out simulations in the presence of a pulling force, which again agreed with our experimental results that the ribosome does not affect the folding trajectory (Fig. 6D and SI Appendix, Fig. S1).
Conclusions
Using both experimental and computational techniques, we conclude that src SH3 folds via the same mechanism as a ribosome-bound nascent chain as it does free in solution. Experimentally, the effect of force, urea, and point mutation on src SH3 unfolding kinetics are the same on and off the ribosome. Simulations on and off the ribosome support the results that the ribosome does not affect the folding pathway. Since in the context of a multidomain protein, src SH3 can fold/unfold rapidly while C-terminal domains are still being translated, for this domain, and others like it, an understanding of cotranslational folding can harness the vast amount of knowledge on its folding free in solution.
Our studies are uniquely suited to probe the effect of the ribosome on the folding transition state, which when combined with studies focused on other features unique to cotranslational folding will allow a better understanding of the folding process of nascent chains in the cell. We find that src SH3 does not fold until the entire domain sequence has reached the mouth of the ribosome exit tunnel. There are examples of other domains that can partially fold before their entire sequence is exposed. FRET experiments suggest that the α-helical N-terminal domain of HemK forms a compact, nonnative state before its entire sequence is outside the exit tunnel (16). Additionally, subunits of bacterial luciferase assemble cotranslationally before the entire LuxB subunit is fully outside the exit tunnel, suggesting the interface required for assembly can fold independently of the C terminus of luxB (48). Finally, some spectrin domains fold via a different pathway on the ribosome if partly folded N-terminal structures are stable enough to fold in isolation of the C terminus (17). All of these proteins (HemK, luxB, and spectrin domains) are primarily α-helical, and so contain short-range contacts. On the other hand, src SH3 is composed predominantly of β-strands, which require long-range contacts that may prohibit folding before the entire domain sequence is outside the ribosome. This long-range contact order may limit the ability of the ribosome to alter the folding pathway via vectorial folding. Still, the ribosome can affect protein folding via chemical and coulombic interactions with the protein once it has emerged from the exit tunnel. These interactions are sufficient to alter the stability and kinetics of some stalled RNCs (13, 14) but, for src SH3, are not sufficient to alter the folding transition state.
Recently, an arrest peptide based assay has been used to interrogate cotranslational folding of another small β domain: the Ig domain, titin I27 (19). This involved monitoring the ability of constructs with different linker lengths to create sufficient force to overcome the SecM stalling sequence and continue translating. This force has been directly correlated with cotranslational folding. These studies, together with cryo-EM, indicate that I27 folds in the mouth of the ribosome. By computing Φ-values from simulations of I27 folding in the mouth of the ribosome, they find a good correlation to experimental Φ-values determined off the ribosome. Thus, despite the different experimental observables, this work comes to a conclusion similar to ours: the ribosome does not affect the folding pathway of this small domain.
The ribosome does not appear to play a significant role in determining the transition state for folding of the src SH3 and I27 domains. By comparing our result with literature data for multidomain proteins, we can speculate that it plays a more dominant role in altering protein-folding pathways for larger multidomain proteins. Due to the vectorial nature of folding, the N-terminal domains of a multidomain protein can fold while the C terminus is still sequestered. In firefly luciferase, the initial folding of the N-terminal domain has been shown to provide a scaffold that greatly speeds up folding of the native protein (30). Vectorial folding can also prevent aggregation-prone intermediates from forming, as in HaloTag, where an aggregation-prone intermediate observed in in vitro refolding experiments is not observed in cotranslational folding experiments (15).
The effect of vectorial folding on multidomain proteins highlights the importance of translation rates. The length of time that the N-terminal domains are outside of the ribosome exit tunnel before the C terminus of the protein emerges determines how long the isolated N-terminal domain has to fold. Indeed, codon usage, which can alter translation rates, can affect protein folding efficiency and therefore downstream cellular processes (24–26, 29). If our observation that the ribosome does not greatly alter folding rates for small domains is general, perhaps knowledge of the folding rates determined in vitro for individual protein domains together with codon translation rates can be used to assess whether or not an individual domain can fold before the protein is fully translated (24).
In sum, chemomechanical unfolding, together with MD simulations of RNCs, have proven to be complementary techniques for probing folding pathways on the ribosome. Because folding on the ribosome is very complex, employing these techniques to study other proteins and other aspects of cotranslational folding, like the presence of molecular chaperones, will help shed more light on this process.
Materials and Methods
Ensemble kinetic data were collected on a BioLogic SFM-400/MOS 200 stopped-flow fluorescence system, and optical trap experiments were conducted using an optical tweezers instrument described previously (49, 50). Additional details of free protein and RNC sample preparation and experiments can be found in SI Appendix. Simulations employed a course-grained structure-based model (51), where each amino acid of src SH3 or ribosome proteins is represented by one bead, and RNA residues are represented by 3 beads (see SI Appendix for additional details on these computational studies).
Supplementary Material
Acknowledgments
We thank Lisa Alexander, Daniel Goldman, Avi Samelson, and Madeleine Jensen for guidance regarding experimental procedures; Brendan Maguire for help with protein and ribosome purification; and Jay Goodman for help with sample preparation. We thank the S.M. laboratory for helpful discussions. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the NIH (https://hpc.nih.gov/). This research was supported by grants from the National Science Foundation (MCB 1616591) and the NIH (R01GM050945) (to S.M.) and an NIH fellowship (F32GM110940) (to E.J.G.). P.T. and R.B.B. were supported by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH. S.M. is a Chan Zuckerberg Biohub investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.V.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810517115/-/DCSupplemental.
References
- 1.Gershenson A, Gierasch LM. Protein folding in the cell: Challenges and progress. Curr Opin Struct Biol. 2011;21:32–41. doi: 10.1016/j.sbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarkar M, Smith AE, Pielak GJ. Impact of reconstituted cytosol on protein stability. Proc Natl Acad Sci USA. 2013;110:19342–19347. doi: 10.1073/pnas.1312678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman I, Gelman H, Tai J, Gruebele M. The extracellular protein VlsE is destabilized inside cells. J Mol Biol. 2014;426:11–20. doi: 10.1016/j.jmb.2013.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sosnick TR, Barrick D. The folding of single domain proteins—have we reached a consensus? Curr Opin Struct Biol. 2011;21:12–24. doi: 10.1016/j.sbi.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guinn EJ, Jagannathan B, Marqusee S. Single-molecule chemo-mechanical unfolding reveals multiple transition state barriers in a small single-domain protein. Nat Commun. 2015;6:6861. doi: 10.1038/ncomms7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guinn EJ, Marqusee S. Exploring the denatured state ensemble by single-molecule chemo-mechanical unfolding: The effect of force, temperature, and urea. J Mol Biol. 2018;430:450–464. doi: 10.1016/j.jmb.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jagannathan B, Marqusee S. Protein folding and unfolding under force. Biopolymers. 2013;99:860–869. doi: 10.1002/bip.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldwin RL. Temperature dependence of the hydrophobic interaction in protein folding. Proc Natl Acad Sci USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright CF, Steward A, Clarke J. Thermodynamic characterisation of two transition states along parallel protein folding pathways. J Mol Biol. 2004;338:445–451. doi: 10.1016/j.jmb.2004.02.062. [DOI] [PubMed] [Google Scholar]
- 10.O’Brien EP, Brooks BR, Thirumalai D. Molecular origin of constant m-values, denatured state collapse, and residue-dependent transition midpoints in globular proteins. Biochemistry. 2009;48:3743–3754. doi: 10.1021/bi8021119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr Opin Struct Biol. 2010;20:33–45. doi: 10.1016/j.sbi.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Cabrita LD, et al. A structural ensemble of a ribosome-nascent chain complex during cotranslational protein folding. Nat Struct Mol Biol. 2016;23:278–285. doi: 10.1038/nsmb.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaiser CM, Goldman DH, Chodera JD, Tinoco I, Jr, Bustamante C. The ribosome modulates nascent protein folding. Science. 2011;334:1723–1727. doi: 10.1126/science.1209740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samelson AJ, Jensen MK, Soto RA, Cate JH, Marqusee S. Quantitative determination of ribosome nascent chain stability. Proc Natl Acad Sci USA. 2016;113:13402–13407. doi: 10.1073/pnas.1610272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Samelson AJ, et al. Kinetic and structural comparison of a protein’s cotranslational folding and refolding pathways. Sci Adv. 2018;4:eaas9098. doi: 10.1126/sciadv.aas9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holtkamp W, et al. Cotranslational protein folding on the ribosome monitored in real time. Science. 2015;350:1104–1107. doi: 10.1126/science.aad0344. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson OB, et al. Cotranslational folding of spectrin domains via partially structured states. Nat Struct Mol Biol. 2017;24:221–225. doi: 10.1038/nsmb.3355. [DOI] [PubMed] [Google Scholar]
- 18.Nilsson OB, et al. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 2015;12:1533–1540. doi: 10.1016/j.celrep.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian P, et al. Folding pathway of an Ig domain is conserved on and off the ribosome. Proc Natl Acad Sci USA. 2018;115:E11284–E11293. doi: 10.1073/pnas.1810523115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Javed A, Christodoulou J, Cabrita LD, Orlova EV. The ribosome and its role in protein folding: Looking through a magnifying glass. Acta Crystallogr D Struct Biol. 2017;73:509–521. doi: 10.1107/S2059798317007446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight AM, et al. Electrostatic effect of the ribosomal surface on nascent polypeptide dynamics. ACS Chem Biol. 2013;8:1195–1204. doi: 10.1021/cb400030n. [DOI] [PubMed] [Google Scholar]
- 22.Farías-Rico JA, Ruud Selin F, Myronidi I, Frühauf M, von Heijne G. Effects of protein size, thermodynamic stability, and net charge on cotranslational folding on the ribosome. Proc Natl Acad Sci USA. 2018;115:E9280–E9287. doi: 10.1073/pnas.1812756115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu K, Rehfus JE, Mattson E, Kaiser CM. The ribosome destabilizes native and non-native structures in a nascent multidomain protein. Protein Sci. 2017;26:1439–1451. doi: 10.1002/pro.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma AK, O’Brien EP. Non-equilibrium coupling of protein structure and function to translation-elongation kinetics. Curr Opin Struct Biol. 2018;49:94–103. doi: 10.1016/j.sbi.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson GN, Clark PL. Quality over quantity: Optimizing co-translational protein folding with non-“optimal” synonymous codons. Curr Opin Struct Biol. 2016;38:102–110. doi: 10.1016/j.sbi.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 27.Jacobs WM, Shakhnovich EI. Evidence of evolutionary selection for cotranslational folding. Proc Natl Acad Sci USA. 2017;114:11434–11439. doi: 10.1073/pnas.1705772114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodnina MV. The ribosome in action: Tuning of translational efficiency and protein folding. Protein Sci. 2016;25:1390–1406. doi: 10.1002/pro.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sander IM, Chaney JL, Clark PL. Expanding Anfinsen’s principle: Contributions of synonymous codon selection to rational protein design. J Am Chem Soc. 2014;136:858–861. doi: 10.1021/ja411302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frydman J, Erdjument-Bromage H, Tempst P, Hartl FU. Co-translational domain folding as the structural basis for the rapid de novo folding of firefly luciferase. Nat Struct Biol. 1999;6:697–705. doi: 10.1038/10754. [DOI] [PubMed] [Google Scholar]
- 31.Goldman DH, et al. Ribosome. Mechanical force releases nascent chain-mediated ribosome arrest in vitro and in vivo. Science. 2015;348:457–460. doi: 10.1126/science.1261909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin J, et al. Genetically encoded short peptide tag for versatile protein labeling by Sfp phosphopantetheinyl transferase. Proc Natl Acad Sci USA. 2005;102:15815–15820. doi: 10.1073/pnas.0507705102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kay BK, Thai S, Volgina VV. High-throughput biotinylation of proteins. Methods Mol Biol. 2009;498:185–196. doi: 10.1007/978-1-59745-196-3_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ismail N, Hedman R, Schiller N, von Heijne G. A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat Struct Mol Biol. 2012;19:1018–1022. doi: 10.1038/nsmb.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guinn EJ, Kontur WS, Tsodikov OV, Shkel I, Record MT., Jr Probing the protein-folding mechanism using denaturant and temperature effects on rate constants. Proc Natl Acad Sci USA. 2013;110:16784–16789. doi: 10.1073/pnas.1311948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 38.Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 39.Best RB, Hummer G, Eaton WA. Native contacts determine protein folding mechanisms in atomistic simulations. Proc Natl Acad Sci USA. 2013;110:17874–17879. doi: 10.1073/pnas.1311599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 41.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 42.Lam AR, et al. Parallel folding pathways in the SH3 domain protein. J Mol Biol. 2007;373:1348–1360. doi: 10.1016/j.jmb.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 43.Ding F, Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Direct molecular dynamics observation of protein folding transition state ensemble. Biophys J. 2002;83:3525–3532. doi: 10.1016/S0006-3495(02)75352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuravlev PI, Hinczewski M, Chakrabarti S, Marqusee S, Thirumalai D. Force-dependent switch in protein unfolding pathways and transition-state movements. Proc Natl Acad Sci USA. 2016;113:E715–E724. doi: 10.1073/pnas.1515730113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Best RB, Hummer G. Microscopic interpretation of folding ϕ-values using the transition path ensemble. Proc Natl Acad Sci USA. 2016;113:3263–3268. doi: 10.1073/pnas.1520864113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riddle DS, et al. Experiment and theory highlight role of native state topology in SH3 folding. Nat Struct Biol. 1999;6:1016–1024. doi: 10.1038/14901. [DOI] [PubMed] [Google Scholar]
- 47.Sánchez IE, Kiefhaber T. Origin of unusual phi-values in protein folding: Evidence against specific nucleation sites. J Mol Biol. 2003;334:1077–1085. doi: 10.1016/j.jmb.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 48.Shieh YW, et al. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science. 2015;350:678–680. doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- 49.Smith SB, Cui Y, Bustamante C. Optical-trap force transducer that operates by direct measurement of light momentum. Methods Enzymol. 2003;361:134–162. doi: 10.1016/s0076-6879(03)61009-8. [DOI] [PubMed] [Google Scholar]
- 50.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 51.Karanicolas J, Brooks CL., 3rd The origins of asymmetry in the folding transition states of protein L and protein G. Protein Sci. 2002;11:2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.