Significance
The handedness of cells, also known as cell chirality, has become widely accepted with the determination of cell chirality using micropatterning techniques. While reliable, this system only works with the cells that can attach, subsequently polarize, and migrate on a 2D protein-coated substrate, which does not necessarily recapitulate the 3D cellular environment. With a unique biomaterial-based approach, we now can determine the chirality of less adhesive and/or less polarized cells in a 3D setting. Thus, it becomes much easier for researchers from various backgrounds, including embryogenesis, epithelial biology, and cancer biophysics, to study chirality. Our discovery will boost a fast-growing field of research: cell chirality in development and disease.
Keywords: cell chirality, left–right asymmetry, cell polarity, tissue morphogenesis, biomaterial
Abstract
Our understanding of the left–right (LR) asymmetry of embryonic development, in particular the contribution of intrinsic handedness of the cell or cell chirality, is limited due to the confounding systematic and environmental factors during morphogenesis and a ack of physiologically relevant in vitro 3D platforms. Here we report an efficient two-layered biomaterial platform for determining the chirality of individual cells, cell aggregates, and self-organized hollow epithelial spheroids. This bioengineered niche provides a uniform defined axis allowing for cells to rotate spontaneously with a directional bias toward either clockwise or counterclockwise directions. Mechanistic studies reveal an actin-dependent, cell-intrinsic property of 3D chirality that can be mediated by actin cross-linking via α-actinin-1. Our findings suggest that the gradient of extracellular matrix is an important biophysicochemical cue influencing cell polarity and chirality. Engineered biomaterial systems can serve as an effective platform for studying developmental asymmetry and screening for environmental factors causing birth defects.
Almost all vertebrates have an asymmetric body plan, a deviation from which often leads to severe malformations (1, 2). In recent years, increasing evidence has suggested that embryonic and organ-specific left–right (LR) asymmetries, such as hindgut and genitalia rotation in Drosophila and symmetry breaking in pond snails (3–6), can arise from the LR bias at a cellular level, also termed cell chirality (7, 8). In addition, this cellular asymmetry has been demonstrated in various models, including early asymmetry in Caenorhabditis elegans (9, 10), the chiral properties of Xenopus egg cortex (11, 12), asymmetric distribution of chirality related proteins at the early developmental stages of different animals (13), and migratory biases of cultured cells in vitro (12, 14–17). However, cell chirality is poorly understood in developing embryos, despite its scientific and clinical significance, due to complexities in imaging and molecular assays when dealing with animal models and confounding systematic and environmental factors that influence data explanation and hinder mechanistic findings. Therefore, it is of great importance to establish a biomimetic system for LR symmetry breaking that truly recapitulates 3D multicellular chiral morphogenesis.
Cell chirality is a fundamental property of the cell, and the universality was not widely regarded until the recent use of microfabricated 2D in vitro systems (16, 18–20), including the 2D microcontact printing developed by us. In these systems, the cells were confined in a narrow area that allows the cells to exhibit their chiral biases in various formats, including cytoskeleton dynamics, cell migration, and multicellular biased alignment. With these new tools, cell chirality was found to be phenotype-dependent and related to the cross-linking of formin-associated actin bundles. Despite these advances in the understanding of cell chirality on 2D substrates, there are concerns about whether a 2D platform can fully mimic the 3D cellular environment in native tissue. Specifically, cells inside a 3D extracellular matrix have narrowed integrin use, enhanced cell motility, and colocalized adhesion proteins, activating different signaling pathways (such as Wnt) compared with those on 2D substrates (21, 22). Indeed, 3D cell cultures are well documented to better recapitulate the native in vivo environment compared with 2D cell cultures, especially for epithelial cells that are relevant for LR asymmetry in development.
In this study, we used the Madin-Darby canine kidney (MDCK) cells, one of the most widely used epithelial cell lines seen in various in vitro studies of tissue morphogenesis, and examined cell chirality in a 3D environment. We quantify the chiral rotational behavior of epithelial cells between two hydrogel layers during their self-assembly into hollow spheroids and reveal an actin cross-linking–dependent cytoskeletal mechanism of cellular chirality.
Results
MDCK Cells Encapsulated Between Matrigel Layers Develop into Organized Luminal Microspheroids.
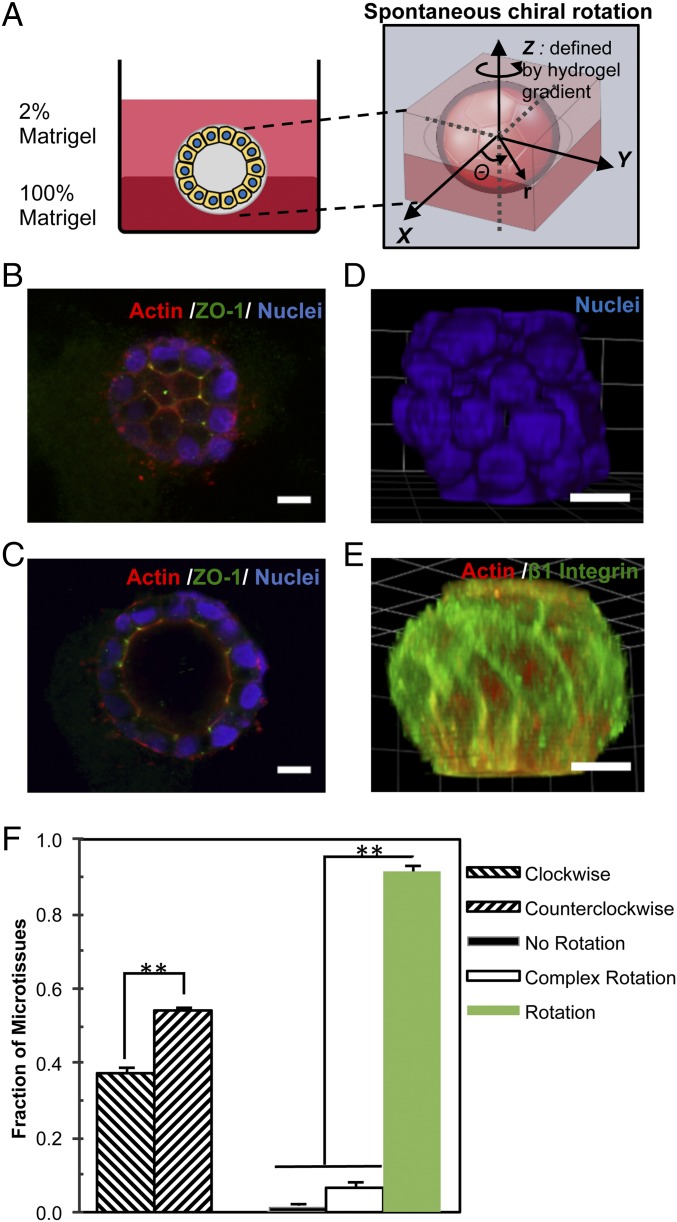
To establish an in vitro 3D system for recapitulating chiral morphogenesis of epithelial tissue during embryonic development, we embedded MDCK epithelial cells (∼6,000 cells per cm2) between two layers of Matrigel: a base layer of 100% Matrigel and a top layer of 2% Matrigel (Fig. 1A) (23–25). The purpose of using the bilayered Matrigel is twofold: to create a flat interfacial plane for cell imaging and to generate a hydrogel gradient that defines a z-axis uniformly across the hydrogel interface, allowing for consistent polar orientation of the cells (Fig. 1A). This configuration is important because the direction of the z-axis must be predefined for the cells to exhibit chirality during rotation at the interface (x-y plane). As expected, the embedded individual cells divided and formed dense microtissues initially and later hollow spheroids with a distinct lumen structure (SI Appendix, Fig. S1 A and B). Confocal microscopy revealed an organized architecture composed of a monolayer of MDCK cells with a hollow lumen at the center of the spheroid. The central lumen was developed after 5 d of culture and remained present through day 10. On the top surface of microspheroids, the cells exhibited a polygonal shape as indicated by actin staining, with strong ZO-1 expression at the vertices between cells (Fig. 1B). In the cross-section of the middle region, stronger actin expression was observed on the apical inner surface (Fig. 1C) and integrin β1 and laminin V on the basal outer surface, indicating a strong apicobasal polarity (SI Appendix, Fig. S1C). In addition, the centrosomes were located closer to the apical surface than the nuclei, but no clear cilia structures were detected with day 5 microspheroids (SI Appendix, Fig. S1D). Through 3D reconstruction of confocal Z-stack images, the orderly arrangement of nuclei and β1 integrin can be readily appreciated and shows a clear directional bias (Fig. 1 D and E). In addition to the apical surfaces, significant accumulation of actin filaments was found between cells and on the basal surfaces of microspheroids, especially at the bottom side where the cells interacted with the 100% Matrigel layer (SI Appendix, Fig. S2 and Movies S1 and S2), while microtubules were distributed primarily toward the lateral and basal surfaces of the cells (SI Appendix, Fig. S2).
Fig. 1.
Spontaneous rotation of self-organized microspheroids is chiral. (A) Schematic depicting the 3D cell chirality assay for epithelial microspheroids. Individual epithelial cells were embedded between a 100% Matrigel base layer and a 2% Matrigel top layer and then allowed to proliferate and form microspheroids containing a lumen after 5 d of culture. The spontaneous chiral rotation of a multicellular hollow spheroid about the z-axis is defined by the hydrogel gradient. (B) Confocal imaging of MDCK microsphere surface at day 5 shows a honeycomb-like assembly of cells with ZO-1 localized at the nodes. (C) A confocal cross-section through the middle of the microspheroid reveals a hollow lumen. (D and E) Cell alignment along the middle region of the microtissue coils around the structure toward the poles at the top and the bottom of the spheroid in 3D reconstructions of confocal imaging. (F) The microspheroids undergo spontaneous in-plane (x-y) rotation, with a bias toward the CCW direction. (Scale bars: 10 µm.) **P < 0.01.
MDCK Spheroids Exhibit Coordinated and Persistent Rotation That Is Chirally Biased.
We then wondered whether the behavior of these cellular structures was chiral in nature. As observed previously, the self-organized cells twirled together in synchronized collective rotation (23, 24, 26) (SI Appendix, Fig. S3 and Movies S3 and S4). We quantified the spontaneous rotation of more than 400 self-assembled spheroids (Fig. 1F) and found that the majority (93%) rotated coherently in-plane at the interface between hydrogel layers. The rotation had a statistically significant bias (P = 2.4 × 10−5), with 55% in the counterclockwise (CCW) direction and only 38% in the clockwise (CW) direction. The bias in rotational behavior persisted throughout cell phases and remained consistent for >15 h (Movie S4). In contrast, when two layers of Matrigel with the same concentration were used, >60% of the microspheroids underwent out-of-plane rotation (termed complex rotation), and among those rotating in the plane, there was no directional bias (SI Appendix, Table S1). Taken together, our data demonstrate that the self-organized microspheroids exhibited a chiral bias in spontaneous rotation in the graded hydrogel bilayer.
Chiral Bias of Microspheroid Rotation Depends on Actin.
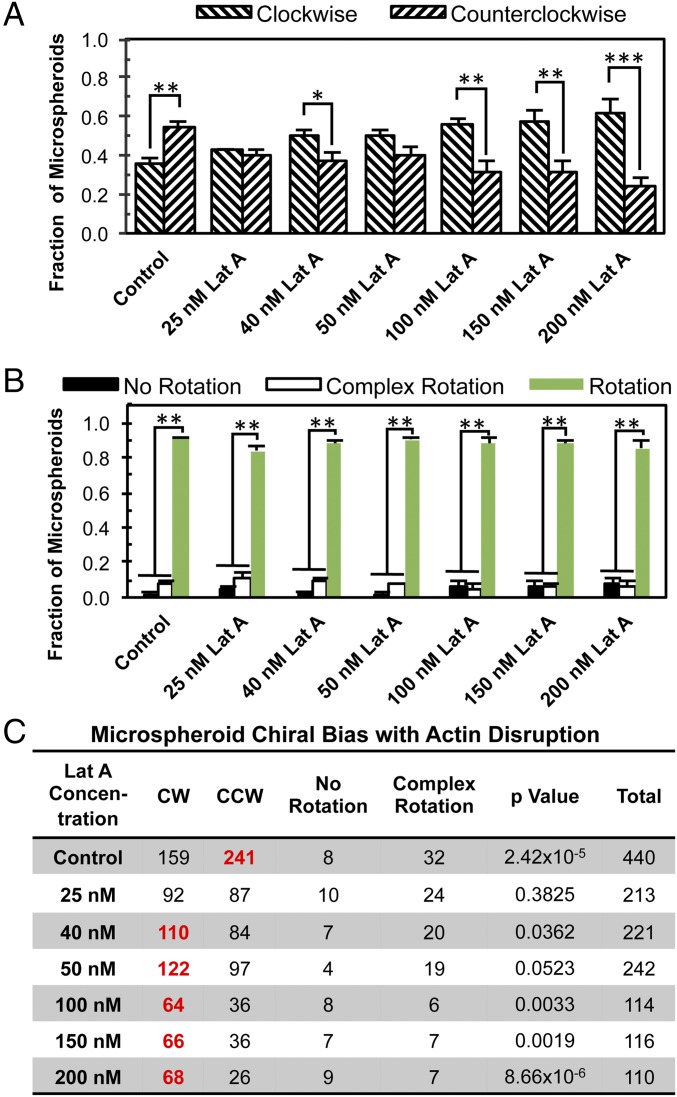
We then investigated whether the cell cytoskeleton determines the biased rotation of the cellular spheroids. Previously, it was found that patterned 2D cell chirality is dependent on actin function (16). Therefore, we posited that functional actin plays a significant role in 3D spheroid rotation. On day 5, after lumen formation, the spheroids were exposed to Latrunculin A (Lat A) for 4 h before time-lapse imaging, which disrupts actin polymerization through sequestration of G-actin monomers and promotion of depolymerization (27). Lat A treatments (40–200 nM) reversed the directional bias of MDCK microspheroids, resulting in predominantly CW rotation (Fig. 2 and Movies S5 and S6). Specifically, under 200 nM Lat A treatment, 62% of the microspheroids rotated CW, and only 24% showed CCW directionality (P = 8.7 × 10−6 for CW vs. CCW). Immunofluorescence imaging showed a slightly irregular lumen structure of Lat A-treated microspheroids with fewer actin filaments at cortical surfaces compared with the control (SI Appendix, Fig. S4A and Movies S7 and S8). Furthermore, we examined another actin-interfering agent, cytochalasin D, and found a similar dependence of cell chirality on actin (SI Appendix, Fig. S5). Although the speed of rotation of microspheroids decreased significantly with 200 nM Lat A treatment, the angular speed did not show a significant difference between CW-rotating and CCW-rotating microspheroids (SI Appendix, Fig. S6A). In comparison, MDCK cells cultured on 2D micropatterned rings were only slightly biased toward CCW alignment (SI Appendix, Fig. S7). However, with the Lat A treatment (25–50 nM), the cells adopted a CW bias, similar to the microspheroids in 3D. Concentrations >50 nM resulted in significant MDCK cell death on 2D substrates, demonstrating a difference in cell viability between 2D and 3D. Similar to what was found on 2D (16), the microtubule-interfering agent nocodazole could not reverse the chirality of 3D rotation, although it did disrupt the bias of rotation (SI Appendix, Fig. S8). The similarity in chirality change between 2D and 3D chirality in response to drug treatment strongly suggests that 3D spontaneous rotation is dictated by a mechanism similar to 2D patterned chirality, and thus it is likely that the driving force behind asymmetrical microspheroid rotation in 3D is also due, at least in part, to the actin cytoskeleton.
Fig. 2.
The 3D rotational direction of hollow microspheroids switched from CCW to CW under Lat A treatment. (A) The fraction of microspheroids undergoing CW or CCW rotation. (B) The fraction of microspheroids undergoing rotation, complex rotation, or no rotation. (C) The corresponding table showing the number of spheroids in different modes of motion. The boldface red numbers in C represent a statistically significant bias toward the specific direction. *P < 0.05; **P < 0.01; ***P < 0.001.
Biased Rotation Occurs in Microtissues with Organized Architecture.
To determine whether the well-organized structure of self-assembled multicellular microspheroids is important for asymmetrical rotation, we formed aggregates of multiple cells. The cells were seeded at a higher density (12,000 cells/cm2), resulting in neighboring cells clustering together and adopting a unique morphology, depending on the size of the aggregate (SI Appendix, Fig. S9 and Movies S9–S12). A large percentage (∼60%) of the aggregates with a spherical geometry underwent spontaneous rotation. Aggregates that had multiple spheroids connected together forming a “raspberry-like” morphology (SI Appendix, Fig. S9) mostly underwent complex rotation. Some 50% of the single-spheroid cell aggregates with >10 cells underwent in-plane spontaneous rotation, much lower than the 90% of hollow spheroids formed from single cells after 5 d of culture. We did not detect a bias in rotational asymmetry in the control single-spheroid aggregates, but there did note an increase in CW rotational motion of the aggregates with disruption of actin polymerization with Lat A, as observed with the hollow spheroids (SI Appendix, Fig. S9C and Movie S13). These data show that a strong rotational chiral bias of cell aggregates can be observed only for well-organized single-spheroid solid aggregates.
Actin-Dependent Chiral Behavior Arises from Single Cells.
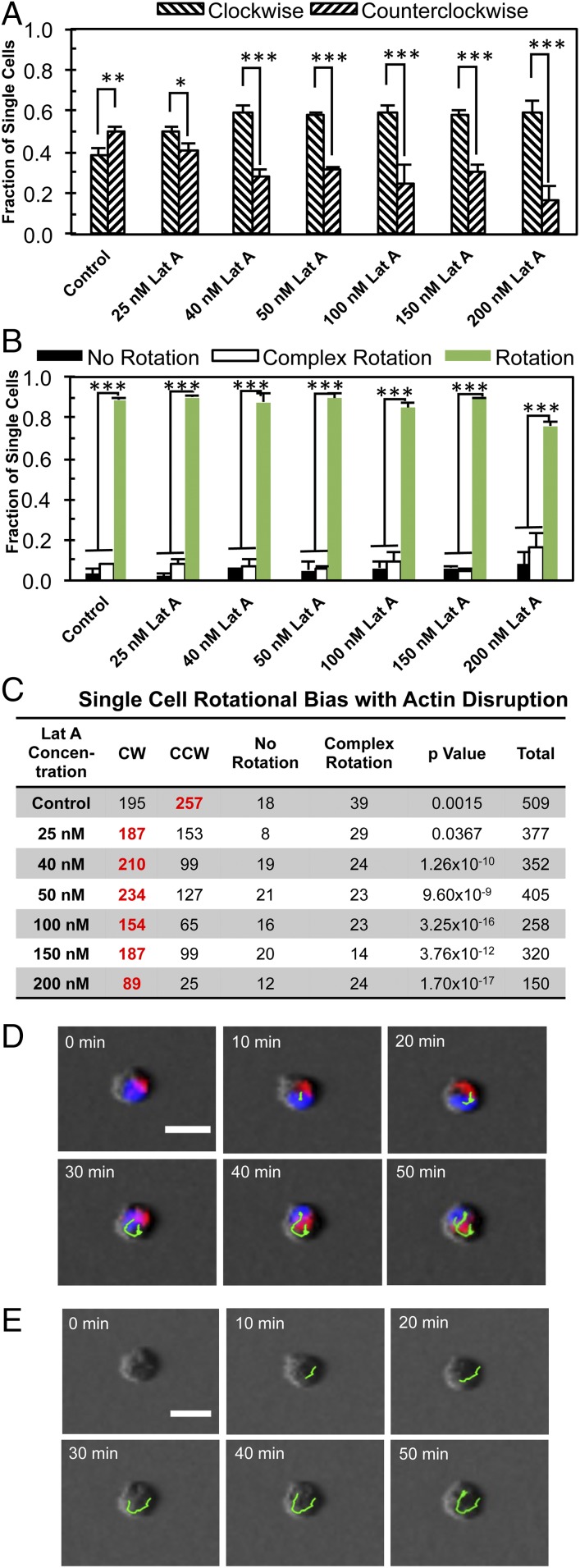
To determine whether the chirality of multicellular structures arises from the collective or individual cells, we investigated the rotation of single cells embedded in the 3D bilayer Matrigel. Immunofluorescence imaging clearly revealed a localization of the cell nucleus toward the bottom, resulting in a teardrop-like morphology with a more rounded shape for the lower cell body compared with the conical top (SI Appendix, Fig. S4B and Movies S14 and S15). We performed phase-contrast live imaging of the cells, along with fluorescence imaging of the nuclei and Golgi apparatuses (Fig. 3 and Movies S16 and S17). Individual MDCK cells underwent rotational motion in 3D (Fig. 3 A–C), as did the cell nuclei (Fig. 3D). Both rotations were slightly biased toward CCW. The disruption of actin polymerization produced a significant increase in instances of CW rotation by single cells (Fig. 3 A–C). The angular velocity of these single cells was only slightly decreased by Lat A treatment (SI Appendix, Fig. S6C). In addition, we also examined the single cell chirality of two other cell types, human mammary cell line MCF 10A and mouse cardiac HL-1 cells, and found that they are CW-biased (SI Appendix, Table S2). Taken together, these data suggest that cells are directionally biased even at a single cell level, and further support the idea that the collective directional rotation of microspheroids is a product of the inherent chirality of individual cells.
Fig. 3.
The 3D rotation of individual cells is CW-biased under Lat A treatment. (A) The fraction of individual cells undergoing CW or CCW rotation. (B) The fraction of cells undergoing rotation, complex rotation, or no rotation. (C) The corresponding table showing the number of cells in different modes of motion. The boldface red numbers in C represent a statistically significant bias toward the specific direction. *P < 0.05; **P < 0.01; ***P < 0.001. (D) Overlay of live fluorescence and phase time-lapse imaging of the cells without Lat A (blue, nuclei; red, Golgi apparatuses), with nuclear movement tracked with green lines. (E) Corresponding phase-contrast images with the cells tracked as shown in green. (Scale bars: 20 μm.)
We also noted that when these single cells proliferated and formed a two-cell system, these two-cell bodies rotated very quickly (SI Appendix, Table S3 and Movies S18 and S19) and exhibited a CCW bias, which can be reversed to a CW bias with 50 nM Lat A treatment. It has been previously demonstrated that two capillary endothelial cells on 2D patterns can undergo spontaneous symmetry breaking, giving a unique “yin-yang” symbol appearance, which also emerged in our 3D systems (26). Indeed, the two-cell system moves much faster, and the rotation is much easier to quantify, allowing for shorter-term time-lapse imaging to determine cell chirality. Therefore, the two-cell stage provides a good opportunity to measure the intrinsic chirality of epithelial cells. Furthermore, the chiral behavior of the cells is dependent on actin function consistently across 2D rings, in 3D microtissues, bicells, and down to the single-cell level.
α-Actinin-1 Regulates the Directional Bias of Cell Rotation.
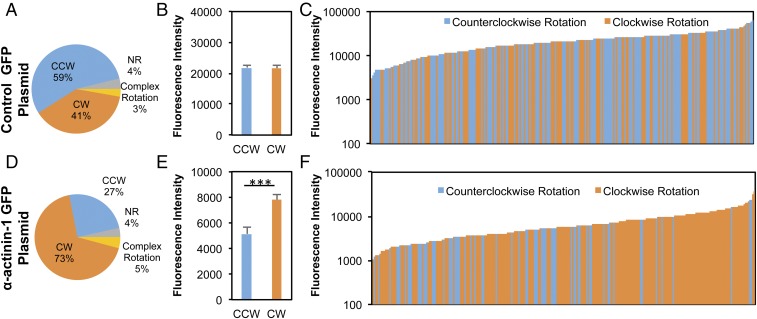
Finally, we proceeded to explore the potential cytoskeletal mechanisms associated with the role of actin in 3D cell chirality. Several previous studies have reported that formin plays a significant role in determining cell chirality (5, 18), and that the α-actinin cross-linking of formin-dependent radial actin fibers can switch the directionality of actin swirling (18). Therefore, we posited that 3D cell chirality might be affected by a similar mechanism. To test this hypothesis, we transfected MDCK cells with pEGFP-N1 α-actinin-1 and observed increases in the number and fluorescence intensity of filopodia-like cell protrusions stained for actin filaments (SI Appendix, Fig. S10 and Movies S20 and S21). The transfected cells were embedded within the bilayer system, and live time-lapse videos were obtained to characterize the rotation of the cells (Fig. 4). The total EGFP fluorescence intensity for individual cells served as a measure of α-actinin-1 expression levels. Overall, overexpression of α-actinin-1 in individual cells demonstrated a CW-dominated bias, in contrast to the CCW dominance among those transfected with control plasmids (73% CW vs. 27% CCW) (Fig. 4 A and D). Among rotating cells, those exhibiting a CW rotational bias had a statistically higher level of fluorescence intensity for α-actinin-1 (P = 5.97 × 10−4) compared with that for the CCW-biased cells (Fig. 4E), in which no difference in fluorescence intensity was noted between CW- and CCW-rotating cells transfected with control plasmids (Fig. 4B). Lower levels of α-actinin-1 expression were generally correlated with a nearly equal distribution of CW- and CCW-rotating cells, and an increasing percentage of CW-rotating cells with higher levels of α-actinin-1 (Fig. 4 C and F). In contrast, the cells transfected with the control plasmid pEGFP-N1 did not exhibit a significant correlation between fluorescence intensity and rotation directionality. These findings indicate that the establishment of single-cell chiral behavior in 3D can be strongly affected by the level of α-actinin-1 expression and its associated actin cross-linking.
Fig. 4.
Directional bias of rotational behavior by individual cells can be regulated by α-actinin-1. (A and D) MDCK cells transiently transfected with pEGFP-N1 α-actinin-1 (D) exhibited a predominantly CW rotation, in contrast to those transfected with the control plasmid, pEGFP-N1 (A). (B and E) Quantification of the total EGFP fluorescence intensity per cell reveals that the CW-rotating MDCK cells exhibited greater average expression of α-actinin-1 compared with the cells identified with CCW rotational motion (E), while there was no significant difference between CW- and CCW-rotating cells transfected with the control plasmid (B). (C and F) Histograms displaying the distribution of CCW and CW cells transfected with pEGFP-N1 (C) or pEGFP-N1 α-actinin-1 plasmids (F) with the increase in fluorescence intensity. Each bar represents an individual CW (orange) or CCW (blue) cell, sorted in ascending order of EGFP fluorescence expression. Cells expressing higher levels of α-actinin-1 expression rotated mostly CW, while there were nearly equal instances of CW and CCW rotation in the cells with lower α-actinin-1 expression. MDCK cells transfected with the control plasmid exhibited a slight bias toward CCW, which did not change with changes in fluorescence intensity. ***P < 0.001.
Here we report a physiologically relevant 3D platform for modeling epithelial chiral morphogenesis of the LR asymmetry of embryonic development, a conserved characteristic of living organisms that could not be easily studied until now. This platform is not intended to fully simulate in vivo environments for LR symmetry breaking, but it provides a simple yet effective system for determining chirality and thereby provides insight into possible regulatory mechanisms of LR asymmetry. We demonstrate that the direction of spontaneous rotation of both self-organized hollow cellular spheroids and individual epithelial cells residing at the interface of two layers of Matrigel is chiral in nature, with a CCW bias that is dependent on functional actin, consistent with the results from 2D ring patterning (16, 28, 29).
In the newly developed 3D system, the chirality of rotational behavior not only was observed as coordinated collective behavior across multicellular spheroids, but also was discerned with individual cells. Therefore, cell chirality likely arises from within the cell before it is manifested and propagated into multicellular chiral structures, such as microspheroids. In particular, individual cells exhibit a clear bias in the rotation of nuclei and cells as a whole. These findings suggest that chirality is not merely a pattern that emerges only in collective cell migration, but a fundamental property of the cell that depends on the chiral nature of the cytoskeleton network, such as actin (14, 15, 18). However, we did find that organized structures such as these hollow microspheroids promote chiral rotation compared with cell aggregates with irregular morphologies, suggesting that the platform needs to control noise at every level so that cell chirality can be observed. Otherwise, the noise, such as cellular random walk or irregular morphology, can easily dominate the behavior of the cells, obscuring cell chirality.
Our results also highlight the actin-dependent mechanisms underlying chiral rotational behavior. It has been demonstrated that the spontaneous rotation of epithelial cells is related to actomyosin contractility (23) and regulated by actin filaments at cellular junctions at the basal sides of the cells (30); however, the chiral nature of the rotation has not been reported or studied previously. In this study, we found localization of both actin filaments and β1 integrin to cell–cell junctions at the basal surfaces of the microspheres, supporting a mechanism based on traction forces between the cells and extracellular matrix for the spontaneous rotation itself. Our study suggests that the chirality of the rotation can be regulated by actin cross-linking. We found that inhibition of actin polymerization reversed the bias rotational motion in multicellular spheroids, pairs of daughter cells (bicells), and single cells in 3D. Actin filaments are chiral structures composed of right-handed intertwined G-actin strands that play an important role in providing structural support to the cell and in various motility functions (31). Actin cross-linking proteins are vital in the dynamic process of actin assembly of bundles and the construction of 3D cytoskeletal networks (32). Our analysis revealed that chiral rotation was regulated by the actin-binding protein α-actinin-1. These findings are in agreement with a previous study by Tee et al. (18), which reported a similar role of α-actinin-1 on chiral actin swirling of human foreskin fibroblasts. Therefore, although the rotation itself is driven by actomyosin related contraction, the chiral bias is possibly regulated by parallel actin structures. This is very similar to what was reported for 2D, in which cell migration requires actomyosin contractility, but LR directional biases relate to other actin structures, such as actin bundles or filopodia-like structures (14, 18). Our findings highlight cytoskeletal mechanisms in the LR bias of 3D epithelial rotational motion.
Actin-related mechanisms have been reported for LR asymmetry in development. In the Xenopus embryo, actin-interfering agents have been shown to induce a large-scale chiral torsion of the actin cortex (11). Likewise, early chiral cell alignment of snails can be regulated by actin-interfering agents, but not by drugs affecting microtubule dynamics (33). Our findings show that disruption of microtubules by nocodazole (0.5–2 µM) significantly enhances the development of complex rotational behavior. Microtubules could play an important role in the capacity of microtissues to detect the gradient of Matrigel layers and to gain z-axis orientation. Disruption of the orientation would in turn cause randomized tilting and potentially inverted orientation, resulting in decreased planar rotation and the obfuscation of chiral bias. A similar effect was seen in vivo in Xenopus after injection of mutant α-tubulin, which caused an increase in heterotaxia (12). In addition, the proteins related to the LR asymmetry were found to be distributed asymmetrically, depending on actin cytoskeleton organization, during early embryo development in frogs and chicks (13). Myosin ID switches the LR polarity and regulates the rotation directionality of Drosophila hindgut (3). Of note, the reversal (instead of randomization) of chirality is rarely found in vertebrate development with genetic mutation, except for the INV mouse mutants of select genetic backgrounds (34). These animals have a 100% reversal of internal organ positioning (35) and altered actin structures (36). Our findings in a 3D system call for further evaluation of cytoskeletal chirality in developmental asymmetry.
In summary, our findings suggest that graded hydrogels of extracellular matrix can provide a niche for microtissues to grow in 3D and provide directional guidance for the cells to orient and exhibit collective chiral behaviors.
Materials and Methods
Cells and Cell Culture.
MDCK epithelial cells (American Type Culture Collection) were cultured in DMEM supplemented with 10% FBS (Sigma-Aldrich), 1% penicillin-streptomycin (Pen Strep; Sigma-Aldrich), and 1% sodium pyruvate (Invitrogen). Cells were maintained at 37 °C in a humidified incubator with 5% CO2. Mammary epithelial cells (MCF 10A, a gift from Jason Herschkowitz, State University of New York at Albany) were cultured in the 1:1 mixed medium of DMEM and Ham’s F-12 (Invitrogen) supplemented with 5% FBS, 1% Pen Strep, 20 ng/mL epidermal growth factor (Sigma-Aldrich), 0.5 mg/mL hydrocortisone (Sigma-Aldrich), 100 ng/mL cholera toxin (Sigma-Aldrich), and 10 µg/mL insulin (Sigma-Aldrich). HL-1 cardiac muscle cells were cultured in Claycomb medium supplemented with 10% FBS, 1% Pen Strep, 1% Norepinephrine (Sigma-Aldrich) and 1% l-Glutamine (Thermo Fisher Scientific).
The 3D Multilayer Matrigel System.
MDCK cells were embedded within a Matrigel bilayer system as described previously (23, 24, 37). Using an eight-chambered glass slide (ibidi), a base layer was formed by coating the bottom of each well with 50 µL of 100% Matrigel (Corning), followed incubation at 37 °C for 25 min to solidify the Matrigel. MDCK cells were then seeded onto the Matrigel base layer. The cell density was 6,000 cells/cm2 for the single-cell study and for the formation of cell spheroids. After 1 h of incubation, the cells attached, and the medium was replaced with fresh cold medium containing 2% Matrigel to construct a top layer, thereby embedding the cells between a bilayer of Matrigel. After another hour, unless stated otherwise, the time-lapse imaging was performed to observe single-cell rotation. For the formation of cell spheroids, the medium was replaced every 2 d, and the culture was maintained for 5–10 d. Cell aggregates were obtained by seeding the cells at a higher density of 12,000 cells/cm2 to allow the cells to aggregate before the analysis of spontaneous rotation was performed.
Morphology and Rotation Analysis.
Phase-contrast time-lapse images were gathered for at least 2 h at 1-min intervals in a humidified incubator at 37 °C with 5% CO2. Time-lapse videos were processed and analyzed with the MTrackJ plugin in ImageJ, and the direction of cellular rotation was categorized as CW, CCW, no rotation, or complex rotation. For complex rotation, the cell rotates out of the interfacial plane or undergoes directional switching. The rotation category was assigned to all in-plane (x-y plane) rotation and encompassed all CW and CCW rotational behaviors.
Microscopy and Immunofluorescence.
Self-organization of cells during microtissue development was observed with a confocal microscope (Zeiss LSM 510 META or Leica TCS SP8 DMi8). Cells were fixed using 4% paraformaldehyde with 1% glutaraldehyde and then stained with DAPI for nuclei, phalloidin for actin filaments, and ZO-1 antibody (Thermo Fisher Scientific) for tight junctions. An anti–α-tubulin-FITC antibody (Sigma-Aldrich) was used to label microtubule structures. An antibody against laminin V (Abcam) was paired with a Golgi apparatus stain, anti-gm130 (Abcam), to mark cellular apical-basal polarity. In addition, the apical-basal polarity was further revealed by a centrosome marker with anti-gamma tubulin antibody, GTU-88 (Abcam), and β1 integrins with anti-integrin β1 (Abcam). Finally, primary cilium structures were characterized by staining for acetylated tubulin (Sigma-Aldrich).
2D Chirality Characterization.
MDCK cultures on ring-shaped micropatterns were prepared as described previously (16, 38). An array of ring-shaped molds was prepared via microfabrication and cast to form polydimethylsiloxane stamps. The stamps were used to microcontact print an adhesive monolayer of octadecanethiol (Sigma-Aldrich) onto gold-coated glass on which fibronectin was applied to promote cell attachment. Nonspecific attachment on the surfaces surrounding the patterns was blocked with 2 mM HS-(CH2)11-EG3-OH (EG3; ProChima Surfaces). On confluency, high-resolution phase-contrast images were gathered and analyzed with a custom-written MATLAB program to determine the chirality of local cell alignment. Analyzed multicellular rings were identified as CW, CCW, or nonchiral.
Pharmacologic Treatments.
To study the role of actin in chiral alignment in 2D and microtissue rotation in 3D, cultures were treated with 25–200 nM Lat A. Actin polymerization was also inhibited with cytochalasin D at 0.2–5 µM (Sigma-Aldrich). The role of microtubules was tested using the depolymerization agent nocodazole at 0.1–2 µM (Sigma-Aldrich).
Plasmid Transfection.
For single cell organelle live imaging, MDCK cells were transfected with CellLight Histone 2B (H2B-GFP) and Golgi-RFP (Life Technologies) following the manufacturer’s instructions. Dynamic F-actin structures were visualized using the TagRFP markers in MDCK cells transfected with pLifeAct. Two plasmids were used to study the role of α-actinin-1 in 3D cell chirality. The plasmid pEGFP-N1 α-actinin-1 was a gift from Carol Otey, University of North Carolina, Chapel Hill, NC (Addgene plasmid 11908), and pEGFP-N1-FLAG was a gift from Patrick Calsou, Institut de Pharmacologie et de Biologie Structurale, Toulouse, France (Addgene plasmid 60360). MDCK cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) and selected using G418 (Sigma-Aldrich). Phase-contrast live cell imaging of individual cells was done at a rate of 5 min per frame, together with still fluorescence images of the cells. Only the fluorescent cells were analyzed for the directional bias of α-actinin-1 overexpression cell rotation. The intensity of fluorescence was measured using ImageJ. The rotational behavior of cells that did not express any fluorescence was also characterized.
Statistical Analysis.
The cellular chirality, the biased affinity toward CW or CCW, was based on cell alignment on 2D ring micropatterns and rotational migration in 3D matrices in which the statistical significance was determined using binomial cumulative distribution. Two-way ANOVA was used to determine the effects of drug treatment and the chirality (CCW vs. CW) on the magnitude of rotation speed, followed by the Tukey honest significant difference post hoc test to detect differences between groups. Differences were deemed significant at P < 0.05 for all statistical tests. Error bars on the histograms represent SE.
Supplementary Material
Acknowledgments
We thank Dr. Sergey Pryshchep for technical assistance at the Microscopy Core, which is partially supported by National Science Foundation (Award MRI-1725984). This work was supported by the National Institutes of Health (Grant OD/NICHD DP2HD083961), National Science Foundation (Award CAREER CMMI-1254656), American Heart Association (Award 13SDG17230047), and March of Dimes (Grant MOD 5-FY14-111). L.Q.W. is a Pew Scholar in Biomedical Sciences (PEW 00026185), supported by the Pew Charitable Trusts.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805932115/-/DCSupplemental.
References
- 1.Aylsworth AS. Clinical aspects of defects in the determination of laterality. Am J Med Genet. 2001;101:345–355. [PubMed] [Google Scholar]
- 2.Levin M. Left-right asymmetry in embryonic development: A comprehensive review. Mech Dev. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Taniguchi K, et al. Chirality in planar cell shape contributes to left-right asymmetric epithelial morphogenesis. Science. 2011;333:339–341. doi: 10.1126/science.1200940. [DOI] [PubMed] [Google Scholar]
- 4.Sato K, et al. Left-right asymmetric cell intercalation drives directional collective cell movement in epithelial morphogenesis. Nat Commun. 2015;6:10074. doi: 10.1038/ncomms10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davison A, et al. Formin is associated with left-right asymmetry in the pond snail and the frog. Curr Biol. 2016;26:654–660. doi: 10.1016/j.cub.2015.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatori R, et al. Left-right asymmetry is formed in individual cells by intrinsic cell chirality. Mech Dev. 2014;133:146–162. doi: 10.1016/j.mod.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Wan LQ, Chin AS, Worley KE, Ray P. Cell chirality: Emergence of asymmetry from cell culture. Philos Trans R Soc Lond B. 2016;371:20150413. doi: 10.1098/rstb.2015.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wan LQ, Ronaldson K, Guirguis M, Vunjak-Novakovic G. Micropatterning of cells reveals chiral morphogenesis. Stem Cell Res Ther. 2013;4:24. doi: 10.1186/scrt172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naganathan SR, Fürthauer S, Nishikawa M, Jülicher F, Grill SW. Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. eLife. 2014;3:e04165. doi: 10.7554/eLife.04165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schonegg S, Hyman AA, Wood WB. Timing and mechanism of the initial cue establishing handed left–right asymmetry in Caenorhabditis elegans embryos. Genesis. 2014;52:572–580. doi: 10.1002/dvg.22749. [DOI] [PubMed] [Google Scholar]
- 11.Danilchik MV, Brown EE, Riegert K. Intrinsic chiral properties of the Xenopus egg cortex: An early indicator of left-right asymmetry? Development. 2006;133:4517–4526. doi: 10.1242/dev.02642. [DOI] [PubMed] [Google Scholar]
- 12.Lobikin M, et al. Early, nonciliary role for microtubule proteins in left-right patterning is conserved across kingdoms. Proc Natl Acad Sci USA. 2012;109:12586–12591. doi: 10.1073/pnas.1202659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu D, et al. Localization and loss-of-function implicates ciliary proteins in early, cytoplasmic roles in left-right asymmetry. Dev Dyn. 2005;234:176–189. doi: 10.1002/dvdy.20509. [DOI] [PubMed] [Google Scholar]
- 14.Tamada A, Igarashi M. Revealing chiral cell motility by 3D Riesz transform-differential interference contrast microscopy and computational kinematic analysis. Nat Commun. 2017;8:2194. doi: 10.1038/s41467-017-02193-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, et al. Polarity reveals intrinsic cell chirality. Proc Natl Acad Sci USA. 2007;104:9296–9300. doi: 10.1073/pnas.0703153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan LQ, et al. Micropatterned mammalian cells exhibit phenotype-specific left-right asymmetry. Proc Natl Acad Sci USA. 2011;108:12295–12300. doi: 10.1073/pnas.1103834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Worley KE, Shieh D, Wan LQ. Inhibition of cell-cell adhesion impairs directional epithelial migration on micropatterned surfaces. Integr Biol. 2015;7:580–590. doi: 10.1039/c5ib00073d. [DOI] [PubMed] [Google Scholar]
- 18.Tee YH, et al. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol. 2015;17:445–457. doi: 10.1038/ncb3137. [DOI] [PubMed] [Google Scholar]
- 19.Chen TH, et al. Left-right symmetry breaking in tissue morphogenesis via cytoskeletal mechanics. Circ Res. 2012;110:551–559. doi: 10.1161/CIRCRESAHA.111.255927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan LQ, Vunjak-Novakovic G. Micropatterning chiral morphogenesis. Commun Integr Biol. 2011;4:745–748. doi: 10.4161/cib.17649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 22.Cukierman E, Pankov R, Yamada KM. Cell interactions with three-dimensional matrices. Curr Opin Cell Biol. 2002;14:633–639. doi: 10.1016/s0955-0674(02)00364-2. [DOI] [PubMed] [Google Scholar]
- 23.Tanner K, Mori H, Mroue R, Bruni-Cardoso A, Bissell MJ. Coherent angular motion in the establishment of multicellular architecture of glandular tissues. Proc Natl Acad Sci USA. 2012;109:1973–1978. doi: 10.1073/pnas.1119578109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H, Lacoche S, Huang L, Xue B, Muthuswamy SK. Rotational motion during three-dimensional morphogenesis of mammary epithelial acini relates to laminin matrix assembly. Proc Natl Acad Sci USA. 2013;110:163–168. doi: 10.1073/pnas.1201141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang S, Brangwynne CP, Parker KK, Ingber DE. Symmetry-breaking in mammalian cell cohort migration during tissue pattern formation: Role of random-walk persistence. Cell Motil Cytoskeleton. 2005;61:201–213. doi: 10.1002/cm.20077. [DOI] [PubMed] [Google Scholar]
- 27.Morton WM, Ayscough KR, McLaughlin PJ. Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 28.Raymond MJ, Jr, et al. Multiaxial polarity determines individual cellular and nuclear chirality. Cell Mol Bioeng. 2017;10:63–74. doi: 10.1007/s12195-016-0467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raymond MJ, Jr, Ray P, Kaur G, Singh AV, Wan LQ. Cellular and nuclear alignment analysis for determining epithelial cell chirality. Ann Biomed Eng. 2015;44:1475–1486. doi: 10.1007/s10439-015-1431-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Squarr AJ, et al. Fat2 acts through the WAVE regulatory complex to drive collective cell migration during tissue rotation. J Cell Biol. 2016;212:591–603. doi: 10.1083/jcb.201508081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satir P. Chirality of the cytoskeleton in the origins of cellular asymmetry. Philos Trans R Soc Lond B. 2016;371:20150408. doi: 10.1098/rstb.2015.0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonet C, Maciver SK, Mozo-Villarías A. The regulatory action of alpha-actinin on actin filaments is enhanced by cofilin. Eur Biophys J. 2010;39:1143–1153. doi: 10.1007/s00249-009-0566-2. [DOI] [PubMed] [Google Scholar]
- 33.Shibazaki Y, Shimizu M, Kuroda R. Body handedness is directed by genetically determined cytoskeletal dynamics in the early embryo. Curr Biol. 2004;14:1462–1467. doi: 10.1016/j.cub.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 34.Oki S, et al. Reversal of left-right asymmetry induced by aberrant Nodal signaling in the node of mouse embryos. Development. 2009;136:3917–3925. doi: 10.1242/dev.039305. [DOI] [PubMed] [Google Scholar]
- 35.Mochizuki T, et al. Cloning of inv, a gene that controls left/right asymmetry and kidney development. Nature. 1998;395:177–181. doi: 10.1038/26006. [DOI] [PubMed] [Google Scholar]
- 36.Werner ME, et al. Inversin modulates the cortical actin network during mitosis. Am J Physiol Cell Physiol. 2013;305:C36–C47. doi: 10.1152/ajpcell.00279.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee GY, Kenny PA, Lee EH, Bissell MJ. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4:359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan LQ, et al. Geometric control of human stem cell morphology and differentiation. Integr Biol. 2010;2:346–353. doi: 10.1039/c0ib00016g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.