Significance
In the United States alone, over 20,000 children are hospitalized each year from parainfluenza virus (PIV)-related illness. By age 10, most children are seropositive to several PIVs, including PIV1 and PIV2, which cause croup, and PIV3, which causes severe lower-respiratory infections. Here we assess whether the prefusion conformation of the F glycoproteins from PIV types 1–4 could induce potent neutralizing responses. We engineered mutations in the F glycoprotein that stabilized its prefusion conformation and immunized mice and nonhuman primates. The prefusion-stabilized PIV F immunogens elicited significantly higher neutralizing titers than the same F immunogens in the postfusion state, both individually and as a quadrivalent PIV1–4 F vaccine, indicating these prefusion-stabilized PIV F glycoproteins to be promising vaccine candidates.
Keywords: antibody, conformational change, structure, vaccine design, virus
Abstract
Parainfluenza virus types 1–4 (PIV1–4) are highly infectious human pathogens, of which PIV3 is most commonly responsible for severe respiratory illness in newborns, elderly, and immunocompromised individuals. To obtain a vaccine effective against all four PIV types, we engineered mutations in each of the four PIV fusion (F) glycoproteins to stabilize their metastable prefusion states, as such stabilization had previously enabled the elicitation of high-titer neutralizing antibodies against the related respiratory syncytial virus. A cryoelectron microscopy structure of an engineered PIV3 F prefusion-stabilized trimer, bound to the prefusion-specific antibody PIA174, revealed atomic-level details for how introduced mutations improved stability as well as how a single PIA174 antibody recognized the trimeric apex of prefusion PIV3 F. Nine combinations of six newly identified disulfides and two cavity-filling mutations stabilized the prefusion PIV3 F immunogens and induced 200- to 500-fold higher neutralizing titers in mice than were elicited by PIV3 F in the postfusion conformation. For PIV1, PIV2, and PIV4, we also obtained stabilized prefusion Fs, for which prefusion versus postfusion titers were 2- to 20-fold higher. Elicited murine responses were PIV type-specific, with little cross-neutralization of other PIVs. In nonhuman primates (NHPs), quadrivalent immunization with prefusion-stabilized Fs from PIV1–4 consistently induced potent neutralizing responses against all four PIVs. For PIV3, the average elicited NHP titer from the quadrivalent immunization was more than fivefold higher than any titer observed in a cohort of over 100 human adults, highlighting the ability of a prefusion-stabilized immunogen to elicit especially potent neutralization.
Human paramyxoviruses and pneumoviruses are widespread pathogens (1–3), cause considerable disease burden (4), and include measles virus (MeV) (5), mumps virus (MuV) (6), respiratory syncytial virus (RSV), metapneumovirus (MPV) (7), and parainfluenza virus types 1–4 (PIV1–4) (Fig. 1A). The paramyxovirus family members PIV1 and PIV3 (genus Respirovirus) are important pediatric pathogens, with lower incidence or disease severity caused by the paramyxovirus family members PIV2 and PIV4 (genus Rublavirus) (8). While effective responses to measles and mumps can be induced by live attenuated viral vaccines (9), licensed vaccines for RSV, MPV, and PIV1–4 have not been obtained using the same approach (10–13). Entry by these viruses utilizes the viral fusion (F) glycoprotein (14, 15), a type 1 fusion machine that transitions between a metastable prefusion conformation and a stable postfusion conformation (Fig. 1B) to merge virus and cell membranes. Structure-based stabilization of RSV F in its prefusion conformation has been shown to induce high levels of RSV-neutralizing activity in vaccine animal models (16–20) and in a recent clinical trial.* While prefusion F stabilization has been proposed as a general means of inducing high titer protective responses, such stabilization with the MPV F glycoprotein does not induce improved neutralization titers (21), thus raising questions about the general utility of prefusion F vaccine stabilization for paramyxoviruses. To investigate the utility of prefusion PIV1–4 F vaccination, we utilized available structural information to engineer soluble prefusion-stabilized versions of the F glycoprotein from PIV1, 2, 3, and 4, which we characterized structurally and tested for immunogenicity in mice and rhesus macaques.
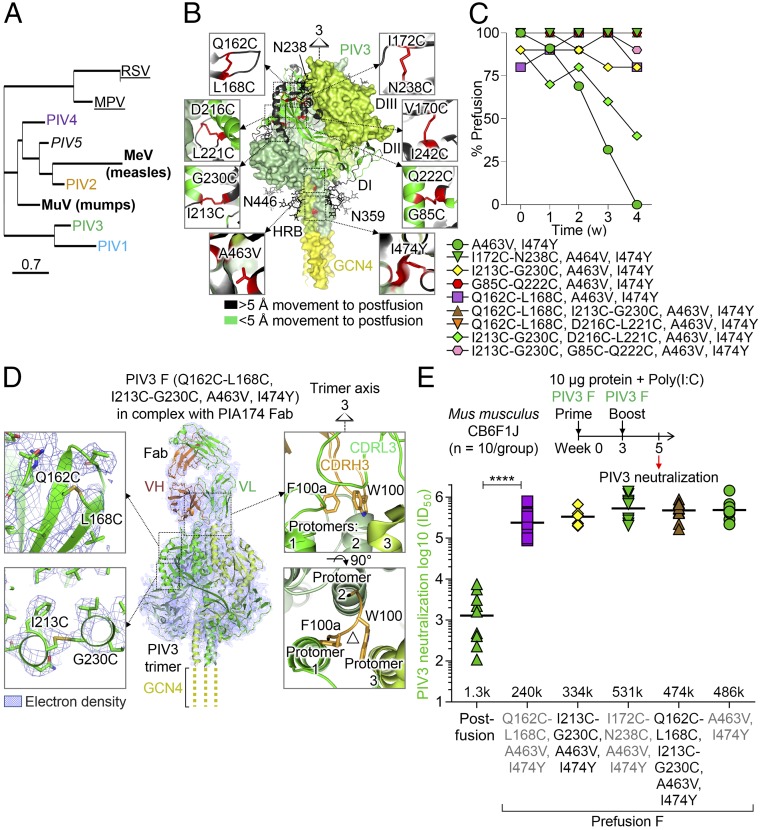
Fig. 1.
Structure-based design of a PIV3 F glycoprotein vaccine that elicits high-titer neutralizing responses. (A) Phylogenetic tree of pathogenic human paramyxovirus F glycoproteins, with human parainfluenzas colored blue, orange, green, and purple for types 1, 2, 3, and 4, respectively, throughout all figures. Viruses for which vaccines have been licensed are shown in bold font, viruses for which prefusion F candidate vaccines have been created are underlined, and the simian PIV5 is shown in italics. (B) Structure-based design of prefusion-stabilizing mutations in PIV3 F showing disulfide and cavity-filling changes in the head and stem, respectively. Model was based on PIV5 prefusion structure (PDB ID 4WSG). Additional PIV F variants are shown in SI Appendix, Fig. S5 B–F. (C) Temporal stability of prefusion F as determined by negative-stain EM during a 4-wk incubation time course in PBS at 37 °C. (D) Cryo-EM structure at 4.3-Å resolution of prefusion PIV3 F stabilized by Q162C-L168C, I213C-G230C, A463V, I474Y mutations and bound to antibody PIA174. The complex was most ordered at the core of the PIV3 F trimer and at its interface with the PIA174 antibody, where the cryo-EM map showed a local resolution of 4.0 Å, calculated using RELION (38). Insets show disulfide bonds (Left) and antibody–apex interactions (Right). (E) Immunogenicity of various PIV3 F variants in CB6F1J mice (10 per group) showing immunization schema (Top), neutralization data (Bottom), and statistical comparison of prefusion-versus-postfusion F immunogens by two-tailed Mann–Whitney U test (****P < 0.0001).
Results
Structure-Based Design of Disulfide Bonds and Cavity-Filling Mutations That Robustly Stabilize the Prefusion PIV3 F Trimer.
As PIV3 causes the most hospitalizations (8), we made it our top priority for vaccine design. We used the crystal structures of the simian prefusion PIV5 F glycoprotein (PDB ID 4GIP, 4WSG) (22) to construct a homology model for the prefusion PIV3 F protein, which consisted of three intertwined monomers contributing to the quaternary assembly of four domains—DI, DII, DIII, and the heptad repeat B (HRB) region—enclosing a large internal cavity. We compared this model of the prefusion PIV3 F to the crystal structure of uncleaved postfusion PIV3 F (PDB ID 1ZTM) (23) to predict regions of the F protein that undergo conformational rearrangements between prefusion and postfusion forms. These regions were the focus of design efforts, which resulted in over 100 prefusion-stabilized PIV3 F variants (Fig. 1B and SI Appendix, Table S1) and which all had the GCN4-trimerization domain appended to the C terminus at residue 481 (SI Appendix, Table S1). These engineered variants were predicted to stabilize the prefusion conformation of the PIV3 F protein and to arrest its transition to the postfusion state. To enable antigenic discrimination of the designed immunogens, we screened B cells for F-directed antibodies capable of neutralizing PIV3 and identified two, antibodies PIA56 and PIA75, which bound postfusion PIV3 F, and two others, antibodies PIA3 and PIA174, which were prefusion F-specific. Antigenic screening of the F designs for binding to PIV3 prefusion and postfusion F-specific antibodies identified variants that expressed as stabilized prefusion trimers. The antigenic analysis showed that introduction of various combinations of six nonnatural disulfide bonds, 172C-238C, V170C-I242C, I213C-G230C, D216C-L221C, Q162C-L168C, and G85C-Q222C, and two cavity-filling mutations, A463V and I474Y, resulted in PIV F glycoproteins that preferentially bound the prefusion-specific neutralizing antibodies PIA3 and PIA174 and not the postfusion F-specific antibody PIA56. (SI Appendix, Fig. S1 A–E). Cavity-filling mutations, A463V and I474Y, were both located within the HRB helical coiled-coil region, whereas the engineered disulfide bonds linked regions that were predicted to be close together in the prefusion-homology model, but to separate in the postfusion state.
Numerous disulfide and cavity-filling prefusion F variants (e.g., I172C-N238C/I474Y-GCN4) showed complete prefusion-conformational fixation over 4 weeks at 37 °C, in contrast to the variant stabilized only with GCN4 (22) (Fig. 1C), with other prefusion-stabilized variants displaying temporal stability, enhanced yield, and/or increased prefusion F physical stability (SI Appendix, Fig. S2 and Table S1).
Structure of a Prefusion-Stabilized PIV3 F Trimer Bound to an Apex-Binding Antibody Reveals Details of Trimer Organization and Stabilizing Mutations.
To provide atomic-level information on the prefusion-stabilized PIV F, we determined the structure of the PIV3 F Q162C-L168C, I213C-G230C, A463V, I474Y variant in complex with the prefusion-specific antibody PIA174 by cryo-EM to an overall resolution of 4.3 Å, as reported according to the FSC0.143 gold-standard criterion (Fig. 1D and SI Appendix, Fig. S2 A–D and Table S2). The resolution of the map progressively decreased towards the more disordered peripheries of the complex, with the lowest resolutions observed for the constant domains of the PIA174 Fab and the tip of the C-terminal HRB (SI Appendix, Fig. S3 A and B). Overall, the cryo-EM reconstruction revealed a single Fab bound at the apex of the prefusion F trimer, reminiscent of HIV-1 prefusion Env trimer apex-specific antibodies PGT145 (24, 25), VRC26.09 (26), or PG9 (27–29).
Similar to the PIV5 F trimer (PDB ID 4WSG), the PIV3 F trimer presented a “tree-like” appearance, with three intertwined monomers forming DI, DII, DIII, and the HRB domains and interconnecting loops (SI Appendix, Fig. S3). The GCN4-trimerization motif and the C-terminal ends of the HRB helices, including the region that contained the I474Y cavity-filling mutation, were disordered and not observed in the cryo-EM reconstruction. One of the cavity-filling mutations (A463V) was placed within the HRB helices and resulted in an increase in van der Waals’ contacts between neighboring helices. The prefusion-stabilizing disulfide I213C/G230C was well-defined in the cryo-EM reconstruction in all three monomers, linking two helices in the heptad repeat C (HRC) DIII domain that lay adjacent to each other in the prefusion structure. The disulfide Q162C/L168C connected two beta strands and, although located in a more disordered region of the DIII domain, was also visible in the cryo-EM reconstruction of the prefusion F PIV3 structure (Fig. 1D), although residues 162 and 168 were separated by 9.9 Å in the postfusion form. Although there were similarities in the overall shapes of the fusion proteins from the different paramyxoviruses, their architecture and the topology of the protein secondary structural elements enclosing the internal cavity differed, as did the mutations that were identified to stabilize prefusion RSV F (15, 17) (SI Appendix, Figs. S1 F–H and S3 G–L).
The prefusion PIV3 F protein in the cryo-EM structure was bound at its apex to neutralizing antibody PIA174, with a single antibody Fab engaging the DIII domain and making contact with all three protomers by using both its heavy and light chains. A total of ∼1,970 Å2 surface area was buried at the Env–antibody interface, with heavy-chain contacts accounting for 68% of this total buried surface area. The heavy chain inserted its 17-amino-acid–long CDR H3 loop into the trimer apex with the hydrophobic W100 and F100a side chains engaged between the three apex protomer alpha helices [Kabat numbering (30) was used for numbering antibody residues]. Although the overall fold and topology of the PIA174-bound PIV3 F protomers was similar to that of prefusion PIV5 F (22) with an rmsd of 2.6 Å (SI Appendix, Fig. S1F), as was the domain organization within the trimer, the interprotomer geometry varied (SI Appendix, Fig. S3 J–L). Overall, these geometric differences in the structure highlight the conformational malleability of the prefusion trimer, along with its ability to undergo conformational adjustments while still maintaining a prefusion topology and fold.
Prefusion PIV3 F Trimers Stabilized with Various Mutations Elicit High-Titer Neutralizing Antibodies in Mice.
To assess the ability of PIV3 prefusion F-stabilized variants to elicit neutralizing antibodies, we immunized CB6F1/J mice with 10-µg doses of postfusion or prefusion PIV3 F glycoprotein variants combined with 10 μg polyinosinic–polycytidylic acid (poly-I:C) adjuvant at weeks 0 and 3 and measured the ability of week 5 sera to prevent PIV3 infection of HEp-2 cells. The PIV3 I213C/G230C/A463V/I474Y-GCN4 prefusion variant elicited PIV3 neutralization titers 250-fold higher than postfusion F (Fig. 1E), and the A463V/I474Y cavity-filling variant elicited neutralizing titers 375-fold higher than postfusion F [geometric mean effective concentrations (EC50 values) of 334,000 and 1,300, respectively]. The double-disulfide variant I213C-G230C/Q162C-L168C/A463V/I474Y (SI Appendix, Fig. S4) elicited neutralizing titers 370-fold higher than postfusion F with an EC50 value of 428,000. Additional PIV3 preF variants showed EC50 values up to 593,000 and similar fold differences with postfusion F (SI Appendix, Fig. S5 A and B). Overall, prefusion PIV3 F stabilized in multiple ways yielded ratios of prefusion F to postfusion F neutralization titers that were substantially higher than observed for RSV, MPV, and other paramyxoviruses.
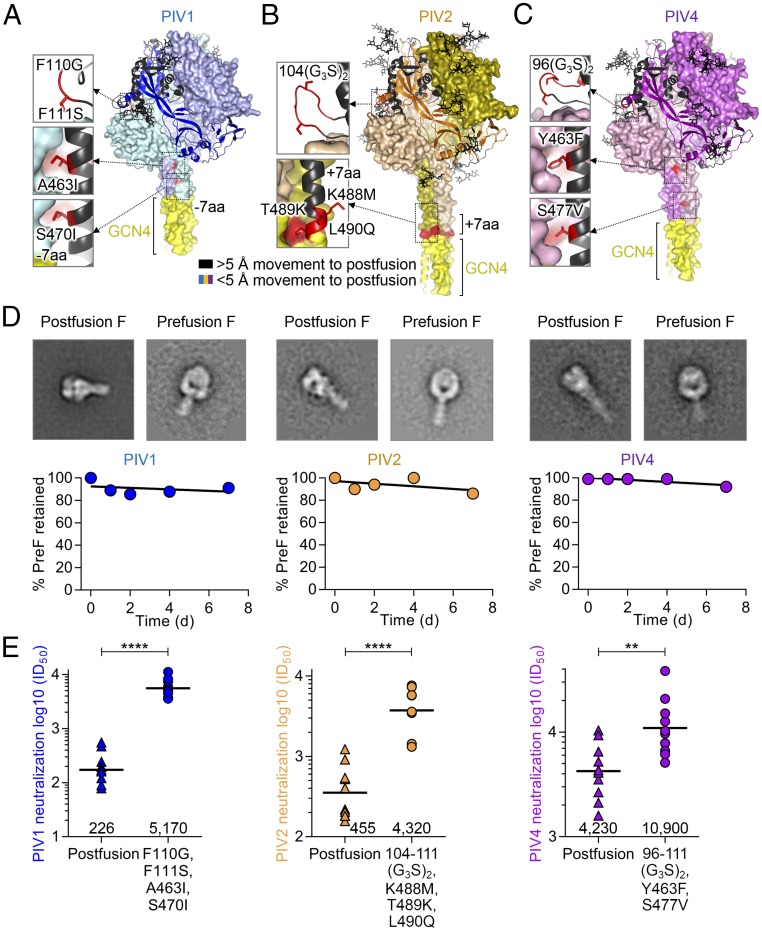
Prefusion-Stabilized PIV1, 2, and 4 F Trimer Immunogens Obtained Through Structure-Based Design Utilizing EM.
Vaccines against PIV1, 2, and 4 are also highly desired, particularly PIV1 and PIV2, which are the primary causes of croup, infect most individuals during infancy, and cause an estimated 14,000 infant hospitalizations (ages 0–4) each year in the United States alone (8). In light of the high neutralization titers induced by PIV3 prefusion F-stabilized immunizations, we used homology design of prefusion F-stabilized variants of PIV1, 2, and 4 (Fig. 2 A–C and SI Appendix, Fig. S6). A strategy similar to that used as for the prefusion PIV3 F design identified regions of high conformational mobility between prefusion and postfusion conformations, which enabled the design of prefusion variants for PIV1, PIV2, and PIV4 (Fig. 2 A–D) that locked the structures in the prefusion state. Since no PIV1, 2, or 4 F-specific antibodies were available to identify antigenically prefusion or postfusion conformation, we used negative-stain EM to assess directly the proportion of prefusion F. The combination of a glycine-rich linker at the F2-F1 cleavage site and stem cavity-filling mutations yielded variants with high prefusion–postfusion ratios as observed with EM micrographs with over 60% prefusion conformation for PIV1 and PIV2 F, while a soluble linker at the F2-F1 cleavage site and a 172C-238C disulfide bond yielded a variant with over 95% of prefusion conformation for PIV4 F (Fig. 2C). To determine the stability of prefusion F PIV1, 2, and 4 designs, we assessed the proportion of prefusion-to-postfusion conversion over time. Prefusion F-stabilized PIV1, 2, and 4 conformations were retained above 85% over the course of a week at physiological conditions and 37 °C (Fig. 2D).
Fig. 2.
Structure-based design of PIV1, 2, and 4 prefusion-stabilized F vaccines based on EM conformational verification. (A–C) Models of PIV1, PIV2, and PIV4, respectively, based on PIV5 prefusion F coordinates (PDB ID 4WSG) showing stabilizing mutations that were used in immunization studies. Glycans are shown in black sticks. Additional stabilization based on transferring antigenically selected PIV3 prefusion-stabilizing F disulfide designs to PIV1, 2, and 4 F is shown in SI Appendix, Fig. S5 B–F. (D) Temporal stability of prefusion F as assessed for 7 d at 37 °C in PBS for prefusion-stabilized HPIV1, 2, and 4 designs. Representative negative-stain electron microscopy averages for prefusion-stabilized F (Right) and postfusion F (Left) of corresponding PIV virus are shown above each plot. (Box dimensions: 30 × 30 nm.) (E) Immunogenicity of recombinant prefusion and postfusion F glycoproteins from PIV1, 2, and 4, showing statistically significant differences in neutralization titers between prefusion and postfusion conformations of the F glycoproteins from all three viruses by two-tailed Mann–Whitney U test (****P < 0.0001, **P < 0.01).
Prefusion-Stabilized PIV1, 2, and 4 F Elicit High-Titer Virus-Neutralizing Responses.
To understand how PIV1, 2, and 4 F conformation impacted elicitation of neutralizing antibodies, we immunized groups of 10 CB6F1/J mice with 10 μg/dose postF or preF PIV 1, 2, or 4 with 50 μg poly-I:C at weeks 0 and 3. Mice elicited a statistically significant improved neutralization titer to all three PIV types when immunized with the prefusion form compared with the postfusion form, with fold improvements for PIVs 1, 2, and 4 of 23, 10, and 2.6, respectively (Fig. 2E).
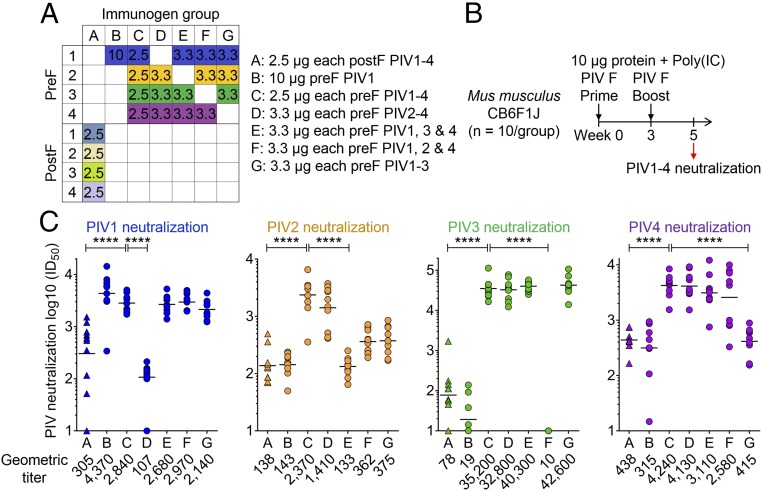
Multivalent Immunization in Mice with Prefusion-Stabilized PIV1–4 F Immunogens Induces Neutralizing Titers Against PIV1–4.
To investigate the ability of combinations of the prefusion-stabilized immunogens to elicit pan-PIV neutralizing antibodies, we immunized mice with monovalent, trivalent, or quadrivalent prefusion F (multivalent components were equimolar) and compared this with quadrivalent postfusion F (Fig. 3 A and B). Since PIV1–4 are related, we assessed whether omitting one of the four prefusion F types in a multivalent immunogen might induce cross-neutralization to the omitted virus type. In mice, using the same procedure as for the monovalent immunization, quadrivalent PIV1–4 prefusion F immunization yielded robust neutralization to all four types with prefusion-to-postfusion F-fold differences of 9.3, 17, 450, and 9.7 for PIV1, 2, 3, and 4, respectively. When one component was omitted from the prefusion F immunogen, titers to the omitted type equivalent to the quadrivalent postfusion F were achieved, while titers to the remaining three viral types were equivalent to that achieved by the quadrivalent prefusion F immunogen (Fig. 3C and SI Appendix, Fig. S7). Last, monovalent immunization with PIV1 prefusion F yielded statistically similar titers in both trivalent or quadrivalent immunizations, despite the three- to fourfold reduced amount of immunized PIV1 F. Thus, for PIV1 prefusion F, there appeared to be little difference in elicited titer in the multivalent context; however, for PIV3 prefusion F, there did appear to be a substantial reduction in the multivalent context (Figs. 1E and 3C). Overall, the prefusion F responses appeared to be type-specific, the level of titers induced by PIV prefusion F immunization in monovalent and quadrivalent formats appeared to also be type-specific, and to achieve pan-PIV immunity, the results indicate a quadrivalent combination of the four PIV1–4 prefusion F immunogens to be required.
Fig. 3.
Multivalent PIV1–4 F immunizations reveal high-titer neutralizing but serotype-specific responses in mice. (A) Immunization matrix of prefusion or postfusion PIV1–4 F glycoprotein mixtures, showing the amounts of each F glycoprotein antigen in the immunogen groups (A–G) with micrograms per immunization shown in the boxes in for each group. (B) Immunization plan for CB6F1J mice (n = 10 per group) with mixed immunogens A–G. (C) Week 5 sera neutralization readout with PIV1–4 virus infectivity inhibition assay showing statistically significant differences between quadrivalent prefusion F from quadrivalent postfusion F in all four viral neutralization assays and statistical significance between the quadrivalent prefusion F group and the trivalent prefusion F groups where F glycoproteins corresponding to the neutralization assay virus were omitted from the immunogen. A two-tailed Mann–Whitney U test was used (****P < 0.0001).
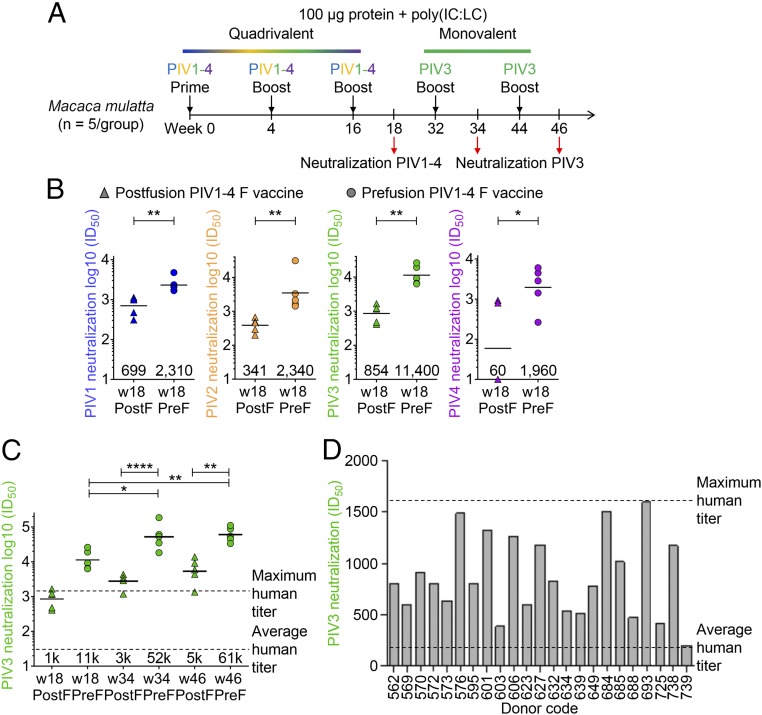
Quadrivalent Prefusion-Stabilized PIV1–4 F Immunization of NHPs Yields Pan-PIV1–4 Neutralizing Responses.
To determine the ability of the quadrivalent PIV prefusion or postfusion F immunization in nonhuman primates (NHPs) to elicit pan-PIV1–4 neutralizing responses, we immunized two groups of five rhesus macaques with 100 μg of either quadrivalent prefusion or postfusion F in combination with poly(IC:LC) (31) three times over 16 wk (Fig. 4A). Statistically significant superior neutralization titers to all four PIV types were achieved by immunizing NHPs with quadrivalent prefusion F compared with the quadrivalent postfusion F (Fig. 4B). Prefusion-to-postfusion F-fold differences were 3.3, 6.9, 13, and 32 for PIV1, 2, 3, and 4, respectively. Thus, immunization with a quadrivalent PIV vaccine elicited neutralizing antibody titers of all four major serotypes of PIV.
Fig. 4.
Quadrivalent immunization with prefusion PIV1–4 F glycoproteins in NHP elicits high-titer neutralizing responses. (A) NHP immunization scheme for quadrivalent prefusion or postfusion PIV1–4 F whereby three immunizations were administered containing the four viral F glycoproteins, followed by two immunizations with only PIV3 F glycoproteins. (B) Neutralization titers for NHP sera using PIV1, 2, 3, or 4 virus following three quadrivalent immunizations at week 18. Statistical significance is observed when comparing prefusion and postfusion groups for all four PIV virus types using a two-tailed Mann–Whitney U test (**P < 0.01, *P < 0.05). Geometric titers are shown above the x axis. (C) Neutralization titers in NHPs at weeks 34 and 46 following two PIV3 F-only boosts. A fivefold increase in prefusion-specific titers and statistically significant differences between prefusion and postfusion groups were observed (****P < 0.0001, **P < 0.01, *P < 0.05). (D) Analysis of 127 human sera tested for PIV3 neutralization showing the top 23 neutralizers for which the highest recorded titer was 1,600.
Prefusion-Stabilized PIV3 F Vaccine Elicits Higher Neutralizing Titers in NHP than Observed in a Cohort of over 100 Healthy Adults.
To provide context for the prefusion F-induced titers by the PIV1–4 quadrivalent vaccine, we assessed the naturally occurring level of titers in a cohort of 127 healthy adults. The average PIV3 titer in the cohort was less than 100, with a highest observed titer of ∼1,600 (Fig. 4D). By contrast, the PIV3 titers after quadrivalent prefusion F immunization were substantially higher, averaging 35,200 in mice and 11,400 in rhesus macaques. As the PIV3 titers were higher in the monovalent context, we tested whether subsequent monovalent immunization could boost titers in the rhesus macaques. Two additional 100-μg immunizations with PIV3 prefusion F resulted in further increases in neutralization titers with the prefusion F PIV3 immunized group, reaching an ID50 value of 60,500 (Fig. 4C).
Discussion
As humans can generate immunity to infection by PIV3 (32), with titers in our cohort averaging ∼100, it seems likely that the high-PIV3 neutralizing titers induced by the quadrivalent vaccine in NHPs would be protective against PIV3 infection. The superior titers induced by prefusion-stabilized F versus postfusion F for PIV1–4 indicate that these viruses are similar to RSV F, for which prefusion F-induced titers were superior to those induced by postfusion F, and, unlike MPV F, for which prefusion and postfusion F immunization generated similar titers (21). Notably, the PIV3 neutralization titers elicited by prefusion-stabilized PIV3 F were much higher than those observed for prefusion-stabilized RSV (16), MPV (21), PIV1, PIV2, or PIV4 F glycoproteins, suggesting the prefusion-stabilized PIV3 F immunogens described here may be highly suitable PIV3 vaccine candidates. It is not clear why prefusion PIV3 F elicits such high neutralization titers; one possibility is that highly neutralization-sensitive epitopes are exposed on the protein immunogen structure that can engage effective counterpart neutralizing antibodies, such as the apex-binding epitope for PIA174 and epitopes similar to site Ø of prefusion RSV F (16). Despite identifying numerous prefusion-stabilizing designs for PIV3 F, all nine combinations of stabilized immunogens elicited similar neutralizing titers, suggesting that stabilization into the prefusion state was the primary criterion for the improved titers and contrasts with prefusion-stabilized RSV F titers, where more graded immunogenicity was observed depending on the prefusion-stabilizing mutations (16).
It may be possible to develop a multivalent vaccine, which combines prefusion-stabilized F immunogens from RSV and PIV1–4; together with licensed vaccines for measles and mumps, such a multivalent vaccine would provide broad-spectrum immunity against pathogenic paramyxoviruses to which humans are exposed throughout life. Multivalent vaccines have been found to be useful against a number of pathogens and are currently licensed for flavivirus vaccines (dengue virus serotypes 1–4) (33), rotavirus vaccines (34), and HPV vaccines (35). Although our work here focused on the use of purified recombinant proteins for vaccination, it seems likely that the prefusion designs can be utilized in genetic contexts, as has been shown for prefusion-stabilized RSV F in the context of a PIV1 vector (36) or as a mRNA vaccine (37). Furthermore, prefusion-stabilized PIV F nanoparticles may be particularly effective. Overall, our results show how prefusion stabilization can substantially increase the titer of virus-neutralizing responses against PIV types, especially type 3. It will be interesting to see if prefusion F vaccine strategies utilized here can succeed with other members of the paramyxovirus family that are considered high risk for pandemic outbreak, such as Nipah and Hendra viruses.
Materials and Methods
Prefusion and postfusion PIV3 F-specific monoclonal antibodies were isolated by comparing B-cell culture supernatant neutralization potency and postfusion PIV3 F ELISA. The design of prefusion-stabilizing PIV3 F mutations was performed based on the prefusion PIV5 F structure and antigenically analyzed using prefusion and postfusion PIV3 F-specific monoclonal antibodies. Prefusion-stabilized F glycoproteins were assessed for prefusion and postfusion conformation by negative-stain EM, and cryo-EM data were collected for the PIA174-prefusion–stabilized PIV3 Q162C-L168C, I213C-G230C, A463V, I474Y complex. CB6F1/J mice were immunized to assess the elicitation of neutralizing antibodies to 10 μg of PIV1–4 F glycoprotein immunogens adjuvanted with Poly-IC. Two groups of five nonhuman primates were immunized with 100 μg prefusion or postfusion PIV1–4 F immunogens adjuvanted with Poly-IC:LC. Donors provided written informed consent for the use of blood and blood components (such as sera), following approval by the Canton Ticino Ethics Committee, Switzerland. Details are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of the Structural Biology Section and the Structural Bioinformatics Core Section of the Vaccine Research Center for helpful comments and members of the Electron Microscopy Group at the New York Structural Biology Center for assistance with data collection. Support for this work was provided by the Intramural Research Program of the Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was also supported in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under Contract HHSN261200800001E. Some of this work was performed at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247), NYSTAR, and the NIH National Institute of General Medical Sciences (GM103310). The work at the Institute for Research in Biomedicine was partially supported by the Swiss Vaccine Research Institute and by the European Research Council (Grant 670955 BROADimmune).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data have been deposited in the Electron Microscopy Data Bank at Protein Data Bank in Europe, www.ebi.ac.uk/pdbe/emdb/ (EMD-9135), and in the Protein Data Bank, www.wwpdb.org (PDB ID 6MJZ).
*Graham BS, RSV Vaccines for The World Conference (RSVVW17), November 30, 2017, Málaga, Spain.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811980115/-/DCSupplemental.
References
- 1.Russell CJ, Simões EAF, Hurwitz JL. Vaccines for the paramyxoviruses and pneumoviruses: Successes, candidates, and hurdles. Viral Immunol. 2018;31:133–141. doi: 10.1089/vim.2017.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jain S, et al. CDC EPIC Study Team Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373:415–427. doi: 10.1056/NEJMoa1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jain S, et al. CDC EPIC Study Team Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372:835–845. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozano R, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128, and erratum (2013) 381:628. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rota PA, et al. Measles. Nat Rev Dis Primers. 2016;2:16049. doi: 10.1038/nrdp.2016.49. [DOI] [PubMed] [Google Scholar]
- 6.Hviid A, Rubin S, Mühlemann K. Mumps. Lancet. 2008;371:932–944. doi: 10.1016/S0140-6736(08)60419-5. [DOI] [PubMed] [Google Scholar]
- 7.Panda S, Mohakud NK, Pena L, Kumar S. Human metapneumovirus: Review of an important respiratory pathogen. Int J Infect Dis. 2014;25:45–52. doi: 10.1016/j.ijid.2014.03.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abedi GR, et al. Estimates of parainfluenza virus-associated hospitalizations and cost among children aged less than 5 years in the United States, 1998-2010. J Pediatric Infect Dis Soc. 2016;5:7–13. doi: 10.1093/jpids/piu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidkin I, Jokinen S, Broman M, Leinikki P, Peltola H. Persistence of measles, mumps, and rubella antibodies in an MMR-vaccinated cohort: A 20-year follow-up. J Infect Dis. 2008;197:950–956. doi: 10.1086/528993. [DOI] [PubMed] [Google Scholar]
- 10.Crowe JE, Jr, et al. Satisfactorily attenuated and protective mutants derived from a partially attenuated cold-passaged respiratory syncytial virus mutant by introduction of additional attenuating mutations during chemical mutagenesis. Vaccine. 1994;12:691–699. doi: 10.1016/0264-410x(94)90218-6. [DOI] [PubMed] [Google Scholar]
- 11.Karron RA, et al. Evaluation of two live, cold-passaged, temperature-sensitive respiratory syncytial virus vaccines in chimpanzees and in human adults, infants, and children. J Infect Dis. 1997;176:1428–1436. doi: 10.1086/514138. [DOI] [PubMed] [Google Scholar]
- 12.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol. 1969;89:405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 13.Weibel RE, et al. Respiratory virus vaccines. VII. Field evaluation of respiratory syncytial, parainfluenza 1, 2, 3, and Mycoplasma pneumoniae vaccines, 1965 to 1966. Am Rev Respir Dis. 1967;96:724–739. doi: 10.1164/arrd.1967.96.4.724. [DOI] [PubMed] [Google Scholar]
- 14.Scheid A, Choppin PW. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology. 1974;57:475–490. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- 15.Aguilar HC, Henderson BA, Zamora JL, Johnston GP. Paramyxovirus glycoproteins and the membrane fusion process. Curr Clin Microbiol Rep. 2016;3:142–154. doi: 10.1007/s40588-016-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McLellan JS, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLellan JS, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–1117. doi: 10.1126/science.1234914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krarup A, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi: 10.1038/ncomms9143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang B, et al. Enhanced neutralizing antibody response induced by respiratory syncytial virus prefusion F protein expressed by a vaccine candidate. J Virol. 2015;89:9499–9510. doi: 10.1128/JVI.01373-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart-Jones GB, et al. A cysteine zipper stabilizes a pre-fusion F glycoprotein vaccine for respiratory syncytial virus. PLoS One. 2015;10:e0128779. doi: 10.1371/journal.pone.0128779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Battles MB, et al. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun. 2017;8:1528. doi: 10.1038/s41467-017-01708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch BD, et al. Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc Natl Acad Sci USA. 2012;109:16672–16677. doi: 10.1073/pnas.1213802109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q, et al. Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat Struct Mol Biol. 2017;24:370–378. doi: 10.1038/nsmb.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JH, et al. A broadly neutralizing antibody targets the dynamic HIV envelope trimer apex via a long, rigidified, and anionic β-hairpin structure. Immunity. 2017;46:690–702. doi: 10.1016/j.immuni.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doria-Rose NA, et al. NISC Comparative Sequencing Program Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker LM, et al. Protocol G Principal Investigators Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julien JP, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci USA. 2013;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McLellan JS, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of Proteins of Immunological Interest. 5th Ed US Dept Health Human Serv, Natl Inst Health; Bethesda: 1991. [Google Scholar]
- 31.Martins KA, Bavari S, Salazar AM. Vaccine adjuvant uses of poly-IC and derivatives. Expert Rev Vaccines. 2015;14:447–459. doi: 10.1586/14760584.2015.966085. [DOI] [PubMed] [Google Scholar]
- 32.Lee MS, et al. Half-life of human parainfluenza virus type 3 (hPIV3) maternal antibody and cumulative proportion of hPIV3 infection in young infants. J Infect Dis. 2001;183:1281–1284. doi: 10.1086/319690. [DOI] [PubMed] [Google Scholar]
- 33.McArthur MA, Sztein MB, Edelman R. Dengue vaccines: Recent developments, ongoing challenges and current candidates. Expert Rev Vaccines. 2013;12:933–953. doi: 10.1586/14760584.2013.815412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonesteller CL, Burnett E, Yen C, Tate JE, Parashar UD. Effectiveness of rotavirus vaccination: A systematic review of the first decade of global postlicensure data, 2006-2016. Clin Infect Dis. 2017;65:840–850. doi: 10.1093/cid/cix369. [DOI] [PubMed] [Google Scholar]
- 35.Kash N, et al. Safety and efficacy data on vaccines and immunization to human papillomavirus. J Clin Med. 2015;4:614–633. doi: 10.3390/jcm4040614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, et al. Attenuated human parainfluenza virus type 1 expressing the respiratory syncytial virus (RSV) fusion (F) glycoprotein from an added gene: Effects of prefusion stabilization and packaging of RSV F. J Virol. 2017;91:e01101-17. doi: 10.1128/JVI.01101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geall AJ, et al. Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakane T, Kimanius D, Lindahl E, Scheres SH. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife. 2018;7:e36861. doi: 10.7554/eLife.36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.