Significance
Archaea have considerable importance in ecology and evolution and have emerging roles in health. However, many of their cellular processes are under active study. Archaea are thought to acquire and inherit adaptive traits solely through mutation. Here, adaptive laboratory evolution of an extremophile trait revealed that an alternative nonmutational process was operative. When genes whose expression had been altered in a heritable manner were replaced by recombination using identical DNA, the evolved traits were changed. This implicated a regulatory role for chromatin proteins and was consistent with an epigenetic-like regulatory mechanism. This finding has evolutionary relevance for the origin of epigenetics, transcriptional regulation, and functional genome architecture.
Keywords: epigenetics, archaea, ALE, recombination, Sulfolobus
Abstract
Epigenetic phenomena have not yet been reported in archaea, which are presumed to use a classical genetic process of heritability. Here, analysis of independent lineages of Sulfolobus solfataricus evolved for enhanced fitness implicated a non-Mendelian basis for trait inheritance. The evolved strains, called super acid-resistant Crenarchaeota (SARC), acquired traits of extreme acid resistance and genome stability relative to their wild-type parental lines. Acid resistance was heritable because it was retained regardless of extensive passage without selection. Despite the hereditary pattern, in one strain, it was impossible for these SARC traits to result from mutation because its resequenced genome had no mutation. All strains also had conserved, heritable transcriptomes implicated in acid resistance. In addition, they had improved genome stability with absent or greatly decreased mutation and transposition relative to a passaged control. A mechanism that would confer these traits without DNA sequence alteration could involve posttranslationally modified archaeal chromatin proteins. To test this idea, homologous recombination with isogenic DNA was used to perturb native chromatin structure. Recombination at up-regulated loci from the heritable SARC transcriptome reduced acid resistance and gene expression in the majority of recombinants. In contrast, recombination at a control locus that was not part of the heritable transcriptome changed neither acid resistance nor gene expression. Variation in the amount of phenotypic and expression changes across individuals was consistent with Rad54-dependent chromatin remodeling that dictated crossover location and branch migration. These data support an epigenetic model implicating chromatin structure as a contributor to heritable traits.
Although mutation is the source of evolutionary change, non-Mendelian mechanisms that confer heritable adaptation are well established. DNA-associated factors, such as DNA methylases and histones, are integral to epigenetic processes. Eukaryotes employ transgenerational histone modification and DNA methylation to diversify cell lineages without accruing mutations (1). Bacteria utilize DNA methylation for transgenerational phenotypic heterogeneity (2) and DNA-binding proteins for gene silencing (3). Such non-Mendelian mechanisms of inheritance are not yet established in archaea. However, many species belonging to the phylum Crenarchaeota possess posttranslationally modified chromatin proteins such as Alba, Cren7, and Sso7D that bind the minor groove of DNA like histones (4–6). These resemble human HMG-box proteins, which influence expression, genome stability, and epigenetic processes (7).
Archaeal chromatin proteins have characteristics that suggest these proteins can have both structural and regulatory function (8). For example, archaeal chromatin proteins engage in architectural functions such as DNA bridging, bending, and wrapping (8), and the crenarchaeote Sulfolobus Lrp protein is classified as a transcription factor but regulates expression of a broad range of genes and binds DNA with relative nonspecificity like a chromatin protein (9). By analogy, bacterial chromatin proteins produce DNA secondary structures that can enhance or suppress access of transcriptional machinery (10). In Sulfolobus, Alba (Sso10b) inhibits transcription, likely by coating or bridging DNA (4). Although archaea have histones, they are not posttranslationally modified (11), but archaeal chromatin proteins in species lacking histones are both acetylated and methylated like eukaryotic histones (4–6). For example, the acetylation state of Alba affects promoter access and transcription in vitro (4), whereas the methylation state of another Sulfolobus chromatin protein, Sso7D, is altered by culture temperature (12). These observations support a potential regulatory role for these chromatin proteins. The high abundance of chromatin proteins in Sulfolobus (1–5% total protein) (5, 6) also suggests they undergo ubiquitous chromosomal binding, which would allow for regulation across a genome-wide scale.
Crenarchaeotes like Sulfolobus may employ alternate systems of transcriptional regulation to compensate for the reduced presence of other common regulatory mechanisms. Archaea universally use simple versions of eukaryotic-like proteins for synthesis, repair, and degradation of informational molecules (13). Archaeal transcriptional regulation is thought to be bacteria-like; however, crenarchaeotes lack some major features; both bacteria and euryarchaeotes utilize two-component regulatory systems, whereas they are absent in crenarchaeotes (14). Bacteria also use polycistronic protein-coding regions, whereas crenarchaeote genomes are mostly monocistronic (15). Although DNA methylation could play a role in archaeal gene regulation, in Sulfolobus, it is only known to participate in a typical type II modification system (16).
The relative simplicity of archaeal systems lends itself to experimentation. For example, the archaeal recombination component Rad54 is orthologous to the eukaryotic protein (17), and archaeal chromatin proteins bind the minor groove like eukaryotic histones. As a result, Rad54-mediated displacement of minor-groove–binding proteins observed in eukaryotes (18, 19) is also predicted to occur in archaea. As a result, targeted displacement of chromatin proteins at loci expected to have transcriptionally relevant patterns could indicate potential regulatory function. Such regulation may occur through the presence and absence of proteins and shifts in their modification state (4–6). However, despite the occurrence of posttranslationally modified chromatin proteins that could emulate eukaryotic histones, epigenetic phenomena have not been reported in these organisms.
Sulfolobus solfataricus is a thermoacidophilic crenarchaeote that grows optimally at pH 3.0 and 80 °C (20). In previous studies, adaptive laboratory evolution was used to generate extremely acid resistant isolates of this organism (21). After several years of passage, three independent and genetically distinct derivative strains were recovered that gained the heritable ability to grow at pH 0.8, representing a 178-fold increase in thermoacidophily. The derived strains were named super acid-resistant Crenarchaeota (SARC). Based on the occurrence of mutations in one SARC lineage, it was hypothesized that the heritable acid-resistance trait resulted from mutation but this turned out to be an incorrect assumption. Instead, as proposed here, an epigenetic-like mechanism governs SARC trait inheritance.
Results
The SARC Phenotype Does Not Result from Mutational Adaptation.
To test the hypothesis that a non-Mendelian process governs trait heritability, genome sequencing was performed for the two SARC lineages that were not analyzed previously (21). Then, the three fully evolved strains and their partially evolved intermediates (SI Appendix, Table S1) were compared with their parental strains to find mutations. In addition, all putative mutations were verified by resequencing. Based on adaptive laboratory evolution (ALE), it was predicted that the evolved strains would acquire adaptive mutations in common genes or pathways (22). However, no mutations were observed in the fully evolved SARC-I strain, whereas SARC-C and SARC-O had 5 and 29 mutations, respectively. Mutations that occurred in partially evolved strains also occurred in their fully evolved derivatives, thereby validating mutation heredity and the traceability of strain passage. However, the two fully evolved SARC strains that exhibited mutations had no overlap in affected genes or their promoter regions (Fig. 1A). None of the affected genes belonged in common metabolic pathways or possessed the same annotated function (SI Appendix, Table S2). Structural and copy number variations were predicted for SARC-I using DELLY (23) and Pindel (24), and their occurrence was tested using PCR, but no variations were found (SI Appendix, Fig. S2A). Although the contribution of mutation to trait acquisition and inheritance cannot be entirely excluded for SARC-C and SARC-O, the complete absence of mutations in SARC-I implicated a nonmutational mechanism for at least this strain. Genome sequencing also found that mutation rates were altered in the SARC strains compared with a control, and this will be discussed in The SARC Strains Exhibit Improved Genome Stability. The absence of a mutational basis for the SARC traits suggested the involvement of an alternate mechanism of adaptation and inheritance. Because this could occur through multiple processes, additional studies were performed involving transcriptomic analysis.
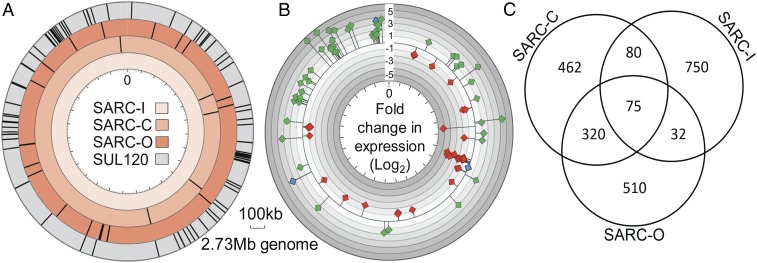
Fig. 1.
Presence and absence of mutations in passaged lines and the SARC transcriptome. (A) During ALE for acid resistance, SARC-C and SARC-O acquired mutations (black lines). SARC-I acquired no mutations. The control strain SUL120 was passaged without acid selection. Mutations were mapped in reference to the SULA genome. (B) Location of genes in the conserved SARC transcriptome. Genes with differential expression (>twofold, P < 0.05) that was conserved in all three SARC strains were mapped to the wild-type genome (SULA). Diamonds indicate genes that were up-regulated (green), down-regulated (red), or targeted for recombination (blue). Distance of diamonds from the axis indicates the average fold change (log2) in expression for the three SARC strains cultured at pH 1.0 compared with the parentals grown at pH 3.0. Log2 2.32 represents a fivefold change. (C) Venn diagram of genes with conserved differential expression patterns in SARC strains. EdgeR differential expression analysis was performed on transcriptomes of SARC strains cultured at pH 1.0 relative to parental strains at pH 3.0. For genes tested for conservation, P < 0.05 in all three SARC strains. Differential expression (>twofold) in the same direction for multiple strains indicated conservation.
The SARC Strains Exhibit Conserved, Heritable Expression Patterns.
If the SARC traits resulted from transient regulatory responses to acid stress and not through a heritable component, it was predicted that expression patterns evident at low pH would not be retained at pH 3.0, a condition that was not selective for the evolved trait. Instead, the distinctive transcriptomes of the SARC strains were retained despite passage without selection. This transcriptomic heritability included the SARC-I strain, which lacked mutations. RNA-sequencing (RNA-seq) analysis (conducted in biological replicate) identified a set of genes that had conserved expression changes in all of the SARC strains, referred to as the “SARC transcriptome” (Fig. 1B and SI Appendix, Table S3). Expression of 64 of these genes was altered between two- and fivefold, and 11 genes were altered more than fivefold. Three genes (SULA_0674, SULA_2620, and SULA_2869) had a >fivefold average expression change that was heritable in all three strains and, in addition, in their partially evolved intermediates, indicative of phenotypic importance. The occurrence of the SARC transcriptome was unlikely to have occurred randomly (SI Appendix, Supplemental Methods). Each SARC strain also independently acquired altered transcriptomes that were unique or overlapped other strains. There were between 32 and 320 conserved in two of three strains, and between 462 and 750 genes that were uniquely altered for each strain (Fig. 1C). The identities of the other genes in the SARC transcriptome also appeared to be related to acid-resistance mechanisms and fell into four main categories: oxidation resistance, proton extrusion, energy generation, and chromatin proteins (21). An up-regulated Sso7D chromatin protein suggested the involvement of chromatin in the SARC traits.
If the SARC transcriptome resulted from classical regulatory features, such as transcription factors, cytosolic elements, and proximal genome sequence features, it was predicted that evidence for such factors would be found. Instead, all were excluded. The SARC transcriptome was stable and was observed after 15 cell divisions at an optimal growth pH of 3.0. Importantly, the SARC strains passaged without selection for 50 cell divisions also retained the extreme trait when then shifted to low pH media (pH 1.0). Fifteen cell divisions would dilute the original cytosol to 0.003% of the original, whereas 50 divisions would dilute it to 8.8 × 10−14%. This dilution was sufficient to reduce all possible soluble factors that might promote or inhibit gene transcription to confer an apparent pattern of heritability. The conserved SARC transcriptome genes were distributed throughout the genome (Fig. 1B) and thereby implicated a mechanism controlling expression that must act across noncontiguous sequences. However, based on analysis of potential transcription factor–binding motifs and genome environment, this mechanism did not appear to be mediated by a transcription factor or to be a consequence of proximity to other genome features. Genome locations of SARC transcriptome genes did not correlate with those of tRNAs or origins of replication within regions spanning five ORFs upstream or downstream (SI Appendix, Fig. S2B). Additionally, no regions of high GC content were detected within a region 100 bp upstream of the ORF start nor within the ORF itself. To identify potential coregulation, SARC transcriptome genes were tested for gene ontology and functional enrichment using DAVID (25). Genes were clustered into pyruvate synthase/oxidoreductase, transmembrane, and ATP-binding categories, but these categories were not enriched (SI Appendix, Fig. S2C). Genes also were tested for potential common transcription factor–binding sites using MEME motif analysis (26) in comparison with a set of random unaltered genes, whose identified motifs were considered false positives. Although no motifs with e values of ≤0.1 were found (SI Appendix, Fig. S2D), FIMO (find individual motif occurrences) (27) was used to calculate the genomic abundance of low-confidence motifs to avoid false negatives. Because there was a maximum of 75 altered genes in the SARC transcriptome that could be coregulated by a common transcription factor, the number of potential genomic binding sites for a motif was not expected to exceed this number. The least abundant motif of the five identified from the SARC transcriptome genes appeared 328 times in SULA. This exceeded the 75 gene SARC transcriptome and genomic abundance of false positive motifs in the control gene set (SI Appendix, Fig. S2E).
Collectively, these results indicate that the SARC strains acquired heritable transcriptomes whose regulation is inconsistent with mutation, a transient stress response, or common systems of regulation across noncontiguous sequences. An alternative mechanism of regulating gene expression could involve epigenetic-like processes. Because epigenetic mechanisms are also known to influence genome stability, genome sequences were analyzed to determine mutation and transposition rates through comparison with the rates observed in a highly passaged control strain.
The SARC Strains Exhibit Improved Genome Stability.
If an epigenetic-like mechanism was involved in the SARC traits, it was predicted that genome stability of the SARC strains might differ from a control strain passaged without selection. When this was examined, improved genome stability was apparent as a secondary fitness trait arising from the adaptive laboratory evolution process. To determine whether mutation rates in the SARC strains were altered, a global mutation rate had to be determined for S. solfataricus without acid selection. The three fully adapted SARC strains were 120 generations distant from parental strains. Therefore, the parental strain, SULA, was passaged for 120 generations without selection to produce control strain SUL120 (SI Appendix, Table S1), which was then resequenced. In comparison with SUL120, all three fully evolved SARC strains exhibited a large reduction in the amount of transposition of insertion sequence elements, and a large reduction in the amount of forward mutation (summarized in Table 1). Both of these observations were indications of improved genome stability and therefore improved biological fitness. Reduction in transposition rates was notable because high transposition rates have been reported to be a general feature of this species (28).
Table 1.
Summary of mutation and transposition events that occurred during passaging
| Strain | Point mutations | Point mutation rate | Transitions:transversions (ratio) | Transposition events | Transposition rate |
| SUL120 | 141 | 4.35 × 10−7 | 100:41 (2.44) | 23 | 0.19 |
| SARC-C | 5 | 1.54 × 10−8 | 4:1 (4) | 0 | NA |
| SARC-I | 0 | NA | NA | 0 | NA |
| SARC-O | 29 | 8.95 × 10−8 | 23:6 (3.83) | 1 | 0.008 |
The point mutation rate was calculated as mutations per base per cell division (cell divisions, 120; bases, 2.7 Mb). The transposition rate was calculated as transpositions each cell division cycle. NA, rate cannot be calculated from no transpositions.
Two of the three SARC strains had no transpositions. One SARC strain underwent a 23-fold decrease in transposition frequency (1 event) compared with the control SUL120 strain (23 events) (SI Appendix, Table S4). No transposition occurred in the partially evolved strains (50 generations distant from the parental strain) (Table 1). This indicated that low transposition in the SARC strains was not attributable to reversion. In the control, SUL120, both replicative and nonreplicative transposition occurred frequently and some insertion sequence (IS) elements were deleted with no apparent reinsertion.
Despite the apparent lack of transposition activity, transposase expression of some IS element families was highly up-regulated in the SARC strains. Many of the annotated transposases with reliable expression data for all strains were up-regulated ≥fivefold on average (37.5%), whereas none appeared down-regulated. In comparison, 2.9% of genes with reliable expression data for all strains were up-regulated ≥fivefold on average for the SARC strains. The heightened transcription of transposases contrasts with the low transposition rate in the SARC strains. SARC strains contained no extrachromosomal elements that could be targets for transposition.
Mutation frequency was also suppressed in the SARC strains in comparison with the control SUL120 strain (Fig. 1A). In eukaryotic cells, mechanisms of improved genome stability have been ascribed to epigenetic processes (29). SUL120 contained 141 point mutations and 19 indels after 120 generations of passage without stress for a mutation rate of 4.35 × 10−7 mutations per base per cell division. In contrast, the two SARC strains with mutations averaged 17 point mutations per adapted strain, with an average mutation rate of 5.19 × 10−8 per base per cell division (Table 1). This represented an average 8.3-fold reduction in mutation rate for these strains. In contrast, the SARC-I strain had no mutations and therefore represented a more dramatic example of increased genome stability. Of the 34 total mutations in the 2 mutated SARC strains, 14 were nonsynonymous (41.2%), 12 were synonymous (35.3%), and 8 occurred in noncoding regions (23.5%) (SI Appendix, Table S2). Of the 141 mutations in SUL120, 71 were nonsynonymous (50%), 45 were synonymous (31%), 2 were nonsense mutations (3%), and 23 occurred in noncoding regions (16%) (SI Appendix, Table S2). SARC and SUL120 strains had similar mutation category proportions. The Ka/Ks ratios for the SARC strains and SUL120 were 1.17 and 1.6, respectively. Although these ratios are greater than 1 and might suggest that both underwent positive selection for mutations, the absence of mutations in SARC-I indicates that the SARC traits can occur without them.
A role of DNA repair genes could be excluded because 22 relevant genes (30) were not consistently up-regulated in SARC strain transcriptomes (SI Appendix, Table S5). This finding indicated that increased repair of DNA mutation was unlikely to be responsible for reducing the SARC strain mutation frequency, although it is possible that enzyme activity was improved through modification state or ATP availability. Additionally, the higher mutation rate in SUL120 was not a result of decreased selection against mutation in comparison with the SARC strains. SUL120 and SARC-C had equivalent growth rates in optimal pH media (SI Appendix, Fig. S1), indicating that mutations occurred in SUL120 did not reduce overall fitness. As such, likely mechanisms for altered mutation frequencies were reduced mutation formation or improved access of repair machinery. Both of these mechanisms could arise from changes in chromatin structure.
Occurrence of the Nonmutational SARC Phenotype Follows an Epigenetic Model.
The SARC traits did not result from mutation but were heritable in the absence of selection; therefore, an epigenetic-like mechanism could be invoked. Because chromatin proteins in this archaeon are highly abundant, bind the minor groove, and are posttranslationally modified (4, 6), an analogy could be made to the role of eukaryotic histones in gene regulatory processes. For example, the S. solfataricus chromatin protein abundance is consistent with the many sites at which the SARC strain molecular characteristics of mutation and transcriptome appeared. In addition, a regulatory role for posttranslational modification of chromatin proteins is supported by in vivo (12) and in vitro data (4). However, although tools commonly used to study epigenetic mechanisms in eukaryotes, such as chromatin modification mapping, locus-specific proteomics, and epigenome editing, are available, they have yet to be described in archaea. In particular, the high growth temperature of S. solfataricus and the unknown characteristics of its putative epigenetic-like systems preclude the use of methods from unrelated mesophilic systems. Because markerless exchange using homologous recombination (HR) is well established in S. solfataricus (31), it was used here as a tool to observe the effects of locus-specific disruption of native chromatin patterns without introducing genetic changes that could alter transcription. In eukaryotes, minor-groove–binding proteins like histones are displaced by Rad54 HR machinery (18, 19). Because archaea also possess Rad54, displacement of their minor-groove–binding chromatin proteins is predicted during HR. To assess the effects of chromatin protein displacement by HR, HR using marker selection (31) was performed at specific loci with identical DNA alleles (Fig. 2).
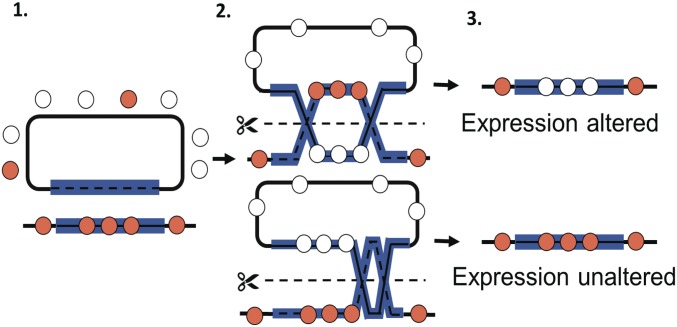
Fig. 2.
Model for displacement of native SARC chromatin proteins by recombination. (1, Left) SARC strains contains a mixture of SARC (orange) and non-SARC (white) chromatin proteins. Chromosomal SARC genes are bound by native SARC chromatin proteins that confer its expression state. Homologous plasmid DNA is naked. (2, Center) During recombination, rad54-mediated branch migration displaces chromatin proteins while replacing native chromatin segments with naïve DNA. In the absence of acid selection, chromatin proteins can repopulate these regions in non-SARC patterns. Depending on the crossover location, different lengths of chromatin can be exchanged. (3, Right) Resulting gene regions have no sequence changes, but are bound fully or partially by reassociated non-SARC chromatin patterns.
If the heritable SARC transcriptome did not result from heritable chromatin patterns that regulated transcription at those loci, it was predicted that HR at those loci would not alter expression or acid resistance. Instead, HR did alter acid resistance and expression when performed at SARC transcriptome loci. The loci that were selected for HR had high expression levels that were thought to confer acid resistance (SARC genes). It was predicted that chromatin protein displacement would change expression patterns and alter the acid-resistance phenotype. Because the degree of HR-mediated DNA replacement depends on the random site of crossover and on the distance of branch migration, the amount of chromatin protein displacement was predicted to vary between recombinant isolates (Fig. 2). Consequently, the intensity of phenotypic and expression shifts would also vary. According to this epigenetic model, HR at a control locus with no expression changes in the SARC strains (non-SARC gene) would not affect phenotype.
Recombinant isolates that underwent HR at SARC genes had significantly different growth rates at low pH compared with those that underwent HR at a non-SARC gene (Fig. 3A; unaveraged replicate data in SI Appendix, Fig. S3). Large changes in the acid-resistance phenotype occurred frequently when the SARC gene SULA_2869 was targeted, where 100% of recombinants had reduced growth rates relative to the parental SARC-I strain. When the next target SARC gene, SULA_2027, underwent HR, the resulting phenotypes were more broadly distributed. Forty percent of isolates had reduced growth rates relative to the parental SARC-I, whereas 50% of isolates were not reduced. This distribution was consistent with the model in which HR crossover location and branch migration can vary and result in different amounts of perturbed chromatin (Fig. 2). Importantly, when a non-SARC gene (SULA_0895) was targeted for HR, no recombinants had altered growth rates. This finding indicated that altered phenotypes in SARC-gene recombinants were not caused by global effects of HR. The frequency at which HR changed the phenotype might vary between the two SARC loci if the contribution of their expression to the SARC phenotype were not equivalent. It was also possible that the chromatin structure at SULA_2869 was more likely to be altered by HR than the structure at SULA_2027.
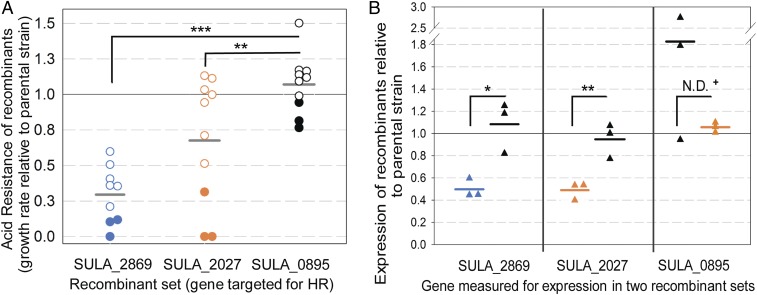
Fig. 3.
Effect of homologous recombination at SARC loci. (A) Acid resistance of isolates that underwent recombination at SARC (blue circles: SULA_2869, orange circles: SULA_2027) or non-SARC (black circles: SULA_0895) genes. Acid resistance is growth rate at pH 1.13 relative to parental SARC-I. Isolates predicted to have altered SARC gene expression based on acid sensitivity (closed circles) were further tested for expression in B. Each point is an average of triplicates. (B) Expression changes at SARC and non-SARC genes after directly undergoing recombination or after recombination occurred at a different gene. Triangle colors (blue: SULA_2869; orange: SULA_2027; black: SULA_0895) correspond to recombinants from A and indicate the identity of those tested for expression. Each point is an average of triplicates. Statistical comparisons were performed using the Student t test. Comparisons marked with + were performed with the Mann–Whitney rank sum test to accommodate inequivalent sample variances. N = 9 (SULA_2869), 10 (SULA_2027) for growth experiments; N = 3 for expression experiments; ***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; N.D., no difference. Horizontal bars indicate the means for sample sets.
It was also predicted that reduced acid resistance resulted from reduced expression of the SARC gene targeted for HR. SARC-gene recombinants with reduced acid resistance did have consistent reductions in expression of the gene targeted for HR (Fig. 3B; unaveraged replicate data in SI Appendix, Fig. S3). Recombinants retaining acid resistance also retained SARC gene expression levels (SI Appendix, Fig. S3). Importantly, reduction in expression at SARC genes was significantly different when the gene was directly targeted for HR compared with when a different gene underwent HR. This again indicated that both phenotypic and expression consequences of HR are locus-specific and did not result from global effects of HR. These data support a model in which the SARC phenotype was acquired and inherited via a mechanism that follows an epigenetic-like process. In this model, patterns of DNA-associated factors confer SARC expression patterns but can be displaced by HR.
Discussion
Trait acquisition and inheritance in archaea has been presumed to occur through a Mendelian process. In the SARC strains, no mutations were found that could cause heritable phenotypes and expression patterns, excluding this mechanism. HR performed at genes with high expression levels thought to be involved in the SARC phenotype (SARC gene) altered both the phenotype and the expression of the target gene. HR at a control gene thought to be uninvolved in acid resistance (non-SARC gene) did not alter SARC phenotype or expression of SARC genes. During eukaryotic HR, Rad54 activity displaces minor-groove–binding histones (18, 19). The observation that the effect of HR on SARC expression and phenotype required targeting of SARC genes was consistent with the prediction that archaeal Rad54 homolog (17) also displaces native SARC chromatin patterns, which then affected expression. Although HR at a non-SARC gene did not alter its expression with statistical significance, some isolates had increased expression. It is possible that suppressive chromatin structures uninvolved in the SARC phenotype were displaced by HR. These data implicated the existence of an epigenetic-like mechanism in archaea that contributes to regulation of acid resistance and likely also to other traits such as genome stability.
Enhanced genome stability reported for the SARC lineages was the second example of improved cellular fitness and followed that of increased thermoacidophily (21). Here, enhanced genome stability was accompanied by survival under much more extreme environmental conditions. However, as an unselected trait, enhanced genome stability was likely to be mechanistically related to the process that provided the SARC transcriptome. One common target of such a mechanism is chromatin structure and composition. Because transcriptomic data did not indicate an increase in DNA repair processes, it was possible that greater genome stability resulted from preventative mechanisms such as epigenetic processes. In eukaryotic cells, chromatin patterns are thought to contribute to regional mutation rates (29) by altering access of repair machinery (32). Because transposition was reduced despite increased transcription of transposases, reduced rates may involve posttranscriptional inhibition. Noncoding RNA interference of transposase transcripts has also been proposed for archaea and Sulfolobus (33).
The apparent involvement of chromatin proteins in transcriptional regulation and inheritance has implications for the evolution of crenarchaeotal transcriptional regulatory systems and for epigenetic systems across domains. For example, regulation by chromatin proteins could compensate for the absence of common regulatory processes in crenarchaeotes. Although the evolutionary history of epigenetic systems is unknown, recent studies propose that eukaryotes arose from an archaeal ancestor (34). It is tempting to speculate that epigenetic processes in eukaryotes originated from archaea. However, because crenarchaeotal epigenetic-like mechanisms are not entirely analogous to eukaryotic histone systems, it is also possible that the process evolved twice. The current work provides a foundation from which to begin future studies on archaeal chromatin proteins and their role in regulating gene expression.
Methods
Archaeal Strains and Cultivation.
Strains of S. solfataricus were cultured, adapted, and isolated from acid-passaged cultures, as described by McCarthy et al. (21). Parental strains were SULA (20, 35), SULG (31), and SULM (31). Partially evolved strains were 50 generations from parental and were SARC-B (formerly SULB) (21), SARC-H, and SARC-N. Fully evolved strains were 120 generations from parental and were SARC-C (formerly SULC) (21), SARC-I, and SARC-O. SULA passaged for 120 generations without acid selection was SUL120 (SI Appendix, Table S1)
Genome and Transcriptome Sequencing.
Genome and transcriptome sequencing was performed for all strains, as described by McCarthy et al. (21) and SI Appendix, Supplemental Methods. Illumina sequencing genome coverage was ≥500 for all samples, read length was 100 bp, and insert size was approximately 300–500 bp. For transcriptome sequencing, clonal, biological duplicates of strains were grown in pH 3.0 for parental strains, pH 1.5 for partially evolved strains, and pH 1.0 for the SARC strains. SARC-I grown at an optimal pH of 3.0 for 15 generations was included as a control. SARC strain libraries were prepared as described previously (21). The SUL120 nonadapted passaging control was sequenced using the Pacific Biosciences (PacBio) RSII sequencing platform and had 120× genome coverage (SI Appendix, Supplemental Methods).
Analysis of Genomic Data.
Mutations were identified via sequence comparisons of derived strain genomes to their respective parental genome using Integrative Genomics Viewer (SI Appendix, Supplemental Methods). For all strains, mutations located within ORFs were further characterized (SI Appendix, Supplemental Methods). The mutation locations for all derived strains were mapped against the SULA genome using Circos (36). Genomic rearrangements were identified by aligning parental and SARC strain sequences using Mauve 2.4.0 (37) (SI Appendix, Supplemental Methods).
Analysis of Transcriptomic Data.
Raw RNA-seq data were processed by removing low quality reads and performing a differential expression analysis on the SARC and parental transcriptomes using the EdgeR Bioconductor package version 3.2.4 (38). Differential expression was measured as fold-change in expression. The transcriptomes of fully evolved SARC strains grown at pH 1.0 and partially evolved SARC strains grown at pH 1.5 were compared with the transcriptomes of their respective parental strains grown at pH 3.0. To identify transient expression patterns resulting from acid stress, acid-adapted SARC-I grown at optimal pH 3.0 for 15 generations was compared with its parental SULG also grown at pH 3.0. Stringent filtering of transcriptomes was used to identify genes with conserved expression in the SARC strains that were likely to contribute to the SARC traits (SI Appendix, Supplemental Methods). The conserved transcriptome was analyzed for regions of high GC content, proximity to tRNA, proximity to origins of replication, functional enrichment, and the existence of potential transcription factor–binding sites (SI Appendix, Supplemental Methods). The log2 fold-change data for SARC transcriptome genes was graphed using the circular graphic software Circos.
Recombination with Phenotypic and Expression Analysis.
Up-regulated SARC genes SULA_2869 and SULA_2027 were selected for HR (SI Appendix, Supplemental Methods). The non-SARC gene, SULA_0895, had unaltered expression in SARC and was a control. SULA_2002 was used for normalization because it had consistent expression across all 10 transcriptomes. The use of the unmutated SARC-I strain for HR avoided potential influence of mutations detected in the other SARC strains.
All markerless exchange recombinant DNA procedures for SARC-I were as described previously (39) (SI Appendix, Supplemental Methods). The target loci of recombinant isolates were sequenced to verify that no mutations occurred that could cause expression changes. The acid-resistance phenotype of recombinants was tested by comparing their growth against a SARC-I control that did not undergo HR in pH 1.13 BST. At this pH value, growth can be observed for most recombinant isolates, whereas the non–acid-adapted parental strain SULG cannot grow. Growth rates were considered reduced when the average of triplicate results was ≤60% that of the SARC-I control. Growth controls at optimal pH were performed to ensure the isolates and controls were otherwise equally fit.
Reduction in acid resistance was predicted to result from altered expression of SARC genes after they underwent HR. Isolates predicted to have and not have expression changes were tested (SI Appendix, Supplemental Methods), in addition to control isolates not expected to alter SARC gene expression. Expression analysis of perturbed gene regions was performed using total cellular RNA extraction and qRT-PCR, as described previously (40) (SI Appendix, Supplemental Methods).
Data Availability.
Genome sequences have been deposited in the GenBank database. Raw DNA-seq data are available in the Joint Genome Institute (JGI) Genome Portal. RNA-seq data are available in the JGI Genome Portal and in the Sequence Read Archive. See detailed accession numbers in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Lutz Froenicke (University of California–Davis Genome Center) for genome sequencing and assembly assistance. We also thank Julien Gradnigo for assistance with data processing. This work was supported by the National Science Foundation Grant Molecular and Cellular Biosciences (MCB)-1517408.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. CP011057 (SULA), CP011055 (SARC-B), CP011056 (SARC-C), CP033235 (SULG), CP033236 (SARC-H), CP033237 (SARC-I), CP033238 (SULM), CP033239 (SARC-N), CP033240 (SARC-O), and CP033241 (SUL120)]. The raw DNA-seq data reported in this paper have been deposited in the Joint Genome Institute (JGI) Genome Portal, https://genome.jgi.doe.gov/portal [accession nos. 1019966 (SULA), 1019969 (SARC-B), 1019972 (SARC-C), 1019975 (SULG), 1019978 (SARC-H), 1019981 (SARC-I), 1019984 (SULM), 1019987 (SARC-N), and 1019990 (SARC-O)]. The raw RNA-seq data reported in this paper have been deposited in the JGI Genome Portal [accession nos. 1019993 (SULA), 1019996 (SARC-B), 1019999 (SARC-C), 1020002 (SULG), 1020005 (SARC-H), 1020008 (SARC-I), 1036511 (SARC-I stress control), 1020011 (SULM), 1020014 (SARC-N), and 1020017 (SARC-O)]. The RNA-seq data reported in this paper have been deposited in the Sequence Read Archive [accession nos. SRX872629 and SRX872630 (SULA), SRX872631 and SRX872632 (SARC-B), SRX712375 and SRX712376 (SARC-C), SRX872634 and SRX872633 (SULG), SRX875205 (SARC-H), SRX712379 and SRX712378 (SARC-I), SRX872636 and SRX872635 (SULM), SRX712413 and SRX712412 (SARC-N), and SRX872640 and SRX872639 (SARC-O)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808221115/-/DCSupplemental.
References
- 1.Richards EJ. Inherited epigenetic variation–Revisiting soft inheritance. Nat Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 2.Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarre WW. Bacterial Chromatin. Springer; Dordrecht, The Netherlands: 2010. H-NS as a defence system; pp. 251–322. [Google Scholar]
- 4.Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- 5.Choli T, Henning P, Wittmann-Liebold B, Reinhardt R. Isolation, characterization and microsequence analysis of a small basic methylated DNA-binding protein from the Archaebacterium, Sulfolobus solfataricus. Biochim Biophys Acta. 1988;950:193–203. doi: 10.1016/0167-4781(88)90011-5. [DOI] [PubMed] [Google Scholar]
- 6.Guo L, et al. Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea. Nucleic Acids Res. 2008;36:1129–1137. doi: 10.1093/nar/gkm1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malarkey CS, Churchill ME. The high mobility group box: The ultimate utility player of a cell. Trends Biochem Sci. 2012;37:553–562. doi: 10.1016/j.tibs.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeters E, Driessen RP, Werner F, Dame RT. The interplay between nucleoid organization and transcription in archaeal genomes. Nat Rev Microbiol. 2015;13:333–341. doi: 10.1038/nrmicro3467. [DOI] [PubMed] [Google Scholar]
- 9.Vassart A, et al. Sa-Lrp from Sulfolobus acidocaldarius is a versatile, glutamine-responsive, and architectural transcriptional regulator. MicrobiologyOpen. 2013;2:75–93. doi: 10.1002/mbo3.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dillon SC, Dorman CJ. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol. 2010;8:185–195. doi: 10.1038/nrmicro2261. [DOI] [PubMed] [Google Scholar]
- 11.Forbes AJ, et al. Targeted analysis and discovery of posttranslational modifications in proteins from methanogenic archaea by top-down MS. Proc Natl Acad Sci USA. 2004;101:2678–2683. doi: 10.1073/pnas.0306575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumann H, Knapp S, Lundbäck T, Ladenstein R, Härd T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Nat Struct Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- 13.Bell SD, Jackson SP. Transcription and translation in Archaea: A mosaic of eukaryal and bacterial features. Trends Microbiol. 1998;6:222–228. doi: 10.1016/s0966-842x(98)01281-5. [DOI] [PubMed] [Google Scholar]
- 14.Wuichet K, Cantwell BJ, Zhulin IB. Evolution and phyletic distribution of two-component signal transduction systems. Curr Opin Microbiol. 2010;13:219–225. doi: 10.1016/j.mib.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wurtzel O, et al. A single-base resolution map of an archaeal transcriptome. Genome Res. 2010;20:133–141. doi: 10.1101/gr.100396.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grogan DW. Cytosine methylation by the SuaI restriction-modification system: Implications for genetic fidelity in a hyperthermophilic archaeon. J Bacteriol. 2003;185:4657–4661. doi: 10.1128/JB.185.15.4657-4661.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haseltine CA, Kowalczykowski SC. An archaeal Rad54 protein remodels DNA and stimulates DNA strand exchange by RadA. Nucleic Acids Res. 2009;37:2757–2770. doi: 10.1093/nar/gkp068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexeev A, Mazin A, Kowalczykowski SC. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol. 2003;10:182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 19.Tamburini BA, Tyler JK. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol Cell Biol. 2005;25:4903–4913. doi: 10.1128/MCB.25.12.4903-4913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rolfsmeier M, Blum P. Purification and characterization of a maltase from the extremely thermophilic crenarchaeote Sulfolobus solfataricus. J Bacteriol. 1995;177:482–485. doi: 10.1128/jb.177.2.482-485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy S, et al. Expanding the limits of thermoacidophily in the archaeon Sulfolobus solfataricus by adaptive evolution. Appl Environ Microbiol. 2015;82:857–867. doi: 10.1128/AEM.03225-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci USA. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rausch T, et al. DELLY: Structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 27.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redder P, Garrett RA. Mutations and rearrangements in the genome of Sulfolobus solfataricus P2. J Bacteriol. 2006;188:4198–4206. doi: 10.1128/JB.00061-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuster-Böckler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 30.Zhang C, et al. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem Soc Trans. 2013;41:405–410. doi: 10.1042/BST20120285. [DOI] [PubMed] [Google Scholar]
- 31.Schelert J, et al. Occurrence and characterization of mercury resistance in the hyperthermophilic archaeon Sulfolobus solfataricus by use of gene disruption. J Bacteriol. 2004;186:427–437. doi: 10.1128/JB.186.2.427-437.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yazdi PG, et al. Increasing nucleosome occupancy is correlated with an increasing mutation rate so long as DNA repair machinery is intact. PLoS One. 2015;10:e0136574. doi: 10.1371/journal.pone.0136574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang TH, et al. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 34.Spang A, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCarthy S, et al. Complete genome sequence of Sulfolobus solfataricus strain 98/2 and evolved derivatives. Genome Announc. 2015;3:e00549-15. doi: 10.1128/genomeA.00549-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: Multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schelert J, Drozda M, Dixit V, Dillman A, Blum P. Regulation of mercury resistance in the crenarchaeote Sulfolobus solfataricus. J Bacteriol. 2006;188:7141–7150. doi: 10.1128/JB.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudrappa D, et al. Identification of an archaeal mercury regulon by chromatin immunoprecipitation. Microbiology. 2015;161:2423–2433. doi: 10.1099/mic.0.000189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequences have been deposited in the GenBank database. Raw DNA-seq data are available in the Joint Genome Institute (JGI) Genome Portal. RNA-seq data are available in the JGI Genome Portal and in the Sequence Read Archive. See detailed accession numbers in SI Appendix, Supplemental Methods.